Abstract
Accurate spatial and temporal expression of gonadotrope-specific genes, such as the gonadotropin-releasing hormone receptor (GnRHR) gene, is critical for gonadotrope maturation. Herein, we show that a specific E-box in the mouse GnRHR promoter binds two group A basic-helix-loop-helix (bHLH) transcription factors. Mutation of this E-box decreases expression in mouse gonadotrope-derived αT3-1 and LβT2 cell lines. Microarray and western blots show that the bHLH transcription factor NeuroD1 is strongly expressed in the gonadotrope progenitor, αT3-1, whereas Mash1 is strongly expressed in the more mature gonadotrope, LβT2. Overexpression of NeuroD1 or Mash1 increases expression of the GnRHR gene or a multimer of the E-box and this increase is lost upon mutation of the E-box. Electrophoretic mobility shift assays reveal that the GnRHR E-box binds NeuroD1 from αT3-1 cells, but binds Mash1 from LβT2 cells. The sequential binding of different members of the group A bHLH transcription factor family to mouse GnRHR E-box 3 as the gonadotrope differentiates may represent a mechanism necessary for proper spatial and temporal expression of the GnRHR during gonadotrope development.
Keywords: Gonadotropin-releasing hormone receptor, bHLH transcription factor, anterior pituitary
1. Introduction
Spatial and temporal expression of specific cohorts of transcription factors are thought to drive differentiation and determine the cell fate decisions necessary for specifying the identity of the five individual endocrine cell types in the anterior pituitary (Scully and Rosenfeld, 2002;Treier et al., 1998; Zhu et al., 2007; Zhu et al., 2006). Appropriate initiation and progression of the gonadotrope cell-specific gene regulation program during anterior pituitary development results in precise spatial and temporal control of expression of the gonadotrope-specific genes. These are activated in a sequential pattern during mouse pituitary development: the α-subunit of the glycoprotein hormones (α-GSU) on embryonic day (e) 10.5, the gonadotropin-releasing hormone receptor (GnRHR) on e13.5, luteinizing hormone (LH) β-subunit on e16.5, and follicle-stimulating hormone (FSH) β-subunit on e17.5 (Japon et al., 1994).
Members of the group A basic-helix-loop-helix (bHLH) family of transcription factors play important roles in cell fate commitment during development in multiple organ systems (Miyata et al., 1999; Morrow et al., 1999; Naya et al., 1997). The 124 mouse bHLH proteins have been classified into six groups from A to F based on DNA-binding specificities and specific structural features (Li et al., 2006). The binding of bHLH heterodimers occurs at regulatory elements termed E-boxes that contain the consensus sequence CAnnTG. Group A proteins, including NeuroD1, Mash1 (Ascl1), and E47 (Pan1), contain a distinct pattern of amino acids (XRX) at sites 5, 8, and 13, and preferentially bind to the motif (5’-CAGCTG-3’). In contrast, group B bHLH proteins, for example, have the 5–8–13 configuration of K/H-X-R which binds a G-Box motif (5’-CACGTG-3’) (Li et al., 2006). The group A bHLH family members utilize their basic region for contacting DNA, while the helix-loop-helix domain allows for dimerization. They cannot bind DNA alone, rather, they must form heterodimers with differentially spliced products of the E2A gene, including the ubiquitously expressed activator E47, to facilitate DNA binding (Murre et al., 1989).
In addition to the actions of individual bHLH proteins in development, temporally distinct patterns of sequential expression of specific bHLH proteins have been shown to be important for cell fate determination and differentiation in retina (Hatakeyama et al., 2001),spinal cord (Lee and Pfaff, 2003), and pancreas (Gasa et al., 2004), among others. Numerous parallels exist between these developmental programs and that of the pituitary. Neurogenic differentiation factor 1 (NeuroD1) expression in the pituitary is initially observed at e11.5,becomes undetectable by e14.5–15.5 (Liu et al., 2001), and remains off in the adult. At some stages, it appears to be restricted to the corticotrope and intermediate lobe (Poulin et al., 2000).In contrast, mammalian achaete-scute homolog 1 (Mash1 or Ascl1) is expressed in the ventral most regions of the developing anterior pituitary around e12; however, expression is only retained in the intermediate lobe by e17.5 (Liu et al., 2001).
NeuroD1-null mice have minor defects in proopiomelanocortin (POMC), α-GSU, and thyroid-stimulating hormone (TSHβ) expression, though no major effects on pituitary development, possibly due to bHLH redundancy (Liu et al., 2001). In addition, NeuroD1 is required for anterior pituitary-specific gene expression when analyzed using in vitro model systems (Poulin et al., 1997). More specifically, NeuroD1 binds an E-box element and activates expression of the corticotrope-specific POMC gene promoter (Poulin et al., 1997). E-box elements have also been described in the α-GSU promoter (Jackson et al., 1993; Jackson et al.,1993), but the proteins binding them are unknown. In Zebrafish, the Mash1 homolog, ascl1a, is critical for pituitary development (Pogoda et al., 2006). Interestingly, it has also been suggested that Mash1 plays a role in the terminal differentiation of thyrotropes, corticotropes, and gonadotropes (Zhu et al., 2006). In vivo, expression of both NeuroD1 and Mash1 is restricted during anterior pituitary development by another bHLH transcription factor Hes1 (Zhu et al.,2006). Although both NeuroD1 and Mash1 are found in the developing anterior pituitary, their roles in gonadotrope-specific gene expression and gonadotrope cell fate specification are unknown.
At present, the model systems most appropriate for investigating expression of the gonadotrope-specific genes in cell culture are the αT3-1 and LβT2 immortalized mouse gonadotrope cell lines (Alarid et al., 1996; Windle et al., 1990). The αT3-1 cell line represents an immature, but committed, gonadotrope of approximately embryonic day 13.5, since it expresses the gonadotrope-specific genes α-GSU, GnRHR and steroidogenic factor 1 (SF-1).The LβT2 cell line recapitulates a more mature gonadotrope of at least embryonic day 17.5,expressing the LH and FSH β-subunits, as well as α-GSU, GnRHR, and SF-1. These model systems have been used extensively to identify and study regulatory elements necessary for both basal and hormonal control of the gonadotrope-specific genes. In point of fact, almost all of the studies defining the regulatory elements in the LH, FSH and GnRHR genes has utilized the αT3-1 and LβT2 cell models (Duval et al., 1997; McGillivray et al., 2005; Norwitz et al., 1999;Pernasetti et al., 2001; Steger et al., 1994).
To further understand temporal progression in the differentiation program of the gonadotrope, we performed Affymetrix microarray analysis of the mRNAs expressed in the αT3-1 versus the LβT2 gonadotrope cell lines and discovered that the bHLH protein NeuroD1 is highly expressed in the αT3-1, while Mash1 is more specific to the LβT2 cells. The mouse GnRHR promoter contains seven putative E-boxes (Resuehr et al., 2007), while other gonadotrope-specific genes, such as LHβ and FSHβ, contain fewer. In particular, the unique E-box element termed E-Box 3 in the GnRHR gene (located at −208/−203 relative to the mRNA start site) is the only one that has the sequence elements that have been shown to preferentially bind group A bHLH family members like NeuroD1, Mash1, and E47. We compare its activities to E-box 4, which has a sequence that is known to preferentially bind members of group B bHLH proteins. Mutation of E-box 3, but not E-box 4, dramatically decreases expression of a GnRHR-Luciferase reporter in both αT3-1 and LβT2 cells. The bHLH proteins NeuroD1 and Mash1,from α T3-1 and LβT2 nuclear proteins respectively, bind to E-box 3 in electrophoretic mobility shift assays (EMSA) and are capable of activating expression of a GnRHR-Luciferase reporter in cotransfection assays, a response that is eliminated by mutation of E-box 3. Thus, regulation of the mouse GnRHR promoter via E-box 3 in αT3-1 versus LβT2 cells by different group A bHLH transcription factors (specifically NeuroD1 and Mash1) indicates that E-box 3 may be exploited by temporally sequential members of the bHLH transcription factor family during gonadotrope differentiation.
2. Materials and methods
2.1. Reporter and expression plasmids
The GnRHR-Luciferase reporter plasmid contains 1.2 kb of the 5′ flanking region of the mouse GnRHR gene cloned into the SmaI/XhoI restriction site of pGL3 (Promega, Madison, WI) as previously described (Albarracin et al., 1994). Site-specific mutations in E-box 3 and 4 consensus binding sites were made using the following oligonucleotides: µE-box 3 (5′-CCTACGATAAAAAGACGGGCCcTCTtCTGAGGGGC-3′) and µE-Box 4 (5’-CTTTCGACCATCAGAATTAGACTCCcAGTtTCCTTCCTCACCTAC-3′); (lowercase letters indicate mutated sequence). Mutagenesis was performed using the QuickChange Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The NeuroD1 expression plasmid has been previously described (Naya et al., 1995). The Mash1 pCLIG vector, generously provided by Dr. Kageyama, was digested with EcoRI to isolate the Mash1 cDNA, which was subsequently ligated into pcDNA3.1 digested with EcoRI (Hatakeyama et al.,2001) to create a pMash1 expression vector. The 4X E-box 3 multimer was constructed by annealing oligonucleotides containing 4 tandem copies of E-box 3 and ligating them into the NheI restriction site of pGL3 upstream of 81 bp of the herpes thymidine-kinase promoter (Coss et al., 2004).
2.2. Cell culture, transient transfections and luciferase and β-galactosidase assays
α T3-1 and LβT2 cells were grown as previously described (Rosenberg and Mellon,2002). The day prior to transfection, αT3-1 and LβT2 cells were plated into 12-well plates at a density of 2×105 cells per well. FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN) was used according to the manufacturer’s protocol. Each well was transfected with 400 ng of luciferase reporter plasmid along with 100 ng of a plasmid containing thymidine kinase controlling β-galactosidase gene expression as a control for transfection efficiency. The cells were harvested 24 hours after transfection. For co-transfection experiments, 200 ng of expression plasmid or the empty plasmid control was also transfected. Cell extracts were prepared and assayed for luciferase and β-galactosidase activity as previously described (Rosenberg and Mellon, 2002). Briefly, cells were washed in PBS, and then lysed with 80 µl of lysis buffer (Galacto-light assay system, Tropix, Bedford, MA). Luciferase and β-galactosidase activities were measured using an E.G. & G. Berthold Microplate Luminometer. Promega SteadyGlo luciferase assay reagent and the Tropix Galacto-light β galactosidase assay system were used according to the manufacturers’ protocols. All experiments were performed in triplicate, and results represent at least three independent replicates. Data are expressed as means ± the standard error of the mean. Thus, all of the transfection data shown represent the means ±SEM of at least three independent experiments each performed in triplicate. Luciferase values were normalized to β-galactosidase values to control for transfection efficiency and those values were normalized to the wild-type reporter gene or the vector control where noted. The means of the ratios of luciferase to β-galactosidase activity for each plasmid were compared by one-way ANOVA using the Tukey-Kramer HSD test. P values less than 0.05 were considered statistically significant. Within each cell line, bars designated with letters (αT3-1) or * (LβT2) are statistically significant as compared to wild-type GnRHR (as in Fig. 1B) or empty expression vector (as in Fig. 3 and Fig. 4).
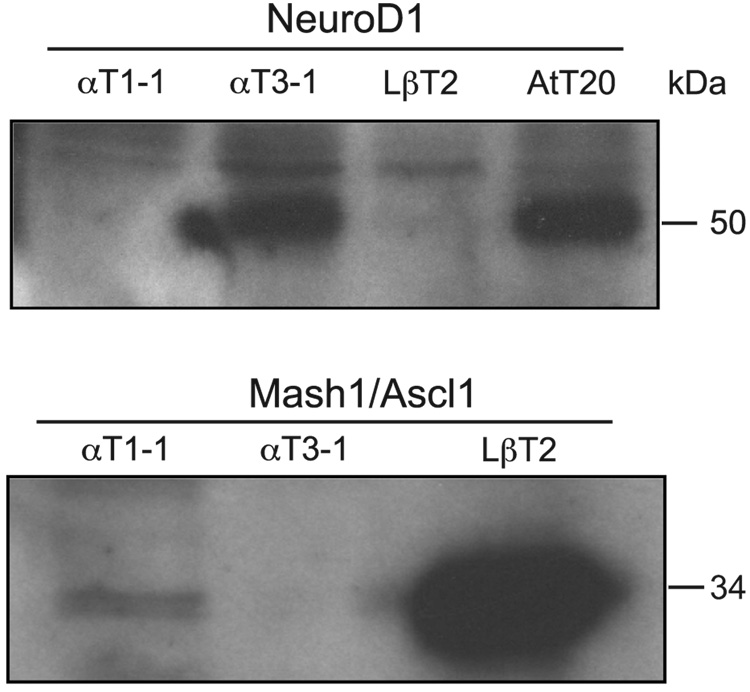
Fig. 1.
The basic helix-loop-helix transcription factors NeuroD1 and Mash1 are differentially expressed in the gonadotrope-derived αT3-1 and LβT2 cell lines. Western blot analysis of total protein from the gonadotrope-derived αT1-1, αT3-1, and LβT2 cell lines was used to confirm findings in the Affymetrix oligonucleotide arrays. Concentrations of protein in lysates were determined by Bradford assay and sample volume adjusted to ensure equal loading.
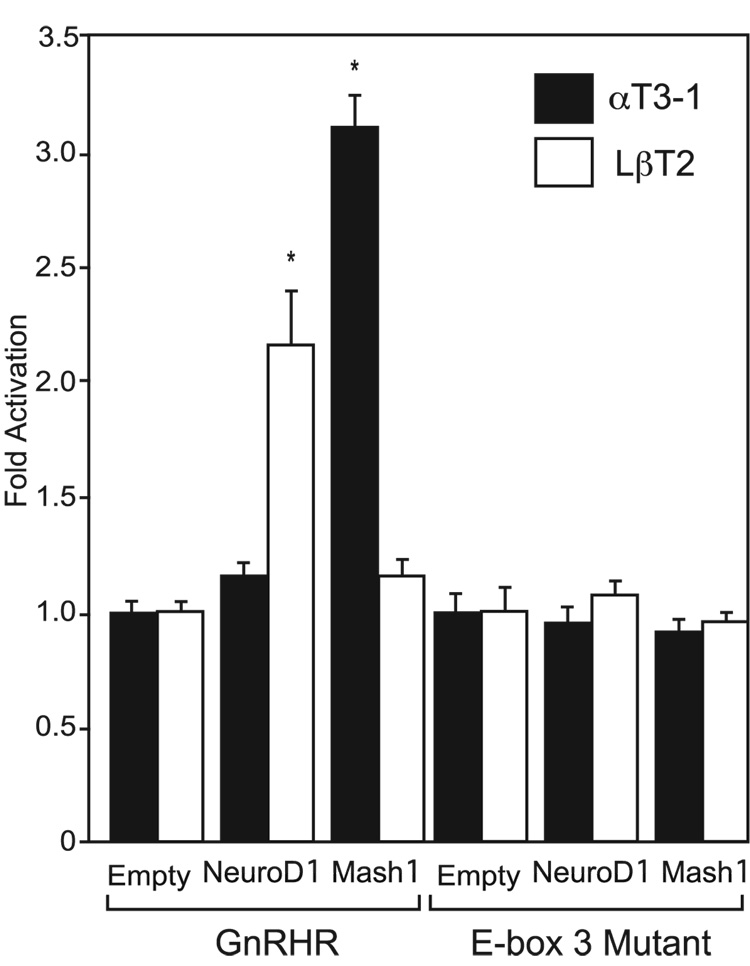
Fig. 3.
Over-expression of NeuroD1 in LβT2 cells or Mash1 in αT3-1 cells activates a wild-type GnRHR-Luciferase reporter gene, but activation is lost upon mutation of E-box 3. NeuroD1 and Mash1 expression vectors or equimolar amounts of an empty expression vector were cotransfected along with the GnRHR-Luciferase reporter gene (GnRHR) or E-box 3 mutant reporter (E-box 3 Mutant) into αT3-1 and LβT2 cells.
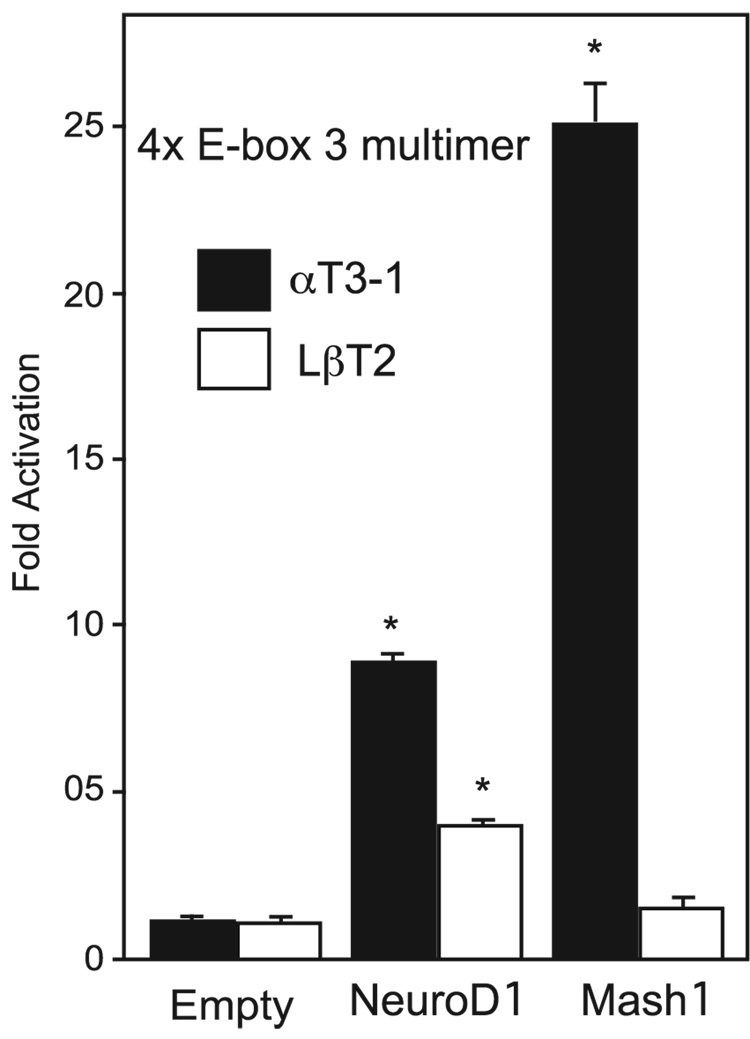
Fig. 4.
Over-expression of the basic helix-loop-helix proteins NeuroD1 or Mash1 are sufficient to increase expression of a 4X E-box 3 multimer reporter gene. αT3-1 and LβT2 cells were transiently transfected with E-box 3 multimer reporter gene and expression vectors for NeuroD1,Mash1, or empty expression vector, as a control.
2.3. RNA Isolation and microarray
Total RNA extraction was carried out using Ultraspec™RNA Isolation System (Biotecx Laboratories Inc. Houston, TX) per manufacturer’s instructions from cells grown in a monolayer on 10 cm plates. Total RNA was then resuspended in DEPC H20 and reprecipitated with 3 M NaOAC, pH 5.2, and then EtOH to eliminate any residual phenol or guanidinium salts. RNA was submitted to the UCSD-VA GeneChip Core facility for analysis using Affymetrix MOE430A chips. Data was analyzed using GeneSpring software (Silicon Genetics, Santa Clara, CA).
2.4. Western blotting
Cells were washed in ice cold PBS and lysed in RIPA buffer containing 20 mM Tris (pH 8.0), 137 mM NaCl, 10% glycerol, 1% NP-40, 0.1% SDS, 0.5% deoxycholate, and 0.2 mM PMSF. Protein concentration in lysates was determined by Bradford assay prior to gel loading to ensure equal protein loading. 6X sample buffer (300 mM Tris-HCl, pH 6.8, 60% glycerol, 30 mM DTT, 6% SDS) was added to yield a final concentration of 1X, lysates were heated to 95°C for 5 min. Samples were subjected to SDS polyacrylamide gel electrophoresis on a 10% gel (acrylamide:bis-acrylamide ratio of 29:1) and electro-blotted to Immobilin (Millipore, Billerica, MA). Membranes were blocked in 5% non-fat dried milk in Tris buffered saline (TBS). Anti-NeuroD1 (SantaCruz Biotechnology sc-1084) and anti-Mash1 (BD Pharmingen 556604) antibodies were used at a 1:1000 dilution with an incubation time of 2 hrs. Blots were washed and then incubated with a 1:10,000 dilution of anti-goat HRP or anti-mouse HRP for 2 hr at room temperature. All blots were washed for 60 min (6 ×10 min) with TBS after secondary antibody and then visualized by chemiluminescence using Pierce SuperSignal reagents (Rockford, IL).
2.5. Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from αT3-1 and LβT2 cells as previously described (Rosenberg and Mellon, 2002). The following probes were annealed, end-labeled using T4 Polynucleotide Kinase (New England Biolabs, Beverly, MA) according to the manufacturer’s protocol, and purified using G25 Probe Quant columns (Amersham, Piscataway, NJ): E-box 3 (5′- ACGGGCCATCTGCTGAGG -3′), Mutant E-Box 3 (5’-ACGGGCtgTCcaCTGAGG-3’),positive control POMC E-box (5′- GGAAGGCAGATGGACGCA-3′), positive control muscle creatine kinase enhancer E-box (MCK: 5’CCCCAACACCTGCTGCCT-3’). Binding reactions were carried out using 2 µg of nuclear protein and 4 fmol of [32P] labeled oligonucleotide in a 10 µl reaction containing 5 mM DTT, 0.025 µg/µl Poly dIdC, and binding buffer (50 mM HEPES pH 7.8, 250 mM KCl, 5 mM EDTA, and 30% glycerol). For competitions, 250 or 500-fold excess of double-stranded unlabeled oligonucleotides were used. After the addition of probe and nuclear protein, binding reactions were incubated 5 minutes on ice prior to electrophoresis on a 5% non-denaturing polyacrylamide gel in 0.25X TBE. For supershift assays, 1 µg of anti-NeuroD1 (Santa Cruz Biotechnology sc-763x), anti-E47 (Santa Cruz Biotechnology sc-763x),anti-Mash1/Ascl1 (Abcam ab 38557), anti-BMAL1 (Abcam ab49421), or normal mouse IgG (Santa Cruz Biotechnology sc-2025) were added to reactions prior to the addition of nuclear protein and incubated on ice for 10 minutes. Gels were run at 250V for approximately 1.5 hours, dried under vacuum, and exposed to autoradiographic film overnight.
3. Results
3.1. The group A basic helix-loop-helix transcription factors, NeuroD1 and Mash1, are differentially expressed in the αT3-1 versus LβT2 gonadotrope-derived cell lines
To better understand the differences in transcription factors expressed in αT3-1 versus LβT2 cells, we used Affymetrix oligonucleotide arrays to characterize gene expression patterns in the gonadotrope-derived cell lines. Comparison of the gene expression profiles between the αT3-1 and LβT2 cell lines reveals that a number of different bHLH proteins such as NeuroD1,Mash1, Id2, and Hes6 are expressed with distinct and somewhat overlapping profiles (data not shown). Our attention was drawn to the group A bHLH proteins NeuroD1 and Mash1 as promising candidates for regulating the mouse GnRHR through group A motif E-box 3 since both transcription factors have previously been shown to be expressed during pituitary development in vivo (Liu et al., 2001). The Affymetrix oligonucleotides array revealed that the αT3-1 cell line expresses 4.5-fold more NeuroD1 mRNA than LβT2 cells (P = 1.6 × 10−17). In contrast, LβT2 cells express 14-fold more Mash1 mRNA as compared to αT3-1 cells (P = 2.5 × 10−17). To verify the expression patterns observed in the microarrays, western blots were performed to compare the levels of NeuroD1 and Mash1 protein between the αT1-1, αT3-1 and LβT2 gonadotrope-derived cell lines. The αT1-1 cell line represents an earlier progenitor than even the αT3-1 cells in that it expresses αGSU, but not GnRHR or SF-1 (Alarid et al., 1996;Windle et al., 1990). Western blots revealed that NeuroD1 protein is much more highly expressed in αT3-1 versus LβT2 or αT1-1 cells, while the corticotrope-derived AtT20 cell extracts serve as a positive control. In contrast, Mash1 protein expression is much greater in LβT2 cells compared to either αT1-1 or αT3-1 cells confirming the microarray findings (Fig. 1).In fact, the differences in expression are much more pronounced in the western blots than the microarrays indicated. NeuroD1 is almost undetectable in LβT2 and Mash1 is almost undetectable in αT3-1, while both are absent or barely detectable in the earliest progenitor, αT1-1, that does not express gonadotrope-specific markers with the exception of αGSU. Thus, the bHLH family members NeuroD1 and Mash1 are differentially expressed in the gonadotrope-derived cell lines.
3.2. Mutation of E-box 3, but not E-box 4, significantly decreases basal activity of the mouse GnRHR promoter
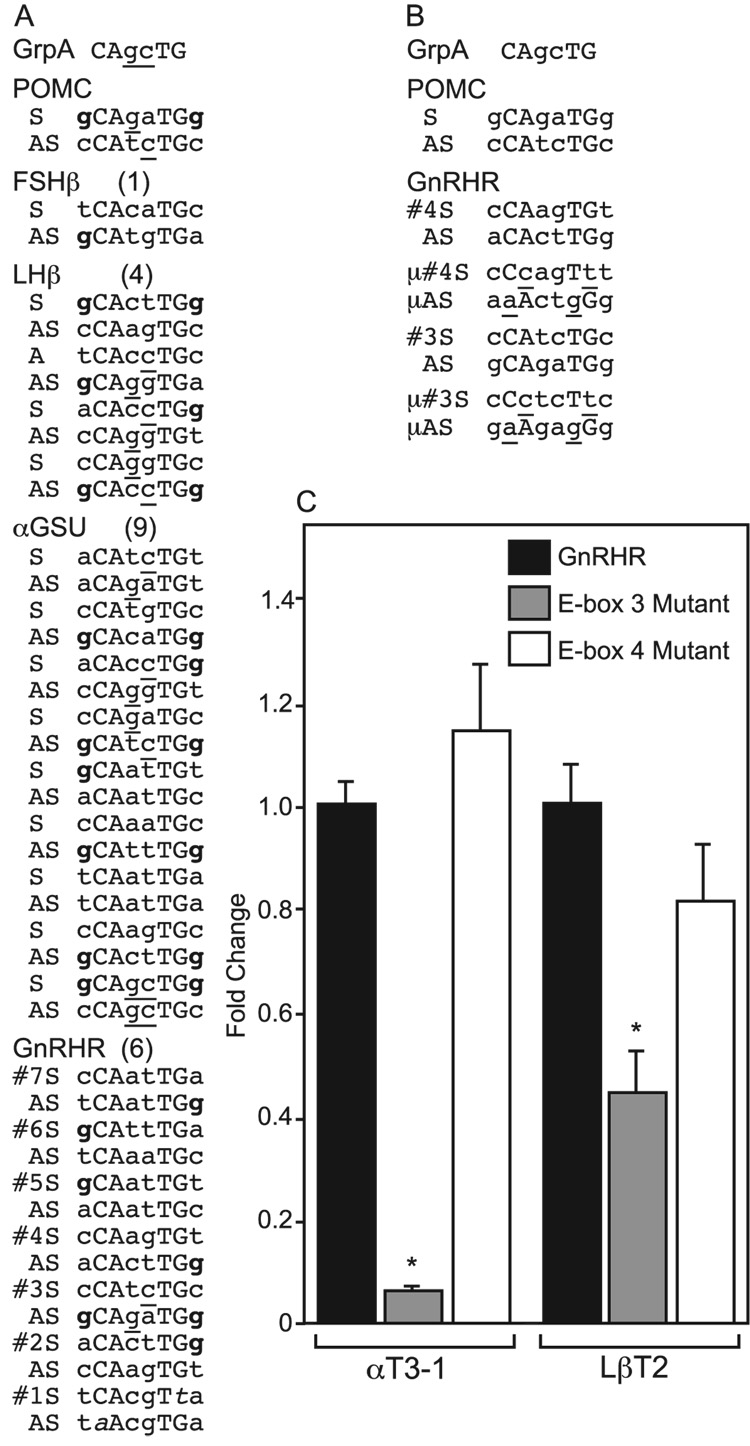
To investigate the function of the bHLH proteins in the differentiation of the gonadotrope lineage, we scanned the first 1000 bp of the promoter regions of the mouse gonadotrope-specific end-target genes, GnRHR, α-GSU, LHβ, and FSHβ. By the simple definition of the sequence requirements for an E-box (CAnnTG), the GnRHR gene has 6, the α-subunit gene has 9, while LHβ gene has 4, and the FSHβ gene has only 1 (Fig. 2A). Fig. 2A shows the sequences of these E-boxes from 5’ to 3’ from both the “coding” and non-coding, i.e, reverse strands (AS). Though the E-box is simply defined as CAnnTG, in fact, the internal and adjacent base pairs play important roles in the specificity of binding. The “E-box” #1 previously noted by Resuehr, et al.(2007), does not fit this definition of an E-box in that it lacks the G of the CAnnTG. As noted above, group A proteins, including NeuroD1, Mash1, and E47, preferentially bind to E-boxes with internal bases of GC as in CAGCTG, while group B bHLH proteins prefer internal bases of CG as in CACGTG (Li et al., 2006). In addition, a consensus Mash1 binding site identified using phylogenetic footprinting also has the internal GC bp (5’-CAGCTG-3’) (Hu et al., 2004),supporting this conclusion. Only 1 E-box in GnRHR, 4 in the α-subunit, 3 in LHβ, and 0 in FSHβ, have at least one of the two central bp in the correct position on at least one strand (underlined in Fig. 2A). Furthermore, an established NeuroD1 binding site (DE2C) from the POMC gene promoter expressed in pituitary corticotrope cells has been studied in detail (Poulin et al., 1997), determining that this protein prefers the two flanking bp to be guanosine nucleotides (5’-GCAGATGG-3’) (bolded in Fig. 2A). Of those E-boxes that have at least one internal bp matching the group A pattern of GC, only 1 E-box in GnRHR (#3), 1 in the α-subunit (the most proximal), 1 in LHβ, and 0 in FSHβ have two flanking bp that match this requirement. In fact, the GnRHR E-box 3 is the only one that is an 8 of 8 bp match (in the antisense strand 5’-GCAGATGG-3’) with the well-characterized NeuroD1-binding POMC E-box, including the important 5’ and 3’ flanking nucleotides. Furthermore, E-box 3 is evolutionarily conserved in the GnRHR genes of many mammalian species. All eight bp are found in the same location in mouse, rat, porcine, and human GnRHR genes, while the other E-boxes in this gene are not conserved outside of rodent species. Thus, we chose to focus on this E-box in the GnRHR gene as a potential target for NeuroD1 and Mash1.
Fig. 2.
Mutation of E-box 3 significantly decreases basal activity of a mouse GnRHR-Luciferase reporter gene in αT3-1 and LβT2 cells. (A) E-box sequences from the proximal 1000 bp of each gene are shown in comparison to the well-characterized POMC E-box and the Group A consensus (GrpA). Within each gene, both the sense (S) and antisense (AS) strands are shown in the 5’ to 3’ orientation. They are ordered from the most distal (top) to the most proximal (bottom) within each gene. The defining bases (CAnnTG) are in capital letters. The internal base pair matches for the Group A bHLH consensus (GrpA, shown at the top) are underlined. The flanking bp matches for to the well-defined NeuroD1 binding site from the POMC gene (shown second) are in bold. (B) Mutated sequences of E-boxes 3 and 4 from the GnRH gene are shown with the mutated bases underlined. (C) The GnRHR-Luciferase reporter either wild-type or containing mutations in either E-box 4 or E-box 3 were transiently transfected into αT3-1 cells or LβT2 cells.
Therefore, we examined the group A E-box, E-box 3 (−208/−203), and compared it to an adjacent group B E-box, E-box 4 (−244/−239) (Li et al., 2006). Initially, we mutated E-boxes 3 and 4 to address their contribution to mouse GnRHR promoter activity (Fig. 2B). Both αT3-1 and LβT2 cells were transiently transfected with a wild-type GnRHR-Luciferase reporter or reporters containing a mutation in either E-box 3 or 4. In transient transfections, mutation of E-box 3, but not E-box 4, in both αT3-1 and LβT2 cells, results in a significant decrease in reporter gene expression (Fig. 2C). Interestingly, while E-box 3 mutation affects expression in both cell lines, its effect is more robust in the αT3-1 cells. For example, E-box 3 mutation decreases expression 95% in αT3-1 cells compared to wild-type reporter gene expression, whereas in LβT2 cells, E-box 3 mutation decreases expression 57%. In contrast, mutation of E-box 4 does not significantly decrease GnRHR-Luciferase expression compared to wild-type reporter. Thus, it appears that E-box 3 is critical for expression of the GnRHR in both αT3-1 and LβT2 cells.
3.3. Over-expression of group A bHLH family members NeuroD1 and Mash1 directly activate a mouse GnRHR-Luciferase reporter gene through E-box 3
To determine whether NeuroD1 or Mash1 is capable of activating transcription of a GnRHR-Luciferase reporter gene, transient transfections were carried out in αT3-1 and LβT2 cells. NeuroD1 and Mash1 expression vectors, or equimolar amounts of an empty expression vector, were co-transfected along with the GnRHR-Luciferase reporter gene into αT3-1 and LβT2 cells (Fig. 3A). Over-expression of NeuroD1 in αT3-1 cells does not alter GnRHR-Luciferase expression, which may be due to high levels of endogenous expression of NeuroD1 in the cell line. In contrast, over-expression of NeuroD1 in LβT2 cells, in which the protein in not highly expressed, activates the GnRHR reporter by 2 fold as compared to empty control (P<0.05). In a similar fashion, over-expression of Mash1 in LβT2 cells has no effect, however in αT3-1 cells, a 3-fold activation occurs. The effect of over-expression of both NeuroD1 and Mash1 in αT3-1 and LβT2 cells was blocked when E-box 3 was mutated (Fig. 3B), indicating that either the other E-boxes cannot bind group A bHLH proteins or that the participation of this specific E-box is required for activation of the mouse GnRHR promoter. As such, E-box 3 can differentially mediate the stimulatory role of group A bHLH transcription factors NeuroD1 and Mash1 in αT3-1 versus LβT2 cells.
3.4. Four tandem copies of mouse GnRHR E-box 3 fused to a minimal promoter are sufficient to mediate bHLH responsiveness
We next addressed whether E-box 3 alone is sufficient to mediate bHLH responsiveness of the mouse GnRHR promoter. To accomplish this, four copies of E-box 3 were fused upstream of a thymidine kinase minimal promoter (4X E-box 3 multimer) and co-transfected into αT3-1 and LβT2 cells with either an equimolar amount of empty vector control, NeuroD1, or Mash1 expression vectors. Co-transfection of NeuroD1 with the 4X E-box 3 reporter in αT3-1 cells results in an 8-fold increase in luciferase expression compared to the empty vector control (Fig.4). However, this effect cannot be observed with a 4X multimer of E-box 4, which is not activated by NeuroD1 in αT3-1 cells (data not shown). Over-expression of Mash1, which is not highly expressed in αT3-1 cells, results in a 25-fold increase in 4X E-box 3 reporter activity. In addition, we tested whether over-expression of NeuroD1 or Mash1 is also capable of activating the 4X E-box 3 reporter in LβT2 cells. The 4X E-box 3 reporter was transactivated by NeuroD1 over-expression in LβT2 cells with a 4-fold increase in activity compared to empty vector control (Fig. 4). Over-expression of Mash1 in LβT2 cells, however, resulted in only a relatively small 1.2 fold increase in activity, which was not statistically significant, again probably due to the high endogenous level of Mash1 in these cells. Thus, E-box 3 is sufficient to mediate responsiveness of the mouse GnRHR promoter to group A members of the bHLH family of transcription factors. Furthermore, these data also indicate that mouse GnRHR E-box 3 can be the target for at least two members of the group A bHLH transcription factor family that are critical for cell fate commitment and present during gonadotrope development.
3.5. NeuroD1, Mash1, and E47, the bHLH dimerization partner, can bind mouse GnRHR E-box 3 In Vitro
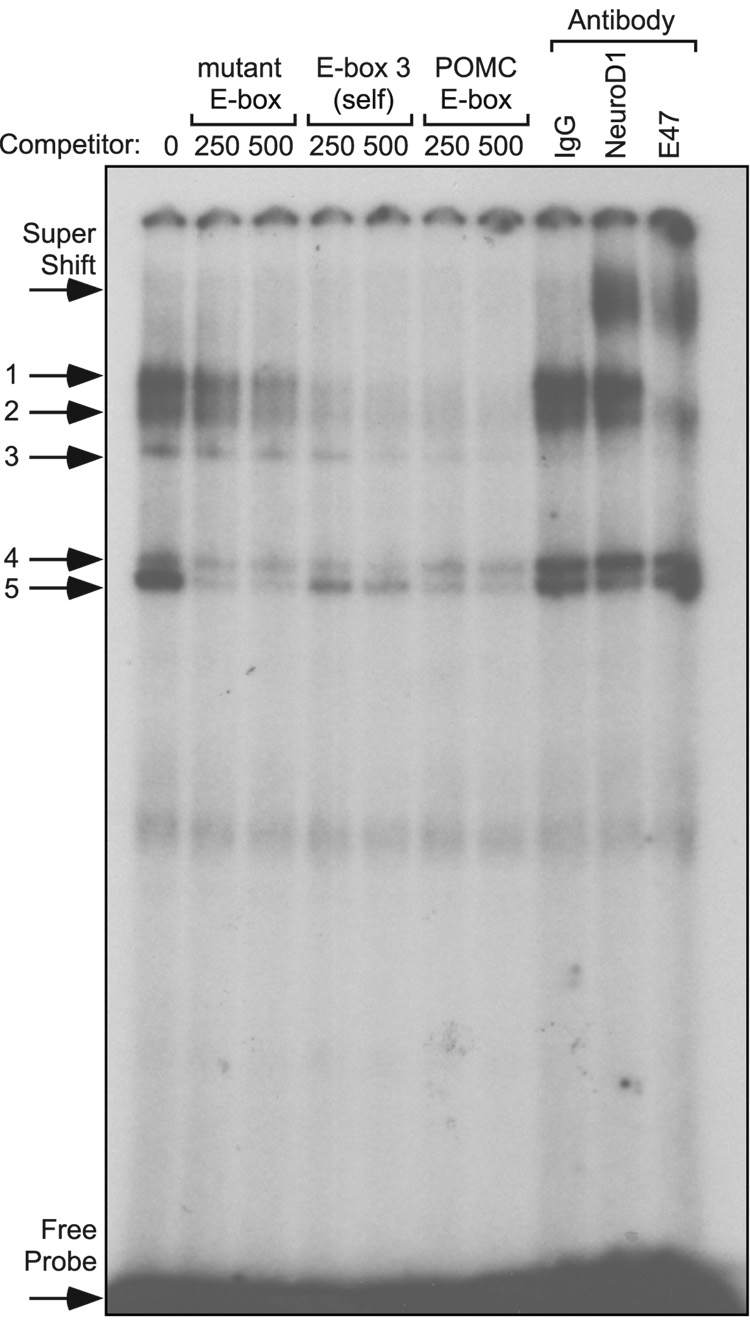
To test whether E-box 3 can bind group A bHLH family member NeuroD1 in combination with the bHLH dimerization partner E47, EMSAs were performed using radiolabeled oligonucleotides encompassing E-box 3 with nuclear extracts from αT3-1 cells. Specificity of the protein complexes was determined by competition with 250- or 500-fold excess of unlabeled oligonucleotides. Competition with either 250- or 500-fold excess of unlabelled oligonucleotides containing a mutated E-box 3 (see Fig. 2B) were weak competitors for protein complexes 1, 2, and 3, while complexes 4 and 5 were competed away, indicating that complexes 4 and 5 are not E-box-specific binding events (Fig. 5). Competition with homologous E-box 3 oligonucleotides eliminates complexes 1, 2, and 3, as did an oligonucleotide containing a characterized E-box from the POMC gene promoter which serves as a positive control (Poulin et al., 1997) (Fig. 5). Consequently, complexes 1, 2, and 3 represent specific binding events at mouse GnRHR E-box 3. To identify the proteins in these complexes, antibodies directed against NeuroD1 and E47, in addition to an equal mass of mouse IgG, were included in the EMSA reactions. Addition of the NeuroD1 antibody shifted radioactivity from complexes 1 and 2, and eliminated complex 3, resulting in the formation of a new complex of markedly decreased mobility (supershift). Inclusion of the antibody directed against the bHLH dimerization partner E47 eliminates complexes 1 and 3, while decreasing intensity of complex 2, and also results in the formation of a new complex of decreased mobility (Fig. 5). In contrast, no specific protein complexes were ever discerned binding to the 5’ E-box oligonucleotide probe, and inclusion of NeuroD1 or E47 antibodies had no effect on the promiscuously competed protein complexes that did bind to this region (data not shown). These results indicate that both NeuroD1 and E47 are capable of interacting with mouse GnRHR promoter E-box 3, and that they may do so as heterodimers, since inclusion of antibodies directed against either of these proteins reduces the intensity of the same protein complexes.
Fig.5.
NeuroD1 and E47 bind to radiolabeled oligonucleotides containing E-box 3 from the mouse GnRHR gene promoter. EMSAs were performed with radiolabeled oligonucleotides encompassing E-box 3 in the 5' flanking region of the mouse GnRHR gene. Radiolabeled oligonucleotide probe was incubated with nuclear proteins from αT3-1 cells. Specific protein/DNA complexes were identified by competition with 250- or 500-fold excess unlabelled oligonucleotides containing a mutated E-box 3 (µE-box), homologous competitor (wild type Ebox 3), or a positive control E-box from the POMC gene (POMC). Identification of the bHLH transcription factors in specific complexes was determined by inclusion of an antibody directed against NeuroD1 or the ubiquitously expressed bHLH heterodimerization partner, E47.
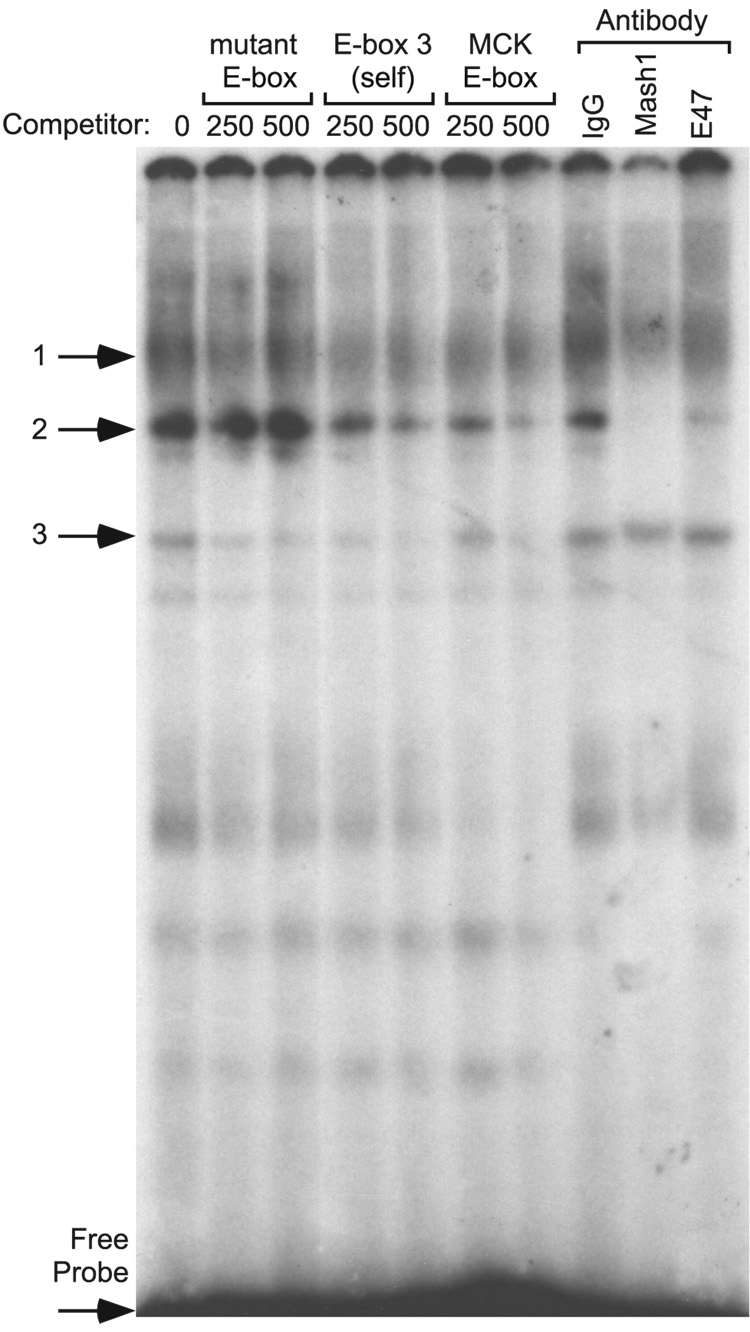
We next investigated the ability of group A bHLH family member Mash1 to bind mouse GnRHR E-box 3. To address this question, nuclear proteins were isolated from LβT2 cells, which express Mash1 at higher levels than αT3-1 cells. While complex 1 appears to be non-specific, oligonucleotides containing mutated E-box 3 competed for complex 3, but were unable to compete for complex 2. However, 250- or 500-fold excess of unlabelled homologous oligonucleotides and muscle creatine kinase enhancer (MCK) oligonucleotides, which have been previously shown to bind Mash1 (Weintraub et al., 1990), both compete for binding of complex 2, indicating a specific binding event. Addition of the Mash1/Ascl1 antibody eliminated radioactivity from complex 2, and inclusion of an antibody directed against E47 reduced the intensity of complex 2 (Fig. 6). These data indicate that both Mash1 and E47 found in LβT2 cells are capable of interacting with mouse GnRHR E-box 3 and may do so as a heterodimer.
Fig. 6.
Mash1 and E47 bind to radiolabeled oligonucleotides containing E-box 3 from the mouse GnRHR gene promoter. EMSAs were performed with radiolabeled oligonucleotides encompassing E-box 3 in the 5' flanking region of the mouse GnRHR gene. Radiolabeled oligonucleotide probe was incubated with nuclear proteins from LβT2 cells. Specific protein/DNA complexes were identified by competition with 250- or 500-fold excess unlabelled oligonucleotides containing a mutated E-box 3 (µE-box), homologous competitor (wild-type E-box 3), or a positive control (MCK). Identification of the bHLH transcription factors in specific complexes was determined by inclusion of an antibody directed against Mash1 or the ubiquitously expressed bHLH heterodimerization partner, E47.
3.6. Binding of NeuroD1 and Mash1 to mouse GnRHR E-box 3 is specific to the αT3-1 and LβT2 cells, respectively
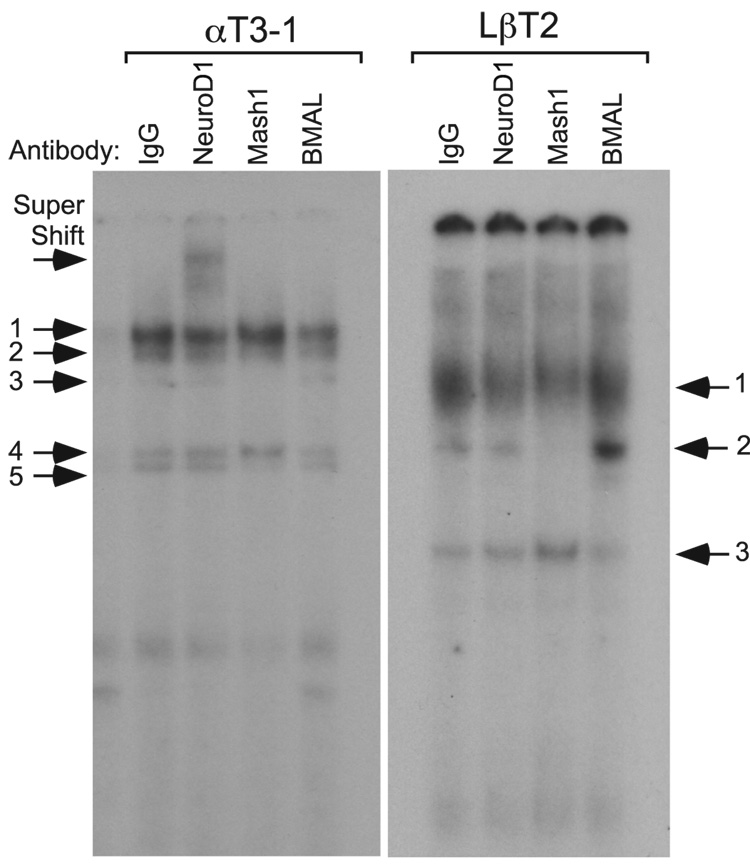
Both western and microarray analysis showed high levels of NeuroD1 in αT3-1 cells and Mash1 in LβT2 cells. We were concerned, however, that the NeuroD1 in LβT2 cells and Mash1 in αT3-1 cells, albeit at lower concentrations, still might be able to bind an E-box 3 probe. In addition, it was important to test whether a group B bHLH transcription factor such as BMAL1/Clock heterodimers could bind to mouse GnRHR E-box 3. To address these questions, EMSA were performed using nuclear proteins from αT3-1 and LβT2 cells and a radiolabeled Ebox 3 probe. Prior to addition of nuclear proteins, NeuroD1, Mash1, or BMAL1 antibodies, or IgG were added to appropriate reactions. Only the addition of NeuroD1 antibody to αT3-1 nuclear proteins results in a supershifted complex, inclusion of either Mash1 or BMAL1 had no effect (Fig. 7). In the converse experiment using LβT2 nuclear proteins, only the inclusion of Mash1 antibody caused an abrogation of the specific binding complex. Consequently, GnRHR E-box 3 is highly specific for the binding of group A bHLH family members NeuroD1 from αT3-1 cells and Mash1 from LβT2 cells versus the one group B bHLH transcription factor tested, BMAL1.
Fig. 7.
The binding of group A bHLH family members NeuroD1 from αT3-1 cells and Mash1 from LβT2 cells to E-box 3 is highly specific and preferential to the binding of group B bHLH family members. EMSAs were performed with radiolabeled oligonucleotides encompassing E-box 3. Radiolabeled oligonucleotide probe was incubated with nuclear proteins from αT3-1 or LβT2 cells. Specificity of binding complexes was determined by inclusion of antibodies directed against NeuroD1, Mash1, and BMAL1, or IgG as a control.
4. Discussion
Programmed cascades of regulatory factors define the differentiation of specific cell types in developing tissues during organogenesis. Sequential expression of specific bHLH proteins is an emerging paradigm for regulating individual genes during differentiation. We have demonstrated that the closely related bHLH factors, NeuroD1 and Mash1 are differentially expressed in the progenitor gonadotrope, αT3-1, versus the more mature gonadotrope, LβT2.Although NeuroD1 and Mash1 are known to be critical for development in multiple systems including the anterior pituitary (Liu et al., 2001; Miyata et al., 1999; Morrow et al., 1999; Naya et al., 1997), their specific role in the gonadotrope gene program has remained poorly characterized. Consequently, examining NeuroD1 and Mash1 in αT3-1 and LβT2 cells sheds light on the role of bHLH proteins in GnRHR expression during gonadotrope development.
We have identified an E-box regulatory element that is critical for developmental expression of the mouse GnRHR gene. E-box 3 can bind and be trans-activated by the group A bHLH transcription factors NeuroD1 and Mash1, in αT3-1 and LβT2 cells, respectively, to activate GnRHR expression. Thus, this E-box may represent an element that is used by different group A bHLH transcription factors during developmental expression of the GnRHR gene as part of the gonadotrope-specific gene program.
Recent work by Resuehr, et al. (2007) showed that the proximal mouse GnRHR gene promoter contains seven putative E-box elements. The authors show that the group C bHLH CLOCK protein can interact with at least one of three E-box elements in the mouse GnRHR gene promoter in ChIP assays (Resuehr et al., 2007); however, the authors did not identify which of the three E-box elements is involved nor address whether the same interactions occur in the more mature LβT2 gonadotrope-derived cell line. Interestingly, work by Li, et al. (Li et al., 2006),describes the nucleotide binding preference for specific groups (A–F) of bHLH proteins. Group A proteins, including NeuroD1, Mash1, and E47 preferentially bind to the motif (5’-CAGCTG-3’); in contrast, group B bHLH proteins bind the motif (5’-CACGTG-3’) (Li et al., 2006).Mouse GnRHR E-box 3 (5’-CAtCTG-3’) is the closest match to a group A binding motif within the proximal 1000 bp the GnRHR gene. Furthermore, this E-box is an 8 of 8 bp match in the antisense strand (AS 5’-gCAGATGg-3’), including the important 5’ and 3’ flanking nucleotides, with an established NeuroD1 binding site (DE2C) from the POMC gene promoter (5’-gCAGATGg-3’) (Poulin et al., 1997) and a 5 of 6 bp match with a consensus Mash1 binding site identified using phylogenetic footprinting (5’-CAGCTG-3’) (Hu et al., 2004). We were also interested in the contribution of group B bHLH binding motifs to mouse GnRHR promoter activity. To address this, we mutated E-box 4 (5’-CAAGTG-3’), which only varies from the consensus group B binding motif (5’-CACGTG-3’) by 1 nucleotide. Mutation of E-box 4 did not significantly attenuate mouse GnRHR promoter activity in either αT3-1 or LβT2 cells suggesting that group B motif E-box 4 is not critical for activity of the promoter. This is in contrast to the results of Resuehr, et al. (2007), in which cis-mutation of every E-box they identified reduced expression in αT3-1 cells. This discrepancy is likely due to the fact that we selectively mutated only the first and last nucleotides of the putative E-box to create precise mutations, while Resuehr, et al., mutated all six nucleotides in the seven E-boxes, which could have affected other binding proteins on adjacent or overlapping sites.
Our reasons were several-fold for pursuing the roles of NeuroD1 and Mash1 in regulation of the mouse GnRHR promoter. First, both NeuroD1 and Mash1 are expressed in the ventral most regions of the developing mouse anterior pituitary and overlap with the approximate time of GnRHR expression in developing gonadotropes (Liu et al., 2001). Second, microarray analysis showed significant differences in expression patterns of NeuroD1 and Mash1 between αT3-1 and LβT2 cells. Western blots confirmed the microarray findings showing higher levels of NeuroD1 in αT3-1 versus LβT2 cells, and greater Mash1 expression in LβT2 cells versus αT3-1 cells.
In over-expression studies, NeuroD1 activates the GnRHR-Luciferase reporter in LβT2 cells by 2 fold, while Mash1 activates the promoter in αT3-1 cells by 3 fold. With mutation of E-box 3, all the responses were eliminated, while mutation of E-box 4 had no effect on basal expression in either cell, confirming the critical role of E-box 3 in mediating the activity of group A bHLH transcription factors. The inability of NeuroD1 over-expression to stimulate expression via E-box 3 in αT3-1, and Mash1 to stimulate in LβT2 cells, is most likely due to high endogenous expression of the proteins in these cell lines. The presence of high levels of a transcription factor often makes it difficult to increase the levels substantially by co-transfection with an expression vector. The same result is seen in over-expression studies with the 4X E-Box 3 multimer, where over-expression of NeuroD1 increases activity of the multimer in LβT2 cells more robustly than in αT3-1, while Mash1 increases activity of the multimer only in LβT2 cells, not in αT3-1 cells. Alternative hypotheses can be put forward for these results. Perhaps limiting amounts of specific forms of bHLH partner proteins present in each cell line could affect heterodimerization partner availability and, therefore, ability to activate transcription. Alternatively, limiting levels of co-activator complexes that differentially associate with these bHLH proteins in their specific cells might create a situation in which all of the key co-activator is bound by the endogenous protein, preventing activation by overexpressed bHLH proteins. Another potential mechanism might be limiting levels or activities of enzymes necessary for posttranslational modification of the bHLH proteins, such that additional bHLH protein is not properly modified to allow activation.
EMSA showed that mouse GnRHR E-box 3 can bind group A bHLH transcription factors from both αT3-1 and LβT2 cells. EMSAs with αT3-1 nuclear proteins produced three specific binding complexes as determined by competition. Furthermore, addition of NeuroD1 or E47 antibodies “supershifted” complexes away from all three of the specific bands. As such, E-box 3 is capable of binding NeuroD1 and appears to do so by heterodimerizing with E47. Using nuclear proteins from LβT2 cells, we also detected one specific binding event with an oligonucleotide probe containing mouse GnRHR E-box 3. Inclusion of antibodies to Mash1 and E47 eliminated or attenuated binding of the specific complex. We conclude that the bHLH transcription factors NeuroD1 and Mash1 bind E-box 3 as heterodimers with the binding partner E47. Finally, crossing the group A bHLH antibodies showed that the binding of NeuroD1 in αT3-1 cells and Mash1 in LβT2 cells is highly specific and preferential to the binding of the one group B bHLH family member tested. What remains to be determined is the role these proteins may play in vivo in regulating or timing GnRHR gene expression during the course of pituitary development.
As previously mentioned, NeuroD1 expression in the pituitary commences at e11.5 and becomes undetectable by e14.5–e15.5 (Liu et al., 2001). Initial activation of GnRHR expression in the developing gonadotrope falls within this same developmental window, which also correlates with the approximate gonadotrope-specific gene expression profile of the αT3-1 cell model (i.e. expression of the α-GSU and GnRHR). Regulation of a pituitary-specific gene by NeuroD1 via an E-box is not without precedent during anterior pituitary development. Work from Lamolet, et. al. (Lamolet et al., 2004), showed that the POMC gene, the expression of which is confined to corticotropes in the anterior pituitary, is regulated by NeuroD1/E47 heterodimers binding an E-box in the gene’s promoter (Poulin et al., 1997). Although NeuroD1 knockout mice show a delay in corticotrope appearance and minor alteration in α-GSU expression, the authors did not examine GnRHR expression (Lamolet et al., 2004).
Presently, the role of Mash1 in anterior pituitary development and the gonadotrope gene program is less clear than that of fellow bHLH family member NeuroD1. Mash1 is expressed in the ventral aspect of the developing mouse anterior pituitary and has been proposed to exert a role in terminal differentiation of the gonadotrope (Zhu et al., 2006). Interestingly, in Zebrafish, the Mash1 homolog, ascl1a, is critical for adenohypophysis formation, and in its absence, all pituitary cell types fail to differentiate (Pogoda and Hammerschmidt, 2007; Pogoda et al., 2006).Since mouse GnRHR expression is initiated around e13.5, it is possible that NeuroD1 is involved in early activation and regulation of the gene. As gonadotrope development progresses and levels of bHLH transcription factors change, E-box 3 is likely exploited by other bHLH proteins such as Mash1. Consequently, cellular programming during gonadotrope development probably requires sequential combinations of transcription factors during different phases of maturation that ultimately activate the gonadotrope-specific genes.
The role of bHLH proteins in the development of several physiological systems has been well characterized (Miyata et al., 1999; Morrow et al., 1999; Naya et al., 1997). Numerous parallels exist between such developmental programs and that of the pituitary, making it tempting to speculate about the potential role these proteins might play in pituitary gonadotrope development as well. A key question yet to be addressed is whether GnRHR expression during development would be delayed or otherwise disrupted in the absence of NeuroD1, Mash1, or both. Indeed, a similar situation has been shown to exist during retina development (Hatakeyama et al., 2001; Inoue et al., 2002). In that case, bHLH family members Math3 (Neurod4) and NeuroD1 are both transiently expressed in developing retinal interneurons or amacrine cells. Loss of Math3 expression alone does not disrupt interneuron development. Loss of NeuroD1 delays differentiation slightly, but does not affect final amacrine cell number in the retina. However, loss of both Math3 and NeuroD1 expression results in complete ablation of the amacrine cell type (Inoue et al., 2002). In addition, sequential patterns of the same bHLH proteins that are required during retina development have also been shown to be important for spinal cord (Lee and Pfaff, 2003) and pancreas (Gasa et al., 2004), among others. Similar sequential patterns of action and functional redundancies clearly exist in anterior pituitary development (Zhu et al., 2007).
In summary, we have shown that E-box 3 in the mouse GnRHR gene proximal promoter is activated by and can bind members of the developmentally critical group A bHLH transcription factor family found in the gonadotrope derived αT3-1 and LβT2 cell lines. E-box 3 may represent a promoter regulatory element that is utilized by sequential group A bHLH transcription factors during gonadotrope development. Understanding the role different bHLH transcription factors play in gonadotrope development will require careful characterization of gonadotrope gene expression in pituitary-specific bHLH knockout mice throughout development, most likely including examination of animals with deletion of multiple bHLH proteins.
Acknowledgments
The authors thank Dr. Kathy Pinson for contributions to the early stages of this research and Susan Mayo for excellent technical assistance. We thank Drs. Varykina Thackray, Kellie Breen-Church, and Rachel Larder for helpful discussions and critical review of the manuscript. The authors also thank Drs. Nicholas J.G. Webster and Mark A. Lawson for assistance with microarray analysis, which was carried out through the UCSD Cancer Center Microarray Core facility.
This research was supported by NIH grant R01 HD020377 (to P.L.M.). This work was also supported by NICHD/NIH through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (P.L.M.).B.D.C. was supported in part by NIH T32 HD007203, J.S.B. was supported in part by NIH T32 GM08666, and A.L.D. was supported in part by NIH T32 GM007198 and a supplement to U54 HD012303. P.L.M. is a member of the Biomedical Sciences Graduate Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- Albarracin CT, Kaiser UB, Chin WW. Isolation and characterization of the 5'-flanking region of the mouse gonadotropin-releasing hormone receptor gene. Endocrinology. 1994;135:2300–2306. doi: 10.1210/endo.135.6.7988412. [DOI] [PubMed] [Google Scholar]
- Coss D, Jacobs SB, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J. Biol. Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval DL, Nelson SE, Clay CM. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol. Endocrinol. 1997;11:1814–1821. doi: 10.1210/mend.11.12.0020. [DOI] [PubMed] [Google Scholar]
- Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang T, Stormo GD, Gordon JI. RNA interference of achaete-scute homolog 1 in mouse prostate neuroendocrine cells reveals its gene targets and DNA binding sites. Proc. Natl. Acad. Sci. USA. 2004;101:5559–5564. doi: 10.1073/pnas.0306988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Barnhart KM, Mellon PL, Gutierrez-Hartmann A, Hoeffler JP. Helix-loop-helix proteins are present and differentially expressed in different cell lines from the anterior pituitary. Mol. Cell. Endocrinol. 1993;96:167–176. doi: 10.1016/0303-7207(93)90107-u. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Barnhart KM, Mellon PL, Gutierrez-Hartmann A, Hoeffler JP. The role of helix-loop-helix proteins in gonadotropin gene expression. In: Lustbader JW, et al., editors. Glycoprotein Hormones: Structure, Function and Clinical Implications. New York: Springer-Verlag, Inc.; 1993. pp. 44–64. [Google Scholar]
- Japon MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J. Histochem. Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- Lamolet B, Poulin G, Chu K, Guillemot F, Tsai MJ, Drouin J. Tpit-independent function of NeuroD1(BETA2) in pituitary corticotroph differentiation. Mol. Endocrinol. 2004;18:995–1003. doi: 10.1210/me.2003-0127. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:7311–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Q, Qiu M, Pan Y, Li Y, Shi T. Identification and analysis of the mouse basic/Helix-Loop-Helix transcription factor family. Biochem. Biophys. Res. Commun. 2006;350:648–656. doi: 10.1016/j.bbrc.2006.09.114. [DOI] [PubMed] [Google Scholar]
- Liu J, Lin C, Gleiberman A, Ohgi KA, Herman T, Huang HP, Tsai MJ, Rosenfeld MG. Tbx19, a tissue-selective regulator of POMC gene expression. Proc. Natl.Acad. Sci. USA. 2001;98:8674–8679. doi: 10.1073/pnas.141234898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–2185. doi: 10.1210/en.2004-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- Norwitz ER, Cardona GR, Jeong KH, Chin WW. Identification and characterization of the gonadotropin-releasing hormone response elements in the mouse gonadotropin-releasing hormone receptor gene. J. Biol. Chem. 1999;274:867–880. doi: 10.1074/jbc.274.2.867. [DOI] [PubMed] [Google Scholar]
- Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang H-J, Miller WL, Mellon PL. Cell-specific transcriptional regulation of FSHβ by activin and GnRH in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, Hammerschmidt M. Molecular genetics of pituitary development in zebrafish. Semin. Cell Dev. Biol. 2007;18:543–558. doi: 10.1016/j.semcdb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, von der Hardt S, Herzog W, Kramer C, Schwarz H, Hammerschmidt M. The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis. Development. 2006;133:1079–1089. doi: 10.1242/dev.02296. [DOI] [PubMed] [Google Scholar]
- Poulin G, Lebel M, Chamberland M, Paradis FW, Drouin J. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 2000;20:4826–4837. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin G, Turgeon B, Drouin J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 1997;17:6673–6682. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resuehr D, Wildemann U, Sikes H, Olcese J. E-box regulation of gonadotropin-releasing hormone (GnRH) receptor expression in immortalized gonadotrope cells. Mol.Cell. Endocrinol. 2007;278:36–43. doi: 10.1016/j.mce.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol.Endocrinol. 2002;16:1280–1298. doi: 10.1210/mend.16.6.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Hecht JH, Mellon PL. GATA-binding proteins regulate the human gonadotropin α-subunit gene in placenta and pituitary. Mol. Cell. Biol. 1994;14:5592–5602. doi: 10.1128/mcb.14.8.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc.Natl. Acad. Sci. USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol. Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- Zhu X, Gleiberman AS, Rosenfeld MG. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol. Rev. 2007;87:933–963. doi: 10.1152/physrev.00006.2006. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]