Abstract
Kinetochores mediate microtubule–chromosome attachment and ensure accurate segregation of sister chromatids. The highly conserved Ndc80 kinetochore complex makes direct contacts with the microtubule and is essential for spindle checkpoint signaling. It contains a long coiled-coil region with globular domains at each end involved in kinetochore localization and microtubule binding, respectively. We have directly visualized the architecture of the yeast Ndc80 complex and found a dramatic kink within the 560-Å coiled-coil rod located about 160 Å from the larger globular head. Comparison of our electron microscopy images to the structure of the human Ndc80 complex allowed us to position the kink proximal to the microtubule-binding end and to define the conformational range of the complex. The position of the kink coincides with a coiled-coil breaking region conserved across eukaryotes. We hypothesize that the kink in Ndc80 is essential for correct kinetochore geometry and could be part of a tension-sensing mechanism at the kinetochore.
Keywords: Nuf2, Spc24, Spc25, Hec1, electron microscopy
Introduction
During mitosis, sister chromatids must segregate properly to maintain the genetic integrity of the two daughter cells. The mitotic spindle, a microtubule-based network originating from opposite poles, captures, aligns, and separates the replicated chromosomes. The spindle-assembly checkpoint ensures that chromosomes do not segregate until they are properly attached.1 A stable attachment between the chromosomes and the mitotic spindle is mediated by the kinetochore, a multiprotein scaffold of ∼100 components that assemble on centromeric DNA (reviewed in Refs. 1–6).
Electron microscopy (EM) of vertebrate kinetochores reveals a trilaminar organization with an electron-dense inner layer composed of centromeric heterochromatin, a less dense middle layer, and an electron-dense outer layer containing microtubule-binding domains.2 In budding yeast, a “point kinetochore” is assembled on ∼150 bp of centromeric DNA and is attached to a single microtubule.7,8 In higher eukaryotes, larger kinetochores assemble on centromeric region spanning over millions of base pairs and bind multiple (15–30) microtubules whose plus-ends terminate within the outer plate.6 Despite these differences in centromeric complexity, most kinetochore components are conserved from yeast to humans,9 and it has been proposed that each of the attachment points in higher eukaryotes might be functionally equivalent to the Saccharomyces cerevisiae point kinetochore.2,10,11
The Ndc80 complex is an essential component of the highly conserved KMN kinetochore network, central to the microtubule–kinetochore interaction.12,13 It is composed of four subunits: Spc24, Spc25, Nuf2, and Ndc80, each containing a globular domain and a long coiled-coil region, which assemble into an ∼57-nm heterotetrameric rod with a globular region at each end (Fig. 1a).14,15 The complex is thought to span from the inner layer, where the Spc24/Spc25 globular heads interact with a still unknown kinetochore receptor, to the outer layer of the kinetochore, where the Ndc80/Nuf2 globular heads interact directly with the microtubules.12,16-19 Recent X-ray crystallography has revealed the structure of the two functional heads of the Ndc80 complex.19,20 The Ndc80 and Nuf2 globular segments fold into calponin homology (CH) domains where conserved, positively charged residues interact with the acidic C-terminal tails of the tubulin dimer.18,19 The interaction with the microtubule is likely to be cooperative12,19 and functionally regulated by phosphorylation of the N-terminal tail of Ndc80p by AuroraB.12,18,19,21
Fig. 1.
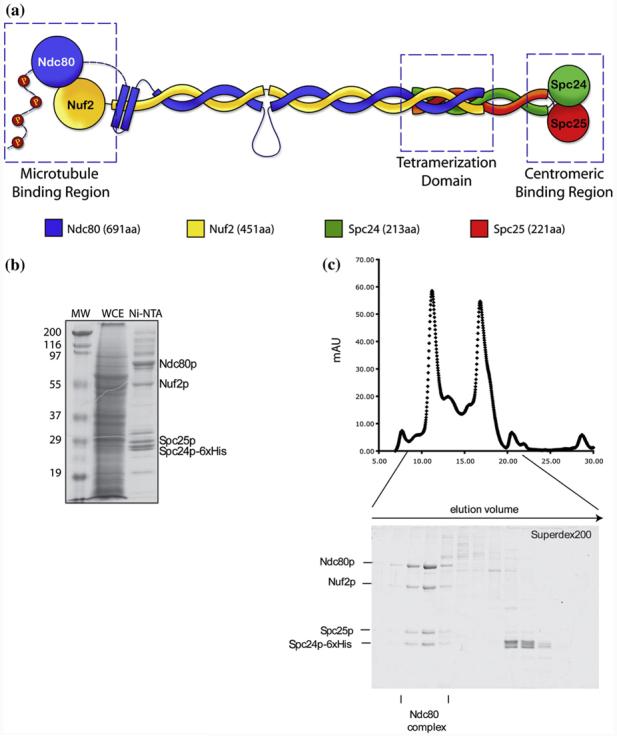
Structural features, reconstitution and purification of the yeast Ndc80 complex. (a) Schematic of the organization of the Ndc80 complex. It is comprised of two heterodimeric subcomplexes, Nuf2/Ndc80 and Spc24/Spc25. Its elongated, rod-like structure is produced by the coiled-coil regions of each subunit, while capped at both ends by two globular domains. The Nuf2/Ndc80 end interacts with the microtubules while the Spc24/Spc25 end interacts with the centromeric region or other kinetochore components. (b) Coomassie-stained gel showing purification of the Ndc80 complex from SF9 insect cells. MW: Molecular weight marker; WCE: whole cell extract; Ni-NTA: eluate from Nickel-affinity chromatography. (c) Size exclusion chromatography of Ni-NTA eluate from A on a Superdex 200 column. The two peaks in the chromatogram correspond to the full 4-protein Ndc80 complex and to a dimer consisting of Spc24p and Spc25p. Coomassie-stained gel of Superdex 200 fractions in the lower panel demonstrates homogeneity of the reconstituted Ndc80 complex. The fractions used for EM examination are marked.
Cross-linking experiments and mass spectrometry data have shed light on the organization of the tetramerization domain and the register of the coiled-coil regions of the different subunits.19,22 These data suggest that the Ndc80/Nuf2 C-terminal coiled-coil and the Spc24/Spc25 N-terminal helical regions contribute to the tetramerization domain by overlapping ∼60 residues of each chain.19,22 They also suggest the presence of an interruption of approximately 50 residues in the coiled-coil region of the Ndc80p that could be functionally important.
Here, we describe the architecture of the yeast Ndc80 complex using negative-staining EM and single-particle image analysis. We identified a kink region present at approximately one-third of the molecule. Classification, alignment, and averaging of the two globular heads allowed us to position the flexible region at a very specific site within the complex and to define the approximate geometrical constraints allowed at the coiled-coil kink. Finally, we discuss our structural findings in terms of possible functional relevance at the kinetochore, for both microtubule attachment and tension sensing.
Results
Reconstitution and organization of the yeast Ndc80 complex as imaged by EM
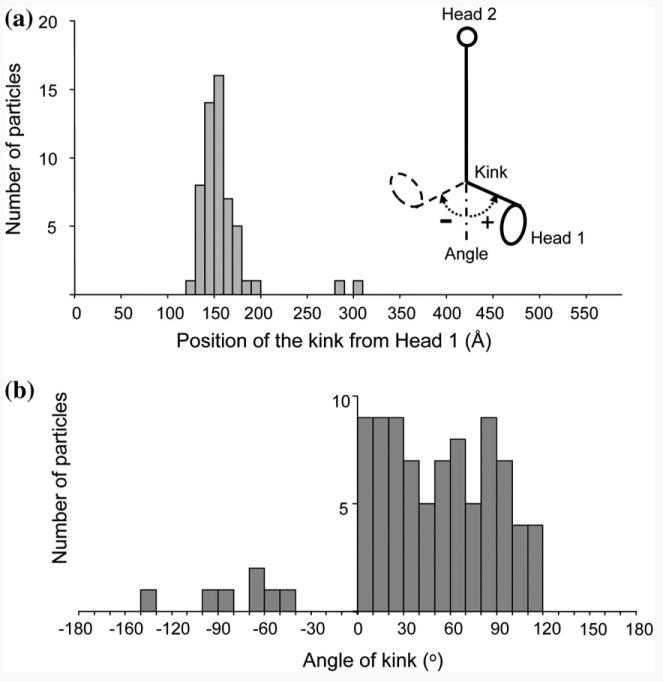
Soluble full-length yeast Ndc80 complex was produced by coexpression in insect cells and was further purified using Ni-NTA affinity chromatography and size-exclusion chromatography (Fig. 1b and c). Fractions containing the Ndc80 molecules were visualized by negative-stain EM (Fig. 2a). The Ndc80 complexes appeared as elongated, rod-like structures with an average length of 558±56 Å (Table 1) and two globular heads at either end of the coiled-coil rod, in agreement with previous analysis by rotary shadowing15 and atomic force microscopy.14 A noticeable kink can be also observed along the rod (Fig. 2b). The kink occurs at a consistent location that is about one-third from one end of the Ndc80 molecule (Fig. 2b; Table 1). We designated the head that is closer to the kink in each molecule as Head 1 and the other one as Head 2 (Fig. 2b, panel 1).
Fig. 2.
Visualization of the yeast Ndc80 complex by electron microscopy. (a) Electron micrograph of negatively stained Ndc80 complexes. The scale bar corresponds to 50 nm. (b) Gallery of individual Ndc80 molecules windowed from the micrographs. The designated Head 1, Head 2 and kink are marked on the molecule in panel 1. The scale bar corresponds to 10 nm.
Table 1.
Measurement of the position of the kink in Ndc80
| Particle number | Position of the kink from Head 1 (Å) | Position of the kink from Head 2 (Å) | Total length of particle (Å) |
|---|---|---|---|
| 1 | 136 | 453 | 589 |
| 2 | 155 | 441 | 596 |
| 3 | 144 | 462 | 606 |
| 4 | 149 | 437 | 586 |
| 5 | 298 | 324 | 622 |
| 6 | 166 | 445 | 611 |
| 7 | 150 | 469 | 620 |
| 8 | 158 | 443 | 601 |
| 9 | 151 | 451 | 602 |
| 10 | 133 | 393 | 526 |
| · | · | · | · |
| · | · | · | · |
| · | · | · | · |
| Average | 160±32 | 411±49 | 558±56 |
The two heads of the Ndc80 complex can be specifically assigned to Ndc80/Nuf2 and Spc24/Spc25 subcomplexes
The images of both heads from 250 Ndc80 molecules were mixed together and analyzed using single-particle methodology. Due to the very small size of these globular domains (∼40 kDa for Ndc80/Nuf2 and ∼20 kDa for Spc24/Spc25), the following particle selection procedure was carried out before our final analysis. As a first step, we performed iterative in-plane rotational and translational alignment and multivariant statistical analysis to generate 50 initial classes for the globular heads (with an average of about 10 particles per class), some of which at this point looked fairly indistinguishable. Of these, 12 classes gave rise to well-defined averages and consisted mainly of images assigned as Head 1, while another 12 classes with good averages included mainly images assigned as Head 2. The particles belonging to the two groups of 12 higher-quality classes were each separated and each group reclassified into just 5 classes. These 10 new class averages served as references for new rounds of alignment and head assignment (Head 1 or Head 2) of the full original dataset of heads. We then checked the assignment of each of the two heads for each Ndc80 molecule. Only Ndc80 molecules with each of their heads assigned to one head type and showing the highest correlation to their corresponding class averages were used in further analysis. This allowed for the selection of 90 “good” Ndc80 molecules. The other complexes either had both heads assigned to the same head type, were outliers within a class, or contributed to poor class averages likely containing poorly stained/preserved molecules. These molecules were not considered for any further analysis.
The five class averages belonging to the Head 1 group show clear differences from the five class averages of the Head 2 group (Fig. 3a and c). Head 1 is clearly larger than Head 2 and appears to have two discernable density domains that line up in an orientation roughly perpendicular to the rod stem. Head 2, on the other hand, seems to have only one density domain that appears to depart at different angles from the coiled-coil rod, likely indicating flexibility at this position.
Fig. 3.
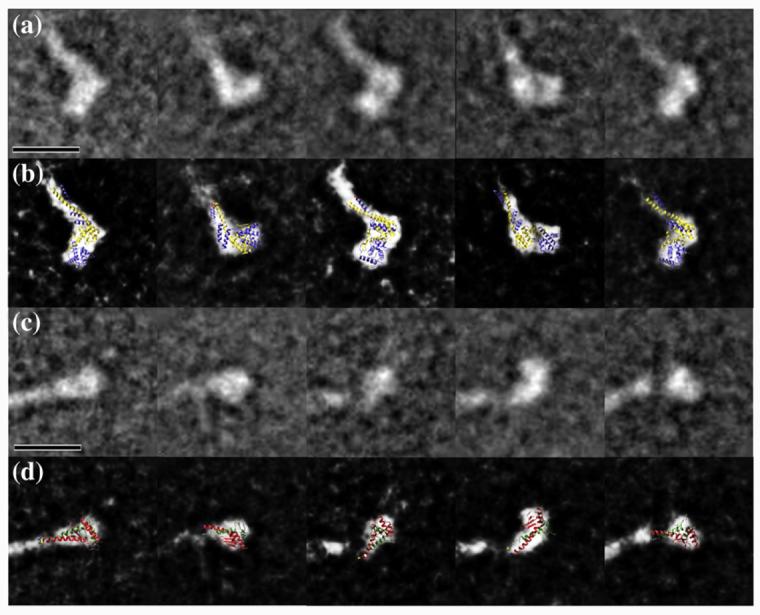
Class averages and docking of the atomic models in Head 1 and Head 2. (a) Five representative class averages of the Head 1. (b) Docking of the atomic model of Ndc80/Nuf2 N-terminal region on the different views represented in the class averages. (c) Five representative class averages of the Head 2. (d) Docking of the atomic model of Spc24/Spc25 C-terminal region on the different views represented in the class averages. In (b) and (d), the threshold of the class averages was adjusted to saturation to show the docking more clearly. The scale bars represent 10 nm.
We compared our two-dimensional class averages with the two globular heads in the atomic models of the globular regions of both an engineered version of the tetrameric human Ndc80 complex19 and the yeast Spc24/Spc25 C-terminal regions18 using manual docking. The atomic model of the Ndc80 and Nuf2 CH domains can be easily fitted in each of the two discrete densities of Head1 seen in the different orientations represented in our five class averages (Fig. 3b). No additional density for the first 113 residues, which are missing in the crystal structure of the human complex, can be seen, suggesting that this region is also flexible in yeast (although we cannot rule out partial proteolysis). On the other hand, the coiled-coil structure that departs from the CH domains in the crystal structure fits well in the stem extending from the density of Head 1, suggesting a significant degree of rigidity for this connection (Fig. 3b). These observations confirm that the overall structure of the Ndc80/Nuf2 globular region is conserved from yeast to humans.
The atomic model of the Spc24/Spc25 globular region can be fitted in the globular density of the class averages for Head 2. In some of the class averages, this domain appears detached from the extended stem, suggesting a more flexible connection at this end of the coiled coil (Fig. 3d) (in agreement with limited proteolysis data14,17).
We next considered the relative orientations of Head 1 and Head 2 within a given Ndc80 molecule by analyzing the possible correlation between the classification assignments of the two heads from each molecule (Table 2). The lack of such correlation indicates that each of the two heads can freely twist within the molecule, independently of the orientation of the other head. Given the relative rigidity in the attachment of the coiled-coil region to the Ndc80/Nuf2 head, this property of the Ndc80 complex is likely due to the flexible regions within the remaining parts of the coiled coil, including its connection with the Spc24/Spc25 globular head.
Table 2.
The numbers of molecules with specific designated heads
| A | B | C | D | E | |
|---|---|---|---|---|---|
| a | 3 | 4 | 5 | 3 | 3 |
| b | 3 | 5 | 3 | 4 | 4 |
| c | 2 | 2 | 6 | 1 | 4 |
| d | 5 | 4 | 4 | 5 | 2 |
| e | 4 | 3 | 4 | 2 | 5 |
A, B, C, D, and E are the five classes for Head 1; a, b, c, d, and e are the five classes for Head 2. The numbers indicate how many Ndc80 molecules have their Head 1 and Head 2 designated at the indicated classes.
The Ndc80 complex contains a flexible kink at a specific site of the coiled-coil rod region
Because the globular Head 1 and Head 2 of each molecule had been assigned, respectively, to the Ndc80/Nuf2 and the Spc24/Spc25 globular ends, we were able to define the location of the kink within the Ndc80 molecule and to measure the kink angle and direction.
We measured the position of the kink in 55 molecules with obvious kinks out of the 90 molecules amenable for head assignment and examined the bending angle of the kink in each molecule. The kink is consistently located at a position 160 Å away from Head 1, which our analysis strongly indicates corresponds to the Ndc80/Nuf2 globular head (Table 1 and Fig. 4a). On the other hand, the angle of the kink falls within a broad range of 0° to 120°, reflecting a remarkable flexibility of the complex at that point (Fig. 4b). Recent data indicated the presence of an ∼50-aa loop insertion in the human Ndc80p coiled-coil region centered at residue 440 and corresponding to residue 475 in yeast.19,22 This loop disrupts the long coiled-coil rod between the two proteins at one-third of its length, in good agreement with the position of the kink in the molecule relative to Head 1 (160 Å/560 Å=2/7).
Fig. 4.
Distribution of positions and angles of the kink in the particles. (a) Distribution of the position of the kink as measured from Head 1 for all the Ndc80 molecules. The schematic shows the architecture of the Ndc80 molecule with the main structural features under consideration marked. The angle and direction of the kink is defined as illustrated. (b) Distribution of the kink angle as measured for all the molecules. Kinks that appear at angles between 0° (when the particle is straight) and 30° were counted together and evenly distributed within this range, because at these angles, the kink is not as clearly defined (the particle tends to appear curved rather than kinked due to the innate flexibility of the coiled coil).
Interestingly, the position and the length of the insertion identified by the cross-linking studies appear to be conserved across different species (with the exception of a few yeasts, such as S. cerevisiae, with a longer insertion) (Fig. 5), suggesting that this loop and the kink that we observed at this position are likely to be functionally important. The presence of predicted secondary structure in this region and the conservation of the sequence further suggest that this region could interact with other proteins, likely kinetochore components (see Discussion).
Fig. 5.
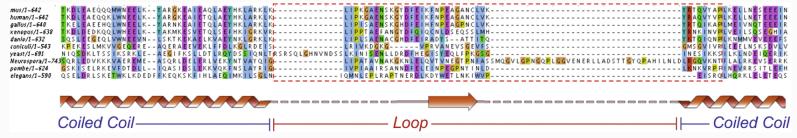
Alignment of the Ndc80 sequences of several organisms around the region of coiled-coil interruption. An interruption in the coiled-coil region of the Ndc80 protein is predicted across species. This loop region introduces a flexible point within the Ndc80 molecule. Within the loop, secondary-structure prediction indicates the presence of a conserved β-strand segment, suggesting the possibility that the region could be relevant for protein–protein interactions.
In a few cases, we observe an additional bend, halfway down the length of the coiled-coil segment connecting the kink to Head2 (Fig. 2c, panel 5), which could correspond to interruptions of the coiled-coil predicted immediately before and after the tetramerization domain.14,19
The kink in the Ndc80 molecule bends like an “elbow” and allows for the Ndc80/Nuf2 globular head to twist within a constrained range
The Ndc80 molecules were absorbed to a carbon support for EM analysis. Through this adsorption process, the coiled coil is “forced” to lie on the plane of the carbon. Therefore, the kink in the Ndc80 complex as so far portrayed corresponds to an in-plane movement. The fairly rigid connection between the Ndc80/Nuf2 head and the coiled-coil defines a boot-like shape for Head 1 (Fig. 3b) that we used to define the in-plane direction of the kink for each molecule (Fig. 4a inset). Interestingly, the large majority of the Ndc80 molecules kink in the same direction (which we defined as having a positive angle; Fig. 4b) so that the kink is like an elbow that only allows bending in one direction.
To further analyze the movement of Head 1 with respect to the kink point, we compared the “off-plane” orientations of the Head 1 class averages based on our docking of the crystal structure into the five distinct two-dimensional class averages. After aligning the five class averages through in-plane translation and rotation, we examined the spatial relationship among the docked atomic models. While the boot-shaped Ndc80/Nuf2 heads corresponding to the five classes align with each other in their top views (or in-plane views), they appear in different out-of-plane rotations in their end-on views (orthogonal to the plane of the carbon) (Fig. 6). Thus, our results indicate the presence of an off-plane twist in addition to the in-plane kink. Importantly, the twist and the kink angles appear uncorrelated, because the kink angle distribution is the same for Ndc80 molecules belonging to each of the five Head 1 classes (i.e., the number of molecules with kink angles larger than 60° is 7, 9, 9, 6, and 6 for the five classes, respectively). Although caution is required due to our limited resolution, the fact that an elbow-like kink can be detected in the images of Ndc80 molecules indicates a constraint range of the twist between Head 1 and the kink point (Fig. 7 and Supplementary Video 1). The twist may be distributed along the coiled-coil segment between the Ndc80/Nuf2 globular domain and the kink point, or it may be constrained to the kink point itself. Our data cannot distinguish between these two possibilities.
Fig. 6.
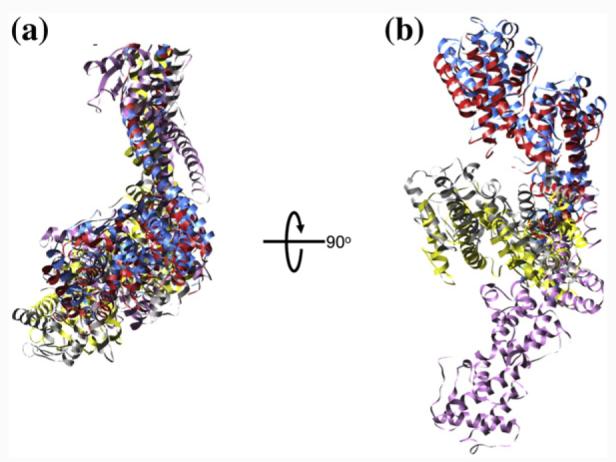
The out-of-plane rotations of the five Head 1 models. The five docked Ndc80/Nuf2 atomic models are aligned by in-plane translation, rotation, and flipping within the plane corresponding to the carbon surface on the EM grid. (a) Top views of the aligned models with the carbon plane parallel with the plane of the page. (b) End-on views of the aligned models with the carbon plane perpendicular to the plane of the page.
Fig. 7.
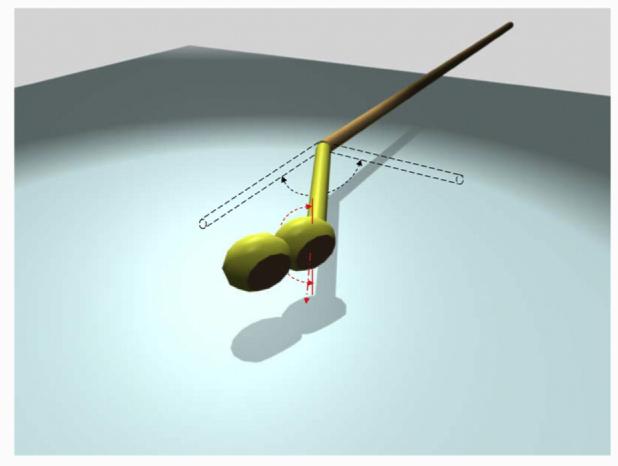
Model describing the Ndc80 molecule's flexibility at the kink and its Head 1's twist. The elbow-like kink bends within a range of 0° to 120°, and the coiled-coil segment connecting Head 1 and the kink point twists in a constraint range of −90° to 90°.
Discussion
The Ndc80 complex is a highly conserved component of the kinetochore. It is one of the constituents of the KMN kinetochore network central to the microtubule–kinetochore interaction. This network also includes the Mis12 complex (MTW1 in yeast) and the KNL-1 protein (Spc105 in yeast). The Ndc80 complex contains a long rod formed by the coiled-coil regions of its four protein subunits and two globular heads at its ends that interact, respectively, with a microtubule and the centromeric region of the chromosome or inner kinetochore components. The length of the rod allows it to span over 500 Å of the outer kinetochore, while its two functional ends are engaged in different interactions.
We have directly visualized a reconstituted yeast Ndc80 complex using EM of negatively stained samples. Our images reveal the presence of an elbow-like kink in a well-defined position within the Ndc80 complex central rod, which adopts a broad range of angles, from that in a fully extended rod to a maximum kink of about 120° (Fig. 7 and Supplementary Video 1). The position of the kink corresponds to the location of a coiled-coil-breaking loop experimentally observed in the human complex.19,22 Sequence analysis shows that this structural feature is conserved across species and points to its functional significance.
We propose that the kink in the Ndc80 coiled coil may be important for correct kinetochore geometry.19 In the “single microtubule” kinetochore of budding yeast, the plus-end of the microtubule (with a diameter of 25 nm) has to interact stably with a centromeric DNA region of only 150 bp, which is possibly organized into a C-loop with a diameter of 15–20 nm.23 Assuming that Ndc80 binds to the microtubule lattice at a fixed angle,12 flexibility in the coiled-coil region may be required to establish the correct geometry of attachment between the kinetochore and the microtubule.
Another likely function for the flexible “elbow” in the Ndc80 coiled coil is a mechanism by which the Ndc80 complex could attach to a microtubule at different angles (Fig. 8a and b). If we envision Ndc80 as a kinetochore reaching “arm”, where the Spc24/Spc25 globular region forms a stable attachment to the centromeric “shoulder” region and where the Ndc80/Nuf2 CH domains act as the “hand” that grabs the microtubule, the kink would serve as a flexible “elbow” that allows the right orientation of the microtubule-binding region to engage the microtubule. Keeping with this analogy, it is interesting that our analysis indicates that the hand is fairly rigidly connected to the forearm, while there is some flexibility in the connection of the upper arm to the shoulder. Interestingly, the crystal structure of the bonsai human complex shows different orientations of their coiled coils with respect to the Spc24/Spc25 globular region 19 in the two molecules present per unit. asymmetric unit.19
Fig. 8.
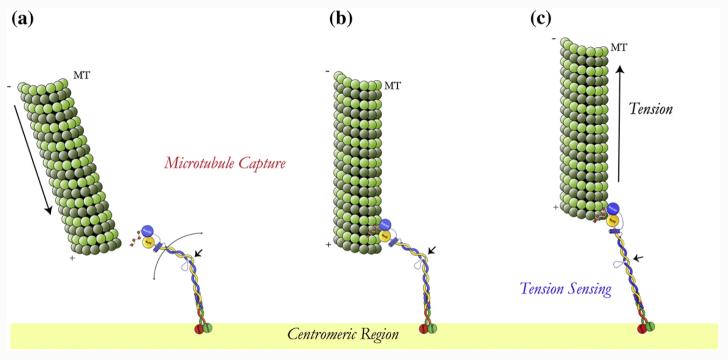
Model for microtubule kinetochore interaction. (a) Ndc80 complex localized at the kinetochore where its flexibility allows it to interact with incoming microtubules in different orientation but with the right polarity. (b) The microtubule-attached Ndc80 complex still undergoes “flexing” while attached to a spindle microtubule. (c) Possible tension-sensing mechanism mediated by the Ndc80 kinked region: as the kinetochore comes under tension, the Ndc80 complex will only exist in the fully extended conformation.
The twist detected in the Ndc80/Nuf2 head around the axis defined by the proximal segment of the coiled coil from the kink would further enhance the capacity to engage incoming microtubules with various orientations. However, the constraint range of the twist and the unidirectional character of the kink would only allow the interaction with microtubules with the right polarity, that is, from only one “correct” spindle pole.
More speculative, but highly attractive, is the possibility that an additional, tension-sensing function could also be related to the presence and conformation of this flexible kinked region in Ndc80 (Fig. 8c). It is easy to imagine that under conditions where both kinetochores in a chromosome are attached to different spindle poles and thus are under tension, the Ndc80 complex will assume a fully straightened conformation, without a kink. Signaling factors could detect either a fully extended or a bent state, perhaps by binding at the kink in only one of these states, initiating a signaling process that defines the activity state of spindle checkpoint kinases. The sequence conservation in the loop (not just its positioning and length) is in agreement with the concept of a specific protein–protein interaction that could read the local environment of the coiled coil. This speculative hypothesis awaits experimental testing.
Materials and Methods
Reconstitution and purification of the Ndc80 complex
Open reading frames encoding Ndc80 subunits were amplified by PCR from yeast genomic DNA using primers with flanking restriction sites. They were inserted into the BamHI/EcoRI (Spc24p) and XhoI/KpnI (Nuf2p) sites of the pFastBacDual plasmid. At the C-terminus of Spc24p, a 6 × His-tag was introduced with the PCR primer. Spc25p was cloned into the BamHI/HindIII site and Ndc80p into the XhoI/NheI site of a second pFastBacDual vector. Bacmid DNA and viral supernatants were generated according to the Bac-to-Bac protocol (Invitrogen). Viral supernatants were amplified three times before expression. SF9 insect cells (1 L) were coinfected with viral supernatants encoding for two subunits each and harvested after 3–4 days. The pellets were washed with phosphate-buffered saline (PBS) and stored at −80 °C. Pellets were resuspended in 100 ml 20 mM Na2HPO4/NaH2PO4, pH6.8, 500 mM NaCl, 20 mM imidazole, and protease inhibitors. They were sonicated and Triton X-100 was added to a final concentration of 0.5%. After centrifugation, the clear supernatant was rotated with nickel-NTAagarose (Qiagen). Beads were washed and proteins were eluted by raising the final imidazole concentration to 200 mM (Supplementary Fig. S1a). The eluate was concentrated (Centricon 30) and 400 μl was loaded onto a Superdex 200 column equilibrated in 20 mM Na2HPO4/NaH2PO4, pH6.8, 500 mM NaCl, and 1 mM ethylenediaminetetraacetic acid (PBS buffer). We collected 0.5-ml fractions (Supplementary Fig. S1b), and peak fractions were snap frozen in liquid N2 and stored at −80 °C.
EM and image analysis
Samples of 2 mg/ml Ndc80 complex were diluted in PBS buffer to 20 μg/ml. Five microliters of sample was deposited onto a glow-discharged copper mesh grid coated with continuous carbon and stained with 2% uranyl acetate. The samples were imaged on an FEI Tecnai 12 electron microscope operated at 120 keV at a magnification of 49 K. The images were collected on a Gatan CCD camera as well as on films that were later digitized on a Nikon Super CoolScan 8000. More than 250 individual particles were windowed from the micrographs using the EMAN Boxer program.24 The two heads of each particle were further boxed and underwent reference-free alignment and classification using the IMAGIC image processing package.25 The best 24 class averages then served as references for further iterative alignment and classification of the heads (for more detail, see Results). The measurement of the particle length and the angle of kink was done using EMAN Boxer program and ImageJ (National Institutes of Health)†.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institute of General Medical Sciences (G.B. and E.N), the Office of Biological and Environmental Research for the U.S. Department of Energy (E.N.), Philip Morris USA Inc., and Philip Morris International (D.G.D.) and by a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft (S.W.). E.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations used
- EM
electron microscopy
- CH
calponin homology
- PBS
phosphate-buffered saline
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/ j.jmb.2008.08.077
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev., Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 3.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore–microtubule interface. J. Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 4.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka TU, Desai A. Kinetochore–microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheeseman I, Desai A. Molecular architecture of the microtubule–kinetochore interface. Nat. Rev., Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 7.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 8.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore–microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, et al. Molecular architecture of the kinetochore–microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Ciferri C, Musacchio A, Petrovic A. The Ndc80 complex: hub of kinetochore activity. FEBS Lett. 2007;581:2862–2869. doi: 10.1016/j.febslet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, et al. Architecture of the human ndc80–hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 15.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl Acad. Sci. USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore–microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 19.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, et al. Implications for kinetochore–microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 21.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 22.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, et al. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 25.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.