Abstract
Layer-by-layer (LbL) assemblies of DNA and polycations on the surface of colloidal templates can be used for gene delivery. Plasmid DNA encoding for secreted alkaline phosphatase (SEAP) was used to deposit LbL films with poly(ethylenimine) (PEI) on the surface of polystyrene and poly(lactide-co-glycolide) microparticles. The formation of LBL films was confirmed by zeta potential analysis and fluorescence and atomic force microscopy techniques. The LbL particles were rapidly internalized in a dose-dependent manner by J774.1 murine macrophages. Transfection activity of the LbL particles was evaluated in J774.1 cells using three different doses (5, 10, 25 particle per cell). The levels of SEAP expression increased with increasing dose but were lower than transfection levels mediated by control PEI/DNA polyplexes at corresponding DNA doses. The LbL particles reported here present a promising platform for delivery of DNA to phagocytic cells.
Keywords: microparticles, PLGA, transfection, layer-by-layer, plasmid DNA, phagocytosis
INTRODUCTION
An exciting engineering feat for the delivery of therapeutic macromolecules is to assemble the various molecular components into a multi-component architecture and then disassemble the components in a controlled fashion at the targeted site. The layer-by-layer (LbL) assembly of multilayer polyelectrolyte films offers such a possibility. The LbL deposition is a film-forming process of placing polyelectrolytes of opposite charges one layer at a time alternately on top of each other (Decher et al., 1992, Mao et al., 1993, Decher, 1997). The high conformity of the LbL films makes them an attractive coating method for surfaces of complex geometries such as microparticles, implantable materials, and biomedical devices. Film thickness and chemical composition of the LbL films can be tuned with nanometer precision.
DNA-containing LbL films are promising advanced materials for both localized and systemic gene delivery (Jewell et al., 2008). DNA has been incorporated into the LbL films on flat substrates (Lvov et al., 1993, Sukhorukov et al., 1996, Pei et al., 2001, Shen et al., 2004, Yamauchi et al., 2005, Jewell et al., 2006, Ren et al., 2006, Blacklock et al., 2007, Lynn, 2007, Lu et al., 2008) as well as on curved surfaces of colloids (Schuler and Caruso, 2001, Vinogradova et al., 2005, Zelikin et al., 2007) and bubbles (Borden et al., 2007). While physicochemical properties of the LbL films have been widely studied, significantly lower number of published studies describes the biological properties of the LbL films (Reibetanz et al., 2006, Ai et al., 2005, Cortez et al., 2007, Cortez et al., 2006, De Koker et al., 2007, Reibetanz et al., 2007). Thin films and coatings that provide sustained DNA release are desirable. The sustained localized release of DNA from thin films and coatings can enhance gene transfer by maintaining an elevated concentration of DNA within the cellular microenvironment (Shea et al., 1999, Luo and Saltzman, 2000, Shen et al., 2004). Both the flat and colloidal templates in principle offer the possibility to deliver a combination of different genes and other therapeutic agents that can be made part of the delivery system (Reibetanz et al., 2006, Reibetanz et al., 2007, Zhang et al., 2007).
Biodegradable PLGA microparticles containing encapsulated DNA have been investigated as possible DNA vaccine delivery systems. However, encapsulation of large hydrophilic DNA molecules in such particles is often inefficient and can lead to damage of the DNA (Wang et al., 2004). Many of the problems can be avoided by surface functionalization of the particles with DNA or polycation/DNA complexes instead of encapsulating DNA inside the particles (Kasturi et al., Trimaille et al., 2003). Compared to adsorbing a single layer of DNA or DNA polyplexes, the LbL process offers higher DNA loading and better control of the loading. In addition, it is possible to encapsulate other active agents in the particle core to augment further the activity of the DNA. Here, we show that multilayer plasmid DNA films deposited on biodegradable PLGA core are readily internalized by murine macrophages in a dose-dependent manner and show moderate levels of prolonged transgene expression.
MATERIALS AND METHODS
Materials
Polystyrene (PS) microspheres, Polybead® with mean diameter 4.67 ± 0.27 μm, were purchased from Polysciences as 2.5% aqueous suspension. Poly(ethylenimine) (PEI, MW 2.5 × 104), deoxyribonucleic acid sodium salt from calf thymus (ctDNA, MW 10–15 × 106), polyvinyl alcohol (PVA, MW 30,000–70,000), fluorescein isothiocyanate (FITC) and 6-coumarin were purchased from Sigma-Aldrich. Acid-terminated poly(DL-lactide-co-glycolide) (PLGA, inherent viscosity ~1.25, monomer ratio 50:50) was purchased from Birmingham Polymers. Sulforhodamine 101 sulfonyl chloride (TexasRed®) fluorescent dye was purchased from Anaspec. All other reagents were purchased from Fischer Scientific. All organic solvents used were of HPLC grade. Plasmid DNA vector containing high-expression secreted human embryonic alkaline phosphatase (SEAP) reporter gene was obtained from Aldevron (Fargo, ND) as 5 mg/mL aqueous solution and used without further purification. Tropix® Phospha-Light chemiluminiscent reporter gene assay system for the detection of the placental alkaline phosphatase was obtained from Applied Biosystems (Bedford, MA). The water used in all experiments was purified using Nanopure Diamond® Ultrapure Water systems (Barnstead International) and had resistivity higher than 18.0 MΩ cm.
Polymer labeling
PEI was fluorescently labeled with FITC and TexasRed in 100 mM sodium carbonate-bicarbonate buffer (pH = 9.0). The reaction was performed at room temperature with stirring overnight. The polymer was purified by dialysis using Pierce® snake skin dialysis membrane (MW cut off 10,000) against water for 48 h. The dialyzed polymer solution was then lyophilized and stored at −20 °C until further use.
Preparation of PLGA microparticles
PLGA microparticles fluorescently labeled with an encapsulated dye 6-coumarin were formulated by oil-in-water (o/w) emulsion and single solvent evaporation technique. Briefly, 75 mg of the acid-terminated PLGA was dissolved in 2.25 mL of methylene chloride and mixed with 250 μg of 6-coumarin in 250 μL of methylene chloride. The solution was then poured into 25 mL of 2.5% solution of PVA and emulsified using a tissue homogenizer (Tissuemizer®) at 10,000 rpm for 3 min. The o/w emulsion was then stirred overnight with a magnetic stirrer until all the organic solvent evaporated. The microparticles were collected by centrifugation at 4,000 rpm for 30 min at 4 °C and washed with 10 mL of deionized water. The centrifugation and washing steps were repeated two more times. Particle size was determined by a calibrated optical microscope. To reduce the size polydispersity, the particles were fractionated by differential centrifugation to obtain particles with a size range of 2–6 μm. PLGA microparticles were redispersed in deionized water and stored at −20 °C until further use.
LbL deposition
To form multilayer films on the surface of microparticles, 1 mL of a 1.5 mg/mL PEI solution (containing 0.5 M NaCl) was added to an aqueous suspension of ~4.5 × 108 of the PS or PLGA particles in 0.6 mL. The polycation was allowed to adsorb on the surface of the colloids for 20 min with intermittent shaking. The dispersion was then centrifuged at 4,500 rpm for 5 min. After centrifugation, the supernatant was removed and 1.6 mL of water was added. The particles were then re-dispersed by ultrasonication for about 40–50 s. Centrifugation and washing steps were repeated three times to ensure complete removal of non-adsorbed polycation. Then, a layer of DNA (either ctDNA on PS or plasmid DNA on PLGA microparticles) was deposited. For this, 1 mL of a 0.5 g/L solution of DNA in 0.2 M sodium acetate buffer (pH = 5.0) was added to the particles and then allowed to adsorb for 20 min. The same procedure of centrifugation and washing was applied as in the case of PEI deposition. This process was continued until the desired number of layers was deposited. The LbL assembly was monitored by fluorescence microscope (Nikon Eclipse TE2000U). Images were analyzed using Metavue® imaging software. The amount of plasmid DNA adsorbed on the surface of the PLGA microparticles was estimated by measuring the decrease in DNA concentration in the supernatant after each deposition step using 260 nm absorbance (A260 = 1.0 corresponds to 50 mg DNA/L). Alternatively, the microparticles were hydrolyzed overnight using 0.1 M NaOH and the DNA concentration was determined by absorbance at 260 nm.
Preparation of hollow DNA/PEI capsules
Hollow DNA/PEI capsules were obtained by dissolving the PS core with tetrahydrofuran (THF). The polyelectrolyte multilayer-coated microparticles were incubated with 0.5 mL of THF overnight. The exposure of the particles to THF was repeated two more times to facilitate complete removal of the colloidal template. The particles were then washed with deionized water three times so that the capsules were free of any organic solvents. The capsules were then dispersed in 1.5 mL water and stored until further use for characterization.
Zeta potential measurement
The zeta potential of the LbL-coated microparticles was determined using a 90 Plus from Brookhaven Instruments Corp. with zeta potential module equipped with 35 mW solid state laser (658 nm). Scattered light was detected at 90° angle and a temperature of 25 °C. About 1.6 mL of a suspension of the charged particles in water was used for the measurements. Zeta potential values were calculated from measured velocities using Smoluchowski equation and results are expressed as mean ± S.D. of 8–10 runs.
AFM characterization
The surface morphology of the hollow capsules and the thickness of the thin films were characterized using atomic force microscopy (AFM). 15 μL of the hollow capsule dispersion was placed on freshly cleaved mica and the sample was air dried for 2–3 h. AFM images were acquired using a Multimode Nanoscope IIIa AFM (Digital Instruments) and J scanner in tapping mode in air. Amplitude, height, and phase images were captured but only height images are shown here. The height images have been plane-fit in the fast scan direction with no additional filtering.
SEM characterization
Scanning electron microscopy (SEM) was used to study the surface morphology of the LbL-coated and PLGA control microparticles. Samples were prepared by placing 25μL of the particle dispersion on freshly cleaved mica substrate and then air dried for 2–3 h. The samples were then coated with gold using a Gold Sputter coater (EFFA® Mark II Coater, Ernst F. Fullam, Latham, NY) for 30 s at 200 mm Hg pressure and 40 mA current. The sputter-coated samples were then imaged using a Hitachi S-2400 SEM operating at an accelerating voltage of 20 keV.
Cellular uptake in J774.1 macrophages
The cellular uptake of the 6-coumarin-labeled LbL-coated PLGA microparticles was studied using fluorescence microscopy and fluorescence-activated cell-sorter (FACS). Murine macrophage cell line J774.1 was obtained from American Type Culture Collection (ATCC, Manassas, VA) and was maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% Penicillin-Streptomycin. For the FACS analysis, the cells were plated in a 12-well plate at a seeding density of 80,000 wells/plate 24 h before incubation. The cells were incubated for 1 and 3 h with three doses of the LbL-coated PLGA microparticles (5, 10, and 25 particles/cell) containing 6-coumarin. After incubation, the cell culture medium was completely removed and the cells were washed once with PBS buffer, trypsinized, resuspended in PBS and centrifuged at 800 rpm for 15–20 min at 4 °C. After centrifugation, the cell pellet was carefully removed and resuspended in 500 μL PBS. The cells were stored on ice during all times and 10,000 events per sample were counted on FACS. 6-Coumarin was excited using the 450 nm line of argon laser and emitted light collected at 490 nm. Background fluorescence and auto fluorescence were determined using untreated cells. Cellular debris showing reduced forward and side-scatter was excluded from the analysis. Histogram plots were constructed using the CellQuest software. Arithmetic mean of fluorescence intensity per cell incorporating fluorescence from each sample was calculated and the results expressed as the mean fluorescence intensity ± S.D. of triplicate samples.
Transfection activity
Transfection efficiency in J774.1 murine macrophage cell line was analyzed using the LbL-coated PLGA microparticles at three different doses (5, 10, and 25 particles/cell) and compared with control PLGA particles without DNA or PEI/DNA polyplexes at the same DNA doses. All the transfection studies were performed in 48-well plates with cells plated 24 h prior to transfection at the seeding density of 30,000 cells/well. Triplicate samples were used in all the cases. On the day of transfection, the cells were incubated with the LbL-coated microparticles or PEI/DNA polyplexes in 1 mL of DMEM supplemented with 10% FBS. After 3 h of incubation, the transfection mixture was completely removed, washed with sterile PBS and the cells were continued to be cultured in 1 mL of the medium. Samples were drawn every three days and replaced with fresh medium. To measure levels of SEAP reporter gene expression, 100 μL of the sample was diluted with 300 μL of the dilution buffer and heated at 65 °C for 30 min to deactivate endogenous phosphatases and then cooled on ice for another 30 min. 100 μL of the above sample was added to a luminometer tube and to this 100 μL of the assay buffer (including appropriate alkaline phosphatase inhibitor) was added and incubated for 5 min. To this, 100 μL of reaction buffer (including CSPD® substrate) was added and incubated for another 20 min. A single tube Sirius® luminometer (Zylux Corporation) was used to measure the luminescence with the injector set off and measurement taken for 10 s with 2 s delay time. The transfection results are expressed as Relative Light Units (RLU)/s. The results are expressed as mean RLU/sec ± S.D. and where necessary, significant differences between two groups are determined by Student’s t-test. A value of P < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Deposition of DNA-containing LbL films on the surface of microparticles is a straightforward way of preparing particles with defined surface properties and controlled DNA content. Biodegradable particles coated with the LbL films containing plasmid DNA may represent a promising delivery system for DNA vaccines. The possibility of encapsulating additional active agents in the biodegradable core further enhances the potential utility of such systems for a variety of combination drug/gene therapeutic approaches. While physicochemical properties of the LbL-coated particles have been widely investigated, data describing the biological properties of the LbL particles are still somewhat limited (Reibetanz et al., 2007, De Koker et al., 2007, Cortez et al., 2006, Johnston et al., 2006, Cortez et al., 2007, Ai et al., 2005, Reibetanz et al., 2006). To improve our understanding of the interactions of the LbL particles with living cells, the present study focuses on evaluating cellular uptake and transfection activity of PLGA particles coated with LbL films based on plasmid DNA and PEI.
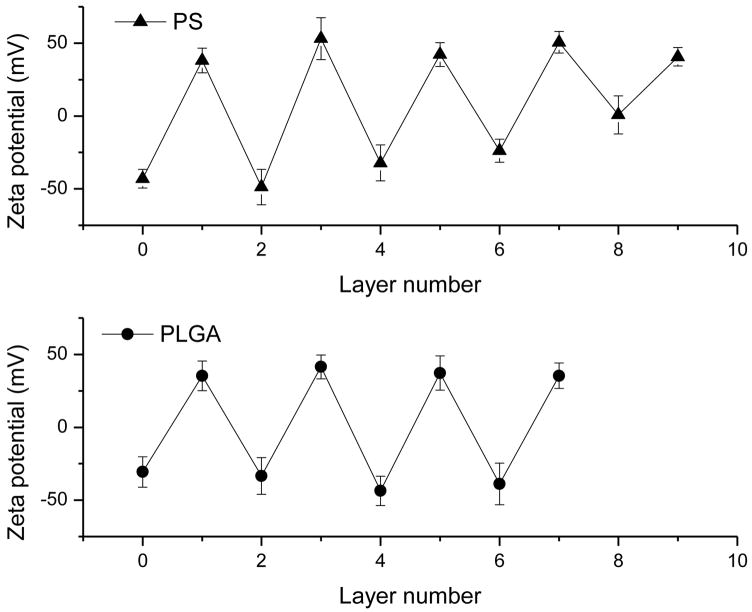
Numerous factors such as DNA size, ionic strength, and pH of solution influence the deposition of the LbL films on colloidal microparticles (Sukhorukov et al., 1998a, Sukhorukov et al., 1998b, Caruso and Mohwald, 1999, Donath et al., 1999). We first used the PS particles and readily available ctDNA to optimize the deposition process before proceeding with the PLGA microparticles containing plasmid DNA/PEI films. ctDNA is often used as a model molecule for plasmid DNA despite its linear topology and typically also a larger size. The LbL deposition was studied by measuring zeta potential of the particles and by monitoring particle size and morphology using optical microscopy. The quality of the layer formation was indicated by alternating zeta potential values as shown in Fig. 1. The charge reversal by DNA was observed until the 8th layer. The zeta potential values of the particles with terminating DNA layers between −25 and −50 mV agree well with previous studies of DNA deposition on microparticles and micro-bubbles (Schuler and Caruso, 2001, Zelikin et al., 2007, Borden et al., 2007). After 8th layer, however, the DNA deposition failed to yield a negatively charged surface. Consequently, we chose to limit the number of PEI/ctDNA bilayers on PS particles to 4.5.
Figure 1.
Zeta potential values as a function of the deposited layer number for the PS (PEI, ctDNA) (top) and PLGA (PEI, pDNA) (bottom) microparticles.
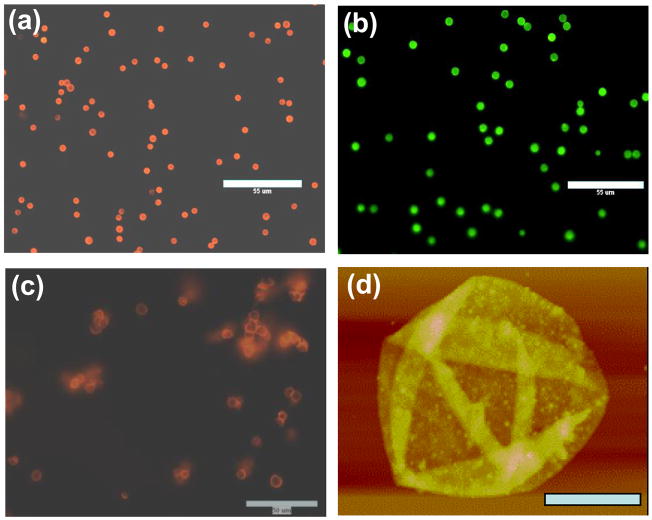
Visualization of the PEI/DNA films adsorbed on PS microparticles was achieved by using fluorescently labeled PEI (FITC-PEI) during deposition and by staining the DNA layers by incubating the particles with ethidium bromide solution after completing the deposition (Fig. 2a–b). The images of PS particles coated with PEI/DNA confirm the presence of both PEI and DNA layers. Figure 2a shows PS microparticles with 3 PEI/DNA bilayers with DNA labeled with ethidium bromide. Subsequent deposition of a polycation layer using FITC-PEI resulted in the appearance of green fluorescence on the particles due to FITC (Fig. 2b). In addition, the particles in Figure 2 show a low level of aggregation, which is often an issue accompanying the formation of LbL capsules containing large DNA molecules (Trimaille et al., 2003). Our results suggest that when aggregation was observed after DNA deposition, sonication of the particle dispersion as well as subsequent deposition of PEI layer effectively reduced the aggregation. It should be noted that while sonication can damage DNA, complexation with polycations protects nucleic acids against ultrasonic fragmentation (McKenzie et al., 2000).
Figure 2.
Microscopic characterization of the LbL films on the PS core after deposition of (a) 3 PEI/DNA bilayers (DNA labeled with ethidium bromide, bar 55 μm) (b) 3 bilayers of PEI/DNA and one layer of FITC-PEI (FITC fluorescence, bar 55 μm). Images of hollow PEI/DNA capsules consisting of 4.5 bilayers after dissolution of the PS core: (c) fluorescence micrograph of the capsules with DNA labeled with ethidium bromide (bar 50 μm) (d) AFM image of the hollow capsules (bar 2.5 μm, z-range = 400 nm).
In order to evaluate the thickness of the deposited LbL films, the polymeric core of the particles was dissolved and the resulting hollow capsules of PEI/ctDNA were studied. The decomposition of the particle template to obtain hollow capsules of similar size and shape as the template has been widely reported (Donath et al., 1998, Peyratout and Dahne, 2004, Sukhorukov et al., 2004). Such capsules are promising materials to encapsulate bioactive compounds for in vitro and in vivo delivery (Caruso et al., 2000, Antipov et al., 2001, Tiourina et al., 2001). We employed previously reported method of overnight incubation and sedimentation for the solubilization of the PS core to obtain hollow PEI/DNA capsules. The incubation of the core shell particles in 0.5 mL THF was carried out for 48 h after which the solubilized core was separated as the supernatant. This procedure was repeated two more times to dissolve completely the remaining PS. Three washings with deionized water were then employed for the complete removal of any THF from the capsule dispersion. The washings were carried out in the same manner as in the case of removal of the core, i.e., by incubating the dispersion in deionized water for 48 h and then removing the supernatant. The hollow capsules in solution can be seen under a fluorescence microscope to largely maintain the spherical shape of the template but show a higher degree of aggregation (Fig. 2c). The aggregation is in contrast to capsules made from other polyelectrolyte systems (Vinogradova et al., 2005). The thickness and surface morphology of the hollow capsules was studied by AFM (Fig. 2d). The AFM image confirms the formation of PEI/DNA shell on the PS microparticles. The PEI/DNA capsules collapsed upon drying showing multiple folds. The morphology is similar to that observed by others (Schuler and Caruso, 2001, Gao et al., 2002). AFM height analysis revealed a total thickness of the capsule wall containing 4.5 bilayers of PEI/DNA to be 43.2 ± 6.5 nm. The thickness was measured using sectional height analysis of the height difference between the flat part of the collapsed capsule and the background baseline. Since the total thickness corresponds to two parts of the wall on top of each other, the bilayer thickness was 4.8 nm. This value lies within the range of previously reported LbL films containing DNA (Lvov et al., 1993, Schuler and Caruso, 2001, Blacklock et al., 2007). The grainy wall texture is also consistent with previous reports of DNA-containing LbL films. The absence of large aggregates at the capsule wall is consistent with the stability of the capsules against coagulation up to this number of layers.
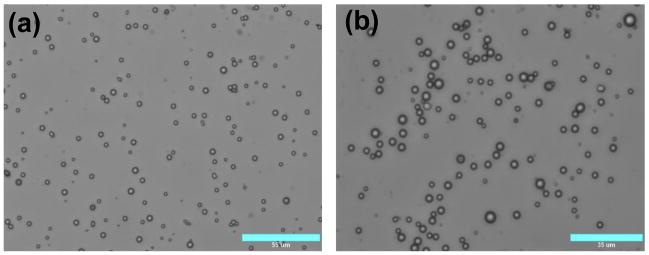
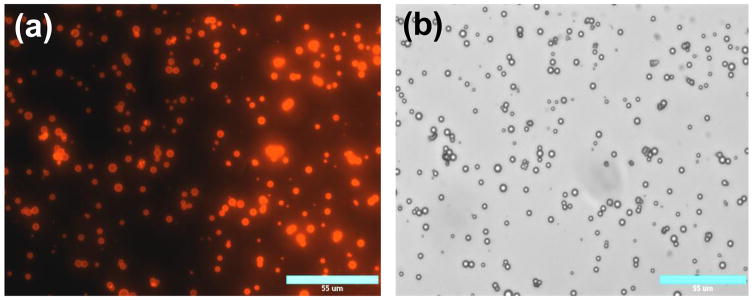
The PS particles have been widely used in the studies of the LbL films because of their physicochemical properties and availability of a wide range of sizes with low polydispersity. However, PS is ill-suited for biomedical applications such as gene delivery because of its poor biodegradability. Therefore, we carried out parallel LbL deposition studies using PLGA microparticles. PLGA is among the most important polymers used in drug and gene delivery due to its biodegradability and biocompatibility (Langer, 1998, Tinsley-Bown et al., 2000). PLGA microparticles were prepared using standard o/w emulsion technique. The o/w emulsion, however, typically results in relatively high size polydispersity. In our case, the size of the PLGA particles ranged from ~600 nm to ~6.5 μm (Table 1, Fig. 3a). When we attempted to deposit PEI/DNA film on the surface of the PLGA particles using the protocol applied to PS particles, excessive aggregation was observed. We hypothesized that the presence of the smallest-sized fraction of the particles was responsible for aggregation due to the combination of the large plasmid DNA size (~ 600 nm contour length) and small nanoparticle size leading to the aggregation (Trimaille et al., 2003). We carried out differential sedimentation experiments in order to reduce the size distribution. The primary goal was to remove particles smaller than 2.0 μm and particles larger than 6 μm in diameter. Table 1 and Fig. 3 show that the number of particles smaller than 2 μm was significantly reduced after fractionation. The content of particles larger than 2 μm increased from 43% to 90% by fractionation.
Table 1.
Size distribution of PLGA microparticles before and after fractionation
| Size range | Before fractionation (average size 1.9 μm) | After fractionation (average size 3.1 μm) |
|---|---|---|
| < 2 μm | 57 % | 13 % |
| 2–3 μm | 29 % | 37 % |
| 3–5 μm | 12 % | 43 % |
| > 5 μm | 2 % | 7 % |
Figure 3.
Microscopic characterization of the PLGA microparticles: (a) unfractionated (bar 55 μm) and (b) fractionated (bar 35 μm) microparticles.
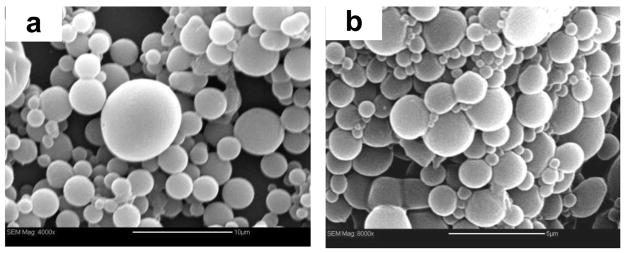
Using the fractionated PLGA particles, the same deposition protocol resulted in significantly less aggregation compared to unfractionated particles. Similar to the LbL deposition on PS particles, zeta potential measurements confirmed the alternating deposition of negatively and positively charged polyelectrolytes on PLGA (Fig. 1b). The zeta potential of bare PLGA particles, −30 mV, indicates that they are less charged than PS particles with −50 mV zeta potential. Although the surface charge of the initial colloidal substrate is important for effective LbL deposition, similar alterations in zeta potential were observed for both PLGA and PS particles and we were able to reproducibly deposit up to 7 layers. SEM was used to determine the effect of PEI/DNA deposition on the surface morphology of the PLGA particles (Fig. 4). No significant changes in the surface morphology were observed.
Figure 4.
Scanning Electron Microscopy micrographs of (a) uncoated and unfractionated PLGA particles (bar 10 μm) and (b) LbL-coated PLGA particles (bar 5 μm).
Since the transfection activity of plasmid DNA is significantly dependent on its dose, we estimated the amount of plasmid DNA adsorbed on the surface of the PLGA particles coated with 3.5 bilayers of PEI/DNA. We assumed 5-μm PLGA particles and 1 g/cm3 DNA density. Based on the results of DNA-containing LbL films on flat substrate (Blacklock et al., 2007) and on PS particles, we assume a typical thickness of the DNA layer to be 4 nm. This is only an estimate since it is possible that the DNA film thickness is different on the PLGA particles. The number of particles per mg of PLGA determined by hemocytometer was ~1.1 × 107 per mg of PLGA particles. We estimate that there is ~10 μg of plasmid DNA adsorbed per mg of PLGA particles containing 3.5 bilayers of PEI/DNA. To verify this estimate, the particles were hydrolyzed in 0.1 M NaOH and DNA content was measured spectrophotometrically. It was determined that 7.9 μg plasmid DNA was adsorbed per mg of PLGA particles with 3.5 PEI/DNA bilayers. These results are lower than those reported by a previous study which found 1.8 μg DNA/106 particles (Reibetanz et al., 2006).
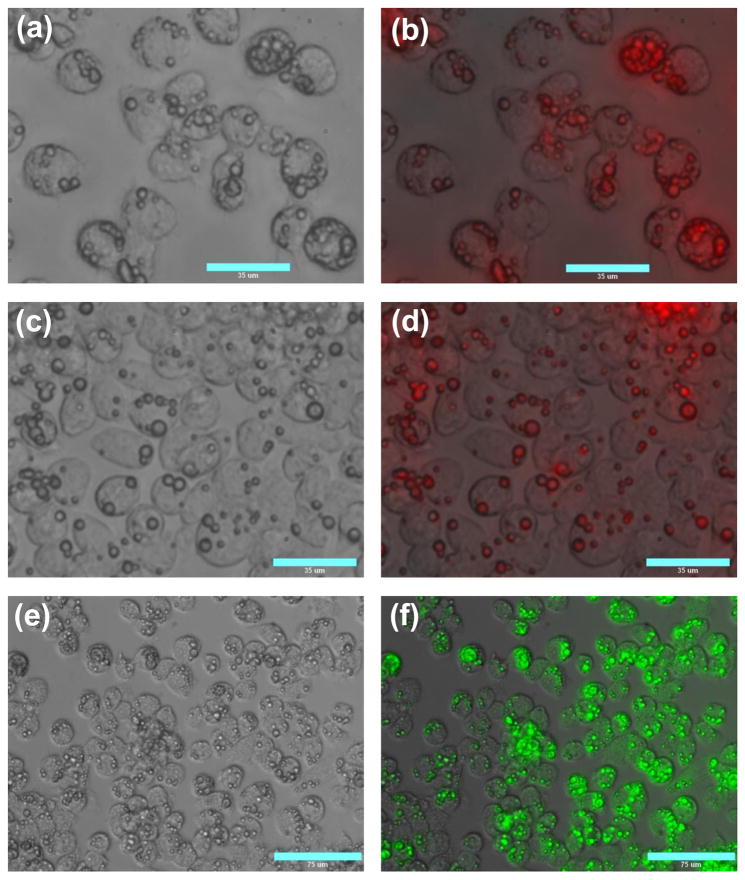
In order to evaluate the intracellular uptake of the PLGA particles with the deposited PEI/DNA film, the particles were fluorescently labeled by encapsulating 6-coumarin in the PLGA core. PEI was labeled with Texas Red to allow imaging of the LbL deposition process (Fig. 5) and to allow intracellular imaging of both the PLGA core and the LbL films. The cellular association of the LbL-coated PLGA particles was studied first by fluorescence microscopy and then by flow cytometry. The LbL-coated particles were readily taken up by J774.1 mouse macrophages (Fig. 6). Increasing the dose of the particles resulted in an increased number of particles per cell. It is clear that virtually all cells (> 95%) internalized at least one particle and some of them internalized as many as 8 particles even at the lowest particle dose used (Fig. 6a). Of the cells that internalized the particles more than 50% of them contained at least 2 particles. Such levels of cellular uptake are significant considering that 5 particle per cell were used in the experiment. The fluorescence micrographs, which were acquired after 8 days of incubation, also show that the red fluorescence of the LbL film remains associated with the PLGA particles (Fig. 6b, c). This observation clearly suggests that the LbL films in the majority of the particles did not disassemble in the intracellular environment. In addition, the results suggest that the PLGA cores remain intact even after 8 days inside the cells (Fig. 6f). Neither the lack of significant disassembly of the PEI/DNA films nor the lack of PLGA degradation is surprising given the known stability of the LbL films and the fact that the PLGA used in this study has been reported to degrade completely in 3–4 weeks (Borodina et al., 2007).
Figure 5.
Microscopic characterization of the LbL deposition on PLGA particles. (a) fluorescence and (b) phase micrographs of the PLGA microparticles coated with 3.5 TexasRed-PEI/DNA bilayers (bar 55 μm).
Figure 6.
Cellular uptake of LbL-coated PLGA particles in J774.1 macrophages. Cells were incubated with 5 (a, b), 10 (c, d), and 25 (e, f) PEI/DNA coated PLGA particles/cell for 3 h and imaged 8 days after incubation (phase images: a, c, e; TexasRed-PEI fluorescence: b, d; PLGA-coumarin fluorescence: f).
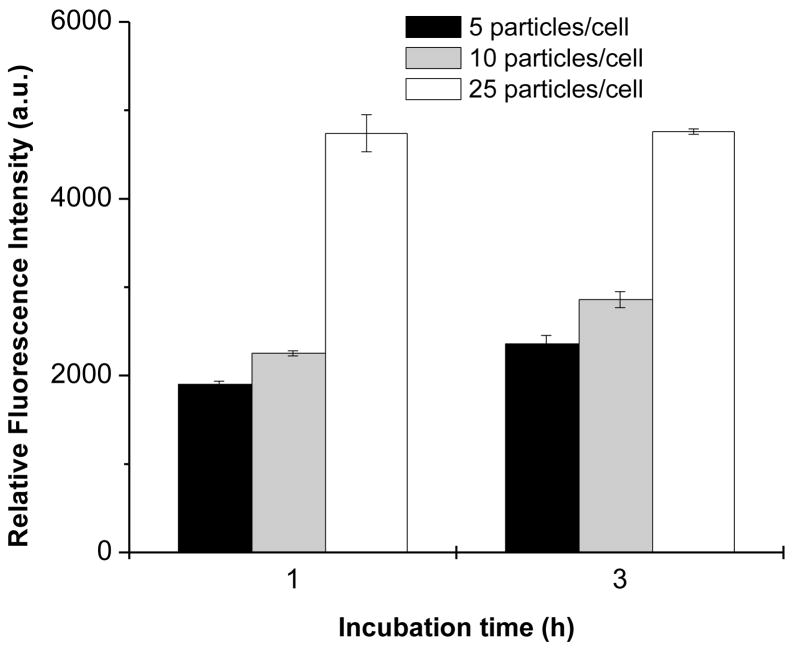
The images in Fig. 6 allowed the determination of the number of particles associated with the cells but they do not provide conclusive evidence that the particles are internalized and not just externally associated with the plasma membrane (Thiele et al., 2003, Hasegawa et al., 2008). Therefore, we used FACS analysis to quantify further the levels of cellular uptake using the same three doses of particles as in Fig. 6. The cells were detached, centrifuged, and washed before the FACS analysis to remove any externally associated microparticles. The cellular uptake was determined at two incubation time points (1 and 3 h) by measuring mean fluorescence intensity of 6-coumarin encapsulated in the PLGA core of the particles (Fig. 7). The results parallel those in Fig. 6 in that only a small difference in cell uptake is observed between the two lower doses (5 and 10 particles/cell) and significant enhancement of uptake is achieved when the highest dose (25 particles/cell) is used. Only small differences were observed between the levels of cellular uptake after 1 and 3 h of incubation, suggesting that the uptake of the LbL-coated particles proceeds rapidly and that most of the uptake occurs within the first hour of incubation. In combination with the images 8 days after the incubation (Fig. 6), these data show that after the initial rapid phagocytosis the particles persist most likely in the phagosomes without a significant degradation.
Figure 7.
Time course of cellular uptake of LbL-coated PLGA particles in J774.1 macrophages. Cells were incubated with increasing dose of PLGA particles labeled with coumarin and coated with 3.5 PEI/DNA bilayers for 1 or 3 h. Cell uptake was quantified by FACS as the average fluorescence intensity of coumarin/cell. The results are shown as mean fluorescence intensity ± SD of three replicates.
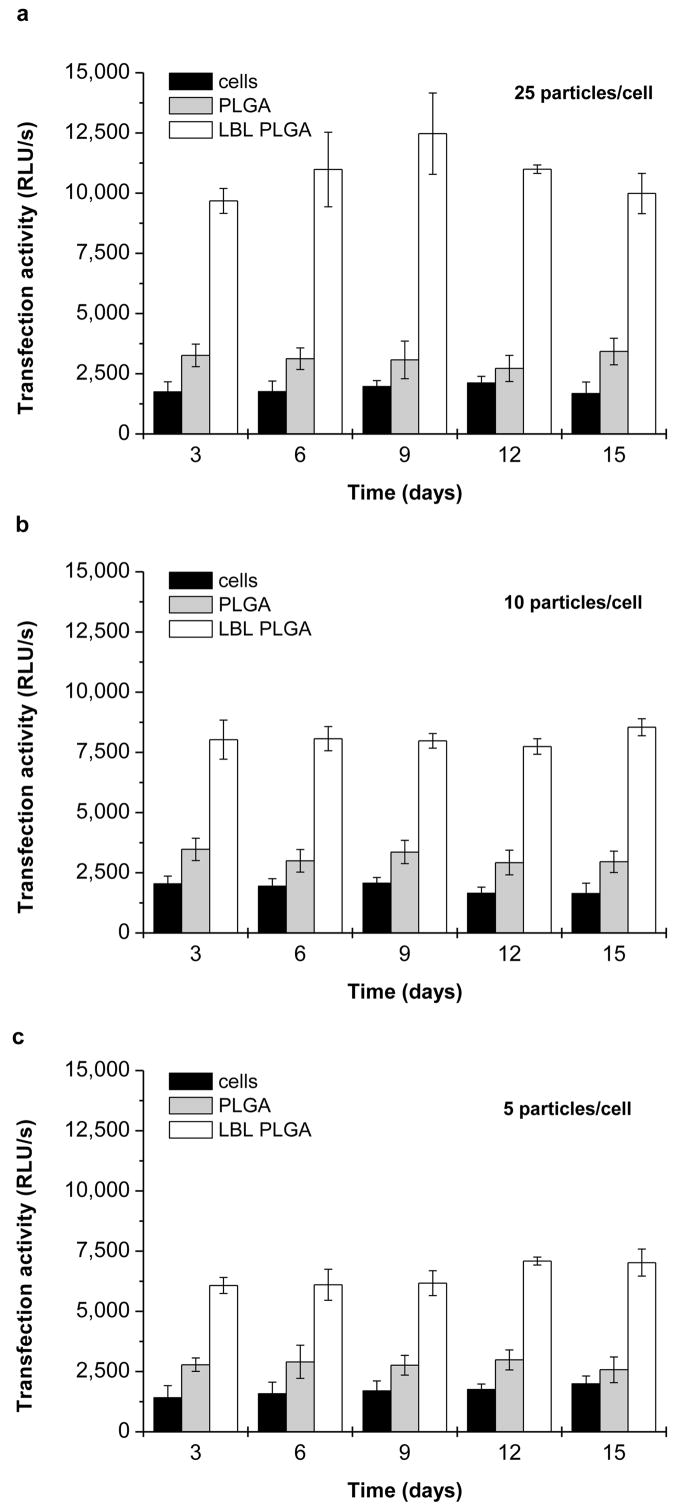
Transfection efficiency of the coated PLGA microparticles at three different doses was then evaluated in the J774.1 murine macrophages (Fig. 8). Untreated cells and cells incubated with non-coated PLGA particles were used as negative controls. Using plasmid DNA encoding for SEAP allows nondestructive sampling over time because the protein is secreted into culture medium. The transfection activity is thus normalized to the initially seeded number of cells. The expression was measured every 3 days up to 15 days, after which the cells could no longer be maintained in culture. The medium was replaced with a fresh cell culture medium at the time of sampling. Thus, each time point represents levels of SEAP expression accumulated over the preceding three days. Overall, the observed transfection levels are strongly dose dependent with even the lowest particle dose mediating transfection activity significantly above the background level (Fig. 8c). The effect of particle dose on transfection results reflects well on the uptake data. It is interesting to note that incubating the cells with control non-coated PLGA particles increased the endogenous alkaline phosphatase background levels, although the statistical significance of this observation is relatively weak (P ~ 0.01–0.05). The differences between the different time points are not significantly different in any of the three tested doses. Thus, the LbL-coated PLGA particles provide sustained levels of transgene expression over the observation period.
Figure 8.
Transfection activity of LbL-coated PLGA particles in J775.1 macrophages. Cells were transfected for 4 h with decreasing dose of PLGA particles coated with 3.5 PEI/DNA bilayers: (a) 25 particles/cell; (b) 10 particles/cell; (c) 5 particles/cell). Levels of SEAP transgene expression were measured every three days in 25 μL of medium sampled from the total volume of 1 mL of the medium. Untreated cells and cells transfected with non-coated PLGA particles were used as controls. Results are expressed as mean RLU/s in 25 μL of media ± SD of three replicates.
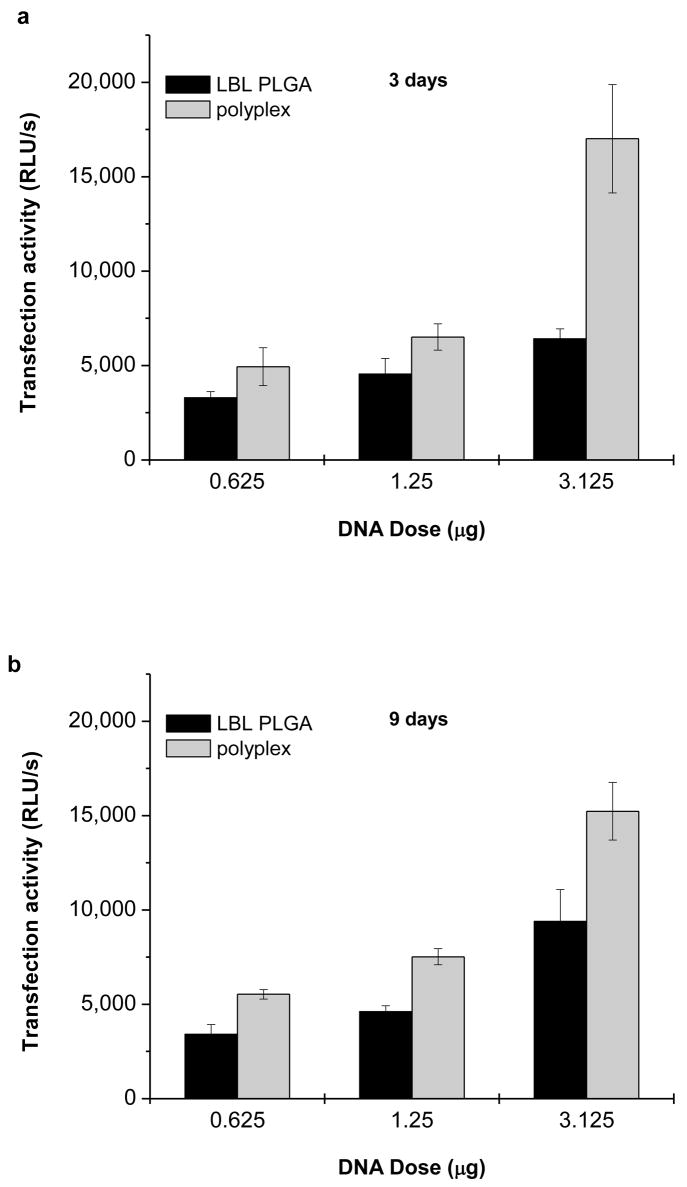
Transfection activity of the PLGA particles coated with 3.5 bilayers of PEI/DNA was then compared with transfection activity of PEI/DNA polyplexes. The polyplexes were formulated at N:P = 8 and used at three different DNA doses that correspond to the doses used in the cell uptake and transfection studies of the LbL-coated particles. Figure 9 offers a direct comparison between the transfection activities of the LbL particles and polyplexes. While the polyplexes were significantly more active at the highest DNA dose used after 3 days, no difference between polyplexes and LbL particles was observed with the two lower DNA doses (Fig. 9a). After 9 days, the difference in the transfection activity decreased suggesting more sustained nature of the transfection mediated by the LbL particles (Fig. 9b). However, it needs to be pointed out that polyplex transfection typically peaks at 24–48 h and thus measuring the transfection at 3 and 9 days probably underestimates the polyplex activity.
Figure 9.
Comparison of transfection activity of LbL-coated PLGA particles and PEI/DNA polyplexes. J775.1 macrophages were transfected for 4 h with different doses of PEI/DNA (N:P = 8) polyplexes and PLGA particles coated with 3.5 PEI/DNA bilayers. Levels of SEAP transgene expression were measured after (a) 3 and (b) 9 days and corrected for background of untreated cells. Results are expressed as mean RLU/s ± SD of three replicates.
The observed transfection activity of the LbL-coated PLGA particles is encouraging for further optimization of these systems for delivery of DNA vaccines. The lack of obvious degradation of the LbL particles suggests that intracellular disassembly of the PEI/DNA films is highly inefficient and is most likely the main barrier to enhanced transfection as suggested previously (Reibetanz et al., 2006). Substituting PEI with a biodegradable polycation is hypothesized to increase the transfection activity of the LbL particles substantially. Furthermore, the effect of the LbL film on the rate of degradation of the PLGA core will have to be investigated and carefully controlled to achieve potential co-delivery of DNA and additional encapsulated active agents to improve the activity of DNA.
Acknowledgments
Financial support from the National Institutes of Health (CA 109711) and the National Science Foundation (CBET 0553533) are gratefully acknowledged. We thank Dr. J. Panyam and Y. Patil for their help with the preparation of PLGA microparticles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AI H, PINK JJ, SHUAI X, BOOTHMAN DA, GAO J. Interactions between self-assembled polyelectrolyte shells and tumor cells. J Biomed Mater Res A. 2005;73:303–12. doi: 10.1002/jbm.a.30289. [DOI] [PubMed] [Google Scholar]
- ANTIPOV AA, SUKHORUKOV GB, DONATH E, MOHWALD H. Sustained release properties of polyelectrolyte multilayer capsules. J Phys Chem B. 2001;105:2281–2284. [Google Scholar]
- BLACKLOCK J, HANDA H, MANICKAM DS, MAO GZ, MUKHOPADHYAY A, OUPICKY D. Disassembly of layer-by-layer films of plasmid DNA and reducible TAT polypeptide. Biomaterials. 2007;28:117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- BORDEN MA, CASKEY CF, LITTLE E, GILLIES RJ, FERRARA KW. DNA and polylysine adsorption and multilayer construction onto cationic lipid-coated microbubbles. Langmuir. 2007;23:9401–9408. doi: 10.1021/la7009034. [DOI] [PubMed] [Google Scholar]
- BORODINA T, MARKVICHEVA E, KUNIZHEV S, MOEHWALD H, SUKHORUKOV GB, KREFT O. Controlled release of DNA from self-degrading microcapsules. Macromol Rapid Comm. 2007;28:1894–1899. [Google Scholar]
- CARUSO F, MOHWALD H. Protein multilayer formation on colloids through a stepwise self-assembly technique. J Am Chem Soc. 1999;121:6039–6046. [Google Scholar]
- CARUSO F, TRAU D, MOHWALD H, RENNEBERG R. Enzyme encapsulation in layer-by-layer engineered polymer multilayer capsules. Langmuir. 2000;16:1485–1488. [Google Scholar]
- CORTEZ C, TOMASKOVIC-CROOK E, JOHNSTON APR, RADT B, CODY SH, SCOTT AM, NICE EC, HEATH JK, CARUSO F. Targeting and uptake of multilayered particles to colorectal cancer cells. Adv Mat. 2006;18:1998. doi: 10.1021/nn700060m. [DOI] [PubMed] [Google Scholar]
- CORTEZ C, TOMASKOVIC-CROOK E, JOHNSTON APR, SCOTT AM, NICE EC, HEATH JK, CARUSO F. Influence of size, surface, cell line, and kinetic properties on the specific binding of A33 antigen-targeted multilayered particles and capsules to colorectal cancer cells. Acs Nano. 2007;1:93–102. doi: 10.1021/nn700060m. [DOI] [PubMed] [Google Scholar]
- DE KOKER S, DE GEEST BG, CUVELIER C, FERDINANDE L, DECKERS W, HENNINK WE, DE SMEDT S, MERTENS N. In vivo cellular uptake, degradation, and biocompatibility of polyelectrolyte microcapsules. Adv Funct Mat. 2007;17:3754–3763. [Google Scholar]
- DECHER G. Fuzzy nanoassemblies: Toward layered polymeric multicomposites. Science. 1997;277:1232–1237. [Google Scholar]
- DECHER G, HONG JD, SCHMITT J. Buildup Of Ultrathin Multilayer Films By A Self-Assembly Process .3. Consecutively Alternating Adsorption Of Anionic And Cationic Polyelectrolytes On Charged Surfaces. Thin Solid Films. 1992;210/211:831–835. [Google Scholar]
- DONATH E, SUKHORUKOV GB, CARUSO F, DAVIS SA, MOHWALD H. Novel hollow polymer shells by colloid-templated assembly of polyelectrolytes. Angew Chem. 1998;37:2202–2205. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2201::AID-ANIE2201>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- DONATH E, SUKHORUKOV GB, MOHWALD H. Submicrometric and micrometric polyelectrolyte capsules. Nachrichten Chem Tech Lab. 1999;47:400. [Google Scholar]
- GAO CY, MOYA S, DONATH E, MOHWALD H. Melamine formaldehyde core decomposition as the key step controlling capsule integrity: Optimizing the polyelectrolyte capsule fabrication. Macromol Chem Phys. 2002;203:953–960. [Google Scholar]
- HASEGAWA T, II, JIMA K, HIROTA K, NAKAJIMA T, MAKINO K, TERADA H. Exact determination of phagocytic activity of alveolar macrophages toward polymer microspheres by elimination of those attached to the macrophage membrane. Coll Surf B. 2008;63:209–216. doi: 10.1016/j.colsurfb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- JEWELL CM, ZHANG JT, FREDIN NJ, WOLFF MR, HACKER TA, LYNN DM. Release of plasmid DNA from intravascular stents coated with ultrathin multilayered polyelectrolyte films. Biomacromolecules. 2006;7:2483–2491. doi: 10.1021/bm0604808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEWELL CM, LYNN DM. Multilayered polyelectrolyte assemblies as platforms for the delivery of DNA and other nucleic acid therapeutics. Adv Drug Deliv Rev. 2008;60:979–999. doi: 10.1016/j.addr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSTON APR, CORTEZ C, ANGELATOS AS, CARUSO F. Layer-by-layer engineered capsules and their applications. Curr Opin Coll Int Sci. 2006;11:203–209. [Google Scholar]
- KASTURI SP, SACHAPHIBULKIJ K, ROY K. Covalent conjugation of polyethyleneimine on biodegradable microparticles for delivery of plasmid DNA vaccines. Biomaterials. 2005;26:6375–85. doi: 10.1016/j.biomaterials.2005.03.043. [DOI] [PubMed] [Google Scholar]
- LANGER R. Drug delivery and targeting. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- LU ZZ, WU J, SUN TM, JI J, YAN LF, WANG J. Biodegradable polycation and plasmid DNA multilayer film for prolonged gene delivery to mouse osteoblasts. Biomaterials. 2008;29:733–741. doi: 10.1016/j.biomaterials.2007.10.033. [DOI] [PubMed] [Google Scholar]
- LUO D, SALTZMAN WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18:893–5. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- LVOV Y, DECHER G, SUKHORUKOV G. Assembly Of Thin-Films By Means Of Successive Deposition Of Alternate Layers Of Dna And Poly(Allylamine) Macromolecules. 1993;26:5396–5399. [Google Scholar]
- LYNN DM. Peeling back the layers: Controlled erosion and triggered disassembly of multilayered polyelectrolyte thin films. Adv Mat. 2007;19:4118–4130. [Google Scholar]
- MAO G, TSAO Y, TIRRELL M, DAVIS HT, HESSEL V, RINGSDORF H. Self-assembly of photopolymerizable bolaform amphiphile mono- and multilayers. Langmuir. 1993;9:3461–3470. [Google Scholar]
- MCKENZIE DL, SMILEY E, KWOK KY, RICE KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjug Chem. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- PEI R, CUI X, YANG X, WANG E. Assembly of alternating polycation and DNA multilayer films by electrostatic layer-by-layer adsorption. Biomacromolecules. 2001;2:463–8. doi: 10.1021/bm0001289. [DOI] [PubMed] [Google Scholar]
- PEYRATOUT CS, DAHNE L. Tailor-made polyelectrolyte microcapsules: from multilayers to smart containers. Angew Chem Int Ed Engl. 2004;43:3762–83. doi: 10.1002/anie.200300568. [DOI] [PubMed] [Google Scholar]
- REIBETANZ U, CLAUS C, TYPLT E, HOFMANN J, DONATH E. Defoliation and plasmid delivery with layer-by-layer coated colloids. Macromol Biosci. 2006;6:153–60. doi: 10.1002/mabi.200500163. [DOI] [PubMed] [Google Scholar]
- REIBETANZ U, HALOZAN D, BRUMEN M, DONATH E. Flow cytometry of HEK 293T cells interacting with polyelectrolyte multilayer capsules containing fluorescein-labeled poly(acrylic acid) as a pH sensor. Biomacromolecules. 2007;8:1927–1933. doi: 10.1021/bm061200r. [DOI] [PubMed] [Google Scholar]
- REN KF, JI J, SHEN JC. Tunable DNA release from cross-linked ultrathin DNA/PLL multilayered films. Bioconjug Chem. 2006;17:77–83. doi: 10.1021/bc050264g. [DOI] [PubMed] [Google Scholar]
- SCHULER C, CARUSO F. Decomposable hollow biopolymer-based capsules. Biomacromolecules. 2001;2:921–6. doi: 10.1021/bm010052w. [DOI] [PubMed] [Google Scholar]
- SHEA LD, SMILEY E, BONADIO J, MOONEY DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17:551–4. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- SHEN H, TAN J, SALTZMAN WM. Surface-mediated gene transfer from nanocomposites of controlled texture. Nat Mater. 2004;3:569–74. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- SUKHORUKOV GB, DONATH E, DAVIS S, LICHTENFELD H, CARUSO F, POPOV VI, MOHWALD H. Stepwise polyelectrolyte assembly on particle surfaces: a novel approach to colloid design. Pol Adv Technol. 1998a;9:759–767. [Google Scholar]
- SUKHORUKOV GB, DONATH E, LICHTENFELD H, KNIPPEL E, KNIPPEL M, BUDDE A, MOHWALD H. Layer-by-layer self assembly of polyelectrolytes on colloidal particles. Coll Surf A. 1998b;137:253–266. [Google Scholar]
- SUKHORUKOV GB, FERY A, BRUMEN M, MOHWALD H. Physical chemistry of encapsulation and release. Phys Chem Chem Phys. 2004;6:4078–4089. [Google Scholar]
- SUKHORUKOV GB, MOHWALD H, DECHER G, LVOV YM. Assembly of polyelectrolyte multilayer films by consecutively alternating adsorption of polynucleotides and polycations. Thin Solid Films. 1996;285:220–223. [Google Scholar]
- THIELE L, MERKLE HP, WALTER E. Phagocytosis and phagosomal fate of surface-modified microparticles in dendritic cells and macrophages. Pharm Res. 2003;20:221–8. doi: 10.1023/a:1022271020390. [DOI] [PubMed] [Google Scholar]
- TINSLEY-BOWN AM, FRETWELL R, DOWSETT AB, DAVIS SL, FARRAR GH. Formulation of poly(D, L-lactic-co-glycolic acid) microparticles for rapid plasmid DNA delivery. J Control Release. 2000;66:229–41. doi: 10.1016/s0168-3659(99)00275-8. [DOI] [PubMed] [Google Scholar]
- TIOURINA OP, ANTIPOV AA, SUKHORUKOV GB, LARIONOVA NL, LVOV Y, MOHWALD H. Entrapment of alpha-chymotrypsin into hollow polyelectrolyte microcapsules. Macromol Biosci. 2001;1:209–214. [Google Scholar]
- TRIMAILLE T, PICHOT C, DELAIR T. Surface functionalization of poly(D, L-lactic acid) nanoparticles with poly(ethylenimine) and plasmid DNA by the layer-by-layer approach. Coll Surf A. 2003;221:39–48. [Google Scholar]
- VINOGRADOVA OI, LEBEDEVA OV, VASILEV K, GONG HF, GARCIA-TURIEL J, KIM BS. Multilayer DNA/poly(allylamine hydrochloride) microcapsules: Assembly and mechanical properties. Biomacromolecules. 2005;6:1495–1502. doi: 10.1021/bm049254t. [DOI] [PubMed] [Google Scholar]
- WANG C, GE Q, TING D, NGUYEN D, SHEN HR, CHEN J, EISEN HN, HELLER J, LANGER R, PUTNAM D. Molecularly engineered poly(ortho ester) microspheres for enhanced delivery of DNA vaccines. Nat Mater. 2004;3:190–6. doi: 10.1038/nmat1075. [DOI] [PubMed] [Google Scholar]
- YAMAUCHI F, KATO K, IWATA H. Layer-by-layer assembly of poly(ethyleneimine) and plasmid DNA onto transparent indium-tin oxide electrodes for temporally and spatially specific gene transfer. Langmuir. 2005;21:8360–8367. doi: 10.1021/la0505059. [DOI] [PubMed] [Google Scholar]
- ZELIKIN AN, BECKER AL, JOHNSTON APR, WARK KL, TURATTI F, CARUSO F. A general approach for DNA encapsulation in degradable polymer miocrocapsules. Acs Nano. 2007;1:63–69. doi: 10.1021/nn700063w. [DOI] [PubMed] [Google Scholar]
- ZHANG J, MONTANEZ SI, JEWELL CM, LYNN DM. Multilayered Films Fabricated from Plasmid DNA and a Side-Chain Functionalized Poly(β-amino Ester): Surface-Type Erosion and Sequential Release of Multiple Plasmid Constructs from Surfaces. Langmuir. 2007;23:11139–11146. doi: 10.1021/la702021s. [DOI] [PubMed] [Google Scholar]