Abstract
Manganese superoxide dismutase (MnSOD) is a primary antioxidant enzyme necessary for the survival of aerobic life. Previously, we demonstrated that specificity protein 1 (Sp1) is essential for the basal transcription of the MnSOD gene. We also identified nucleophosmin (NPM), an RNA-binding protein, as an important co-activator of NF-κB in the induction of MnSOD by cytokine and tumor promoter. Here, using chromatin immunoprecipitation (ChIP) analysis, we demonstrate that Sp1 and NPM interact in vivo to enhance NF-κB-mediated MnSOD induction. Interaction between NPM and Sp1 or NF-κB at the promoter and enhancer of the MnSOD gene in vivo were verified by the presence of the PCR products from the promoter and enhancer elements in the ChIP assay. Unexpectedly, we also found p53, another transcription factor, to be a component of the complex detected by ChIP assay. The presence of p53 in this transcription complex was verified by immunoprecipitation of p53 proteins with antibody to Sp1 in nuclear extracts. Using a vector expressing full-length p53 cDNA, we demonstrated that p53 overexpression suppresses MnSOD mRNA and protein levels. Consistent with the negative role of p53 in the expression of the MnSOD gene, expression of small interfering RNA for p53 leads to an increase of MnSOD mRNA and protein levels. Using ChIP assays and immunoprecipitation, we further demonstrated that p53 interacts with Sp1 to suppress both the constitutive and 12-O-tetradecanoylphorbol-13-acetate-stimulated expression of the MnSOD gene. Inhibition of the MnSOD gene by p53 was abolished when Sp1 sites on the MnSOD promoter were mutated or when the Sp1 protein was reduced by siRNA approaches. Because expression of MnSOD protects against cell death, our findings reveal a previously unrecognized mechanism of p53-mediated cell death and demonstrate an intricate relationship between the positive and negative control of MnSOD expression.
Oxidative stress caused by reactive oxygen species is detoxified by intracellular antioxidant enzymes such as manganese superoxide dismutase (MnSOD), 2 catalase, and glutathione. Among these primary antioxidant enzymes, MnSOD is the enzyme that must function in order for all aerobic organisms to survive. MnSOD is a highly regulated SOD encoded by the sod2 located in human chromosome 6q25.3 (1, 2).
The critical role of MnSOD as a cytoprotective enzyme is illustrated in both MnSOD knock-out and transgenic animal models. For instance, MnSOD knock-out mice develop cardiomyopathy and die within a few days after birth (3). MnSOD knock-out mice treated with a SOD mimetic were protected from systemic toxicity and from neonatal death (4). Conversely, transgenic mice overexpressing human MnSOD experienced less injury resulting from inflammation (5), cardio-toxic drugs (6), and pathological and physiological conditions leading to brain injury (7). The human MnSOD gene is a single-copy gene consisting of five exons interrupted by four introns with a typical splice junction (8). The sod2 from human, bovine, rat, and mouse share more than 90% homology in the coding sequence. The basal promoter of the sod2 has multiple transcription factor binding motifs containing Sp1 and Ap-2 binding sites. Functional studies in different cell lines with different levels of Sp1 and Ap-2 proteins suggest that cellular levels of these proteins differentially regulate the expression of the human MnSOD gene. Transcription factor Sp1 is essential and sufficient, whereas Ap-2 is unnecessary and antagonistic to the constitutive expression of the gene (9). Sp1 is a prototype member of a small family of transcription factors (Sp1, Sp2, Sp3, and Sp4) with homologous functional domains. Members of this family have an inhibitory domain in the N termini and four transcriptional activation domains (A, B, C, and D) (10). Transcriptional domains A and B are required for general and full activation, whereas domain C possesses very low activation (11), and domain D is required for synergistic activation (12). The Sp1 protein is capable of inducing homotypic, Sp1-Sp1 interaction (13) or forming heterotypic interactions with different classes of nuclear proteins such as TATA box-binding protein (TBP) (14), C/EBP (15), and YY1 (16). Because the MnSOD promoter does not contain a TATA or CAAT binding element, transcription of the MnSOD gene is dependent on Sp1-Sp1 interaction.
The sod2 of mice and humans contains enhancer elements in the second intron of the gene (17, 18). Functional analyses of enhancer elements demonstrate that the intronic element consists of NF-κB, C/EBP, and NF-1 transcription factor binding sites. The activation of NF-κB is essential but not sufficient for the induction of MnSOD by cytokines and tumor promoter. NF-κB forms various homo- and heterodimer units among the mammalian subunits of p50, p52, p65 (Rel A), c-Rel, and Rel B. These dimer units in turn bind to a group of NF-κB DNA binding sites with different affinities within the target gene (19, 20).
Although the role of Sp1 in the expression of MnSOD in HepG2 cells is well documented (21), the role of p53 in the expression of MnSOD is unclear. p53 is a transcriptional regulator that plays an important role in suppressing tumor development. In response to stress, p53-mediated function causes two major cellular events; they are cell cycle arrest or apoptotic death, both of which prevent the proliferation of cells containing damaged DNA. It is well established that the apoptotic function is mediated in part by the property of p53 that is a positive transcriptional regulator of proapoptotic genes such as Bax (22). However, it has also been shown that p53 may function as a negative regulator of gene transcription, repressing the expression of the number of genes including MnSOD (23). However, the mechanism by which p53 represses its target gene is unknown. We have previously demonstrated that the tumor promoter TPA enhances MnSOD expression by activation of Sp1 and that NPM serves as a co-activator of NF-κB in the induction of MnSOD by cytokine and TPA. In this paper we report the mechanisms by which p53 mediated MnSOD suppression and demonstrate that p53 plays a negative role on MnSOD gene transcription by forming a complex with Sp1, leading to the reduction of MnSOD gene transcription under both constitutive and TPA-induced conditions.
EXPERIMENTAL PROCEDURES
Cell Culture
The human hepatoma cell line HepG2 was purchased from American Type Culture Collection (Manassas, VA). Cells were grown in 5% CO2 incubator at 37 °C in media consisting of Dulbecco’s modified Eagle’s/Ham’s F-12 (Sigma) supplemented with 10% (v/v) fetal bovine serum (Hyclone Inc., Logan, UT), 200 µm l-glutamine (Invitrogen), and 1% penicillin-streptomycin-neomycin (PSN) antibiotic (Invitrogen).
Reagents
Unless otherwise stated all antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).Anti-β-actin monoclonal antibody and wild-type anti-p53 antibodies were purchased from Sigma and Calbiochem, respectively. Rabbit polyclonal MnSOD antibody, glyceraldehyde-3-phosphate dehydrogenase antibody, and chromatin immunoprecipitation (ChIP) assay kits were purchased from Upstate Biotechnologies (Lake Placid, NY).All chemicals were purchased from Sigma unless otherwise indicated. pMM/p53, a p53 expression vector, was kindly provided by Dr.Yi Sun(University of Michigan).The pCMV4-Sp1 expression vector was obtained from Dr. Jonathan M. Horowitz (North Carolina State University). All siRNAs were purchased from Invitrogen.
Construction of Plasmid
A BamH1 fragment (B7) containing a 3.4-kilobase 5′-flanking region was used to generate the −555 to +24 MnSOD basal promoter by PCR. To create this construct, PCR primers with recognition sequences Kpn1 (upstream) and BglII (downstream) restriction sites were added for subcloning at the upstream of the luciferase reporter gene. To generate intronic enhancer fragment (I2E) (1742–2083) constructs, BamH1-digested 39b λphase DNA, containing the entire human MnSOD gene (8074 bp), was used as the template for PCR amplification with a primer having a BglII artificial restriction site. The PCR product was then ligated to the BglII site of the −555 to +24 basal promoter containing pGL3 vector, which yielded the natural orientation of the gene. The following primer sets were used to generate the aforementioned constructs: forward primer, −555, 5′-CGGGGTACCCGCTGGCTCTACCCTCAGCTCATA-3′; reverse primer,+24, 5′-GGAAGATCTGCCGAAGCCACCACAGCCACGAGT-3′; forward primer, +1742, 5′-GGAAGATCTCGGGGTTATGAAATTTGTTGAGTA-3′; reverse primer, +2083, 5′-GGAAGATCTCCACAAGTAAAGGACTGAAATTAA-3′.
After amplification of the human MnSOD basal promoter (−555 to +24), the construct was independently subcloned into the pGL3 basic vector containing the luciferase gene. PCR-amplified I2E (1742–2083) fragments were directly subcloned downstream of the human MnSOD basal promoter (−555 to +24). pMM/p53, a cDNA clone, was subcloned into pcDNA3.1 expression vector. Sp1 expression vector pCMV4/Sp1 was amplified in Escherichia coli, and DNAs were purified. The presence of cDNA was confirmed by restriction digestion and sequencing.
Site-directed Mutagenesis
Site-specific mutations of Sp1 binding sites in the MnSOD promoter were performed according to our previous report (9), in which the resulting mutated sites were obtained using Chameleon® double-stranded site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions and confirmed by competitive gel-shift analysis. The selection primer corresponding to a change of unique XbaI (underlined sequence) site was 5′-GATCGCCGTGTAATTCTGGAGTCGGGGCGGCCGGCCGCTTC-3′. The mutated primers to target each Sp1-binding site (underlined sequences with the mutated nucleotides in lowercase) were designed as follows: Sp1-M1, 5′-CAGCTCATAGGCCGGCTGaatGGCGCTGACCAGCAGCTAG-3′; Sp1-M2, 5′-CCCCGCCGGCACCCTCAGGaatGGACCGGAGGCAGGGCCTTCG-3′; Sp1-M3, 5′-GTGTACGGCAAGCGCGGGtaaaCGGGACAGGCACGCAGGG-3′; Sp1-M5, 5′-CTTTCTTAAGGCCCGCGaat-GGCGCAGGAGCGGCACTC-3′. Inserting mutations in double complementary nucleotides made sp1-M4. After annealing, the double-stranded DNAs were cloned between SmaI and PvuII restriction sites. Two sets of oligonucleotides were used: 5′-GGGGTTGGGCGCGGCGGGCGCGaaatGGGGCCCGCGGGGGGGGaaatGGttatGCGGTGCCCTTGCGGCGCAG-3′ and 5′-CTGCGCCGCAAGGGCACCGCataaaCCatttCCCCCCCCGCGGGCCCCatttCGCGCCCGCCGCGCCCAACCCC-3′. All mutations in the binding sites were confirmed by DNA sequencing. The internal restriction sites were used to link mutations for the individual sites.
Transient Transfection and Luciferase Assay
HepG2 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum, l-glutamine, and insulin, with no antibiotics. 70–80% confluent cells were transfected with plasmids after a Lipofectamine® transfection protocol as directed by the manufacturer. Cells were co-transfected with 2 µm plasmid DNA constructs containing the I2E or basal promoter of the human MnSOD gene in the pGL3 reporter gene. pRL-TK containing Renilla cDNA was also co-transfected with firefly luciferase reporter vector as an internal control at a concentration of 0.2µm. Twenty-four h after transfection, the cells were washed twice with PBS and incubated in fresh medium. Twelve hours later, transfected cells were trypsinized and replated in a 12-well plate at a density of 1.5 × 105 cells per well. After an additional 24 h, cells were treated with 100 nm TPA. Twelve hours post-treatment, the cells were washed with PBS and lysed in passive lysis buffer. Similarly, p53 expression vector (pcDNA3.1/p53) was co-transfected with MnSOD promoter containing pGL3 reporter vector in subconfluent HepG2 cells. In other experiments, Sp1 expression vector and p53 expression vector or control vectors were co-transfected with MnSOD promoter containing pGL3 reporter vector in HepG2 cells. p53 and Sp1 siRNA were transfected in subconfluent HepG2 cells according to manufacturer’s instructions. Cells were treated with a TPA for 12 h in the same manner as described above. Then the cells were lysed in passive lysis buffer; cell lysates were collected, and the samples were analyzed with the luciferase reporter assay system, in accordance with the manufacturer’s instructions, in a TD-20/20 luminometer.
Nuclear Extract Preparation
Nuclei were isolated from HepG2 cells. A subconfluent monolayer of cells was collected and centrifuged at 100×g for 2 min at 4 °C to obtain cell pellets, which were then re-suspended in buffer A containing 10 mm HEPES (pH 7.9), 1.5mm MgCl2, 10mm KCl, 0.5mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride with the inclusion of protease inhibitors (pepstatin, aprotinin, leupeptin) at a concentration of 1µg/ml. Additionally, the phosphatase inhibitors NaF (5 mm) and Na3VO4 (1 mm) were included. The cell suspension was incubated on ice for 15 min. 12.5 µl of 10% Nonidet P-40 was added, and the mixture was vigorously vortexed for 15 s. The cytoplasmic and nuclear fractions were separated by centrifugation at 17,000 × g at 4 °C for 30 s. Subsequently, the nuclear pellets were re-suspended in buffer B containing 20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, 35% glycerol, 0.5 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, and protease inhibitor (pepstatin, aprotinin, leupeptin) at a concentration of 1 µg/ml and incubated on ice for 20 min. Nuclear proteins in the supernatant fraction were collected by centrifugation at 14,000×g at 4 °C for 2 min.
Immunoprecipitation
Immunoprecipitation studies were carried out with nuclear extracts from HepG2 cells (either vehicle- or TPA-treated) in radioimmune precipitation assay buffer (9.1 mm Na2HPO4, 1.7 mm NaH2PO4, 150 mm NaCl (pH 7.4), 1% (v/v) Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 µg/ml phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin). The antibodies used for immunoprecipitation were goat anti-p50 and mouse anti-p53. One mg of nuclear extract was incubated at 4 °C overnight with 2 µg of corresponding antibodies. After incubation with the antibody, 20µl of protein A/G (Santa Cruz Biotechnology, Santa Cruz, CA) was added to the reaction mixture of the antibody and nuclear extract and rotated 2 h at 4 °C. Immunoprecipitates were collected by centrifugation at 2500× g for 5 min followed by washing 4 times with radioimmune precipitation assay buffers. After the final wash, all the adhering liquids with the protein A/G beads were removed. Samples were then resuspended in the 1× Laemmli buffer, subjected to SDS-polyacrylamide gel electrophoresis (10% gel (w/v)), and transferred onto a nitrocellulose membrane; the immunoprecipitated proteins were then detected by Western blotting.
ChIP Assays
One million HepG2 cells were grown on a 10-cm dish for 24 h and then treated with Me2SO (control) and TPA (100 nm) for 12 h. Histone to DNA cross-linking was performed by adding formaldehyde directly to culture media to a final concentration of 1%. Cells were then washed twice using ice-cold PBS containing protease inhibitors (1mm phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, and 1 µg/ml pepstatin). Cells were pelleted by centrifuging at 2000×g for 4 min at 4 °C and then resuspended in protease inhibitor containing SDS lysis buffer (50 mm Tris-HCl (pH 8.1), 10 mm EDTA, and 1% SDS). Crossed-linked chromatin was sonicated to shear DNA on ice to lengths between 200 to 1000 base pairs. Sonicated, cross-linked chromatin was centrifuged at 13,000 × g for 10 min to remove the insoluble materials. The protein concentration of the supernatant was estimated. An equal amount of soluble chromatin from each sample was then diluted 10-fold in ChIP dilution buffer (16.7 mm Tris, 167 mm NaCl, 1.1% Triton X-100, and 0.01% SDS). The diluted soluble chromatin fraction was pre-cleaned by 20µl of salmon sperm DNA/protein A-agarose beads. Pre-cleaned chromatin was mixed with the appropriate antibodies (p50 (Santa Cruz) 2 µg, NPM (Santa Cruz) 2 µg, and Sp1 (Santa Cruz) 2 µg). Parallel preimmune control precipitation was performed by normal IgG. Antibodies and chromatin were allowed to mix and were rotated overnight at 4 °C and precipitated with the appropriate amounts of (60 µl) salmon sperm DNA/protein A-agarose slurry for 2 h at 4 °C with rotation to collect the antibody/chromatin complex.
Beads were then washed by rotation at 4 °C with 1 ml of buffer in the following order of washing; low salt immune complexes wash buffer (20 mm Tris-HCl (pH 8.1), 150 mm NaCl, 2 mm EDTA, 1%Triton X-100, 1% SDS); high salt immune complex wash buffer (20mm Tris-HCl (pH 8.1), 500mm NaCl, 2mm EDTA, 1%Triton X-100, 1% SDS); LiCl immune complex wash buffer (10 mm Tris-HCl (pH 8.1), 0.25 mm LiCl, 1% deoxycholate, 1% Nonidet P-40, 0.1% SDS); followed by two washes with TE buffer (10 mm Tris-HCl (pH 8.1), 1 mm EDTA (pH 8.1)). The precipitated antibody-chromatin complexes were analyzed by either Western blotting or polymerase chain reaction.
For Western blot analysis, immunocomplexes attached to the beads were boiled for 10 min with 25 µl of 1× Laemmli buffer and then subjected to 10% SDS-polyacrylamide gel electrophoresis. Immunoprecipitated proteins were detected by Western blotting. For PCR amplification, precipitated immunocomplexes were eluted twice for 15 min each at room temperature with 250µl of elution buffer (1% SDS, 0.1 m NaHCO3). Reversal of cross-linking was performed by heating the elution mixture at 65 °C for 4 h in the presence of 20 µl of 5 m NaCl. Proteins were digested with proteinase K by incubating for 1 h at 45 °C in the presence of EDTA. DNA was then recovered by phenol:chloroform extraction and ethanol precipitation followed by resuspension in nuclease-free water for PCR amplification. Immunoprecipitated DNA was analyzed by PCR amplification. For amplification of enhancer (1742–2083) elements of the MnSOD gene, the primer sets were: 5′-CGGGGTTATGAAATTTGTTGAGTA-3′ (forward primer, +1742) and 5′-CCACAAGTAAAGGACTGAAATTAA-3′ (reverse primer, +2083); −154 to +6, 5′-ACAGGCACGCAGGGCACCCCCGGGGTT-3′ (forward primer,−154) and 5′-TCCTGCGCCGCCCGCGGGGCCTTAAGAAA-3′ (reverse primer, −6); −555 to +24, 5′-CGCTGGCTCTACCCTCAGCTCATA-3′ (−555, forward primer) and 5′-GCCGAAGCCACCACAGCCACGAGT-3′ (reverse primer,+24). For Bax promoter, the forward primer was 5′-ACGTCCTGCCTGGAAGCA-3′, and reverse primer 5′-CCCCAGCGCAGAAGGAAT-3′ were used. We also amplified the transcription factor binding of the off-target region of MnSOD gene using forward primer (5′-GGCCTACGTGAACCTGA-3′) and reverse primer (5′-CAATTCACCAGTGCTGGCAA-3′) ranging from +521 to +839.
Electrophoretic Mobility Shift Assay (EMSA)
The consensus double-stranded oligonucleotides of NF-κB sequence 5′-AGTTGAGGGGACTTTCCCAGGC-3′ and Sp1 sequence 5′-ATTCGATCGGGGCGGGGCGAGC-3′ (Promega, Madison, WI) were radioactively labeled with [γ-32P]ATP and T4 polynucleotide kinase. The probes were purified on 20% native polyacrylamide gel and eluted in 600 µl of TE buffer (pH 7.4) containing 10 mm Tris HCl and 1 mm EDTA. The activity of radiolabeled probes was counted and stored at −80 °C. Probes were used within 2 weeks after preparation.
In each reaction, 5 µg of nuclear protein and 6 µl of 5× binding buffer containing 20% (v/v) glycerol, 5 mm MgCl2, 2.5 mm EDTA, 5 mm dithiothreitol, 50 mm Tris-HCl (pH 7.5), 0.25 mg/ml poly(dI-dC), and 50,000 cpm of radiolabeled probes were used. Samples were incubated for 20 min at room temperature. To determine the components of the complex bound to the consensus element after treatment, supershift experiments were performed by adding 1 µg of the primary antibody to the binding reaction and extending the incubation to 1 h at room temperature. DNA-protein complexes were separated from unbound probes on 6% polyacrylamide native gel.
Western Analysis
Nuclear extracts (20 µg) from either control or treated cells were used to analyze the levels of various transcription factors or co-activators. For whole cellular proteins, 50 µg were analyzed by Western blotting. Samples were boiled with 1×Laemmli buffer and then subjected to 10% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The transfer efficiency was assessed by incubation with 0.1% Ponceau solution. The membrane was washed with distilled water until the dye disappeared completely. The membranes were then blocked by 5% nonfat dried milk in TBS-T (10 mm Tris-HCl (pH 7.8), 150 mm NaCl, and 0.05% (v/v) Tween 20) buffer (pH 7.8) for at least 1 h at room temperature. After a short wash with TBS-T buffer, the membranes were incubated in the primary antibody for at least 2 h at room temperature or overnight at 4 °C. The primary antibodies were diluted in TBS-T buffer containing 5% nonfat dried milk in a dilution range of 1000–5000. The membranes were then washed 3 times, each time for 10 min, with TBS-T. The membranes were incubated with the secondary antibody at a dilution range of 2,000–10,000 for 1–2 h at room temperature. The membranes were washed twice with TBS-T buffer for 10 min and once with PBS for 5 min. Proteins were detected using the enhanced chemiluminescence detection system (ECL®, Amersham Biosciences). The Quantity One® Image Analyzer software program (Bio-Rad) was used for quantitative densitometric analysis.
RNA Isolation, cDNA Synthesis, and RT-PCR
Total RNA was isolated from HepG2 cells by single-step guanidium thiocyanate/ phenol:chloroform extraction using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, subconfluent HepG2 cells were washed with ice-cold 1× PBS and harvested by scraping and centrifuging. The cells were suspended in 1 ml of TRIzol reagent and incubated at room temperature for 15 min. Chloroform (0.2 ml) was added to the sample, and the sample was vigorously vortexed and then incubated at room temperature for 10 min. The RNA-containing aqueous phase was extracted after centrifugation at 12,000 × g for 15 min at 4 °C.RNA was precipitated with isopropyl alcohol, and the pellet was washed with 70% ethanol in diethyl pyrocarbonate-treated water. The RNA pellet was air-dried. RNA concentration was measured, separated into aliquots, and stored at −80 °C until used. RNA samples having an A260/A280 ratio of 18.0 or more were used for RT-PCR.
For reverse transcription cDNA was generated using 2 µg of total RNA, oligo(dT) primer, and SuperScript™III first strand reverse transcriptase according to the manufacturer’s instructions (SuperScript™III first strand synthesis for RT-PCR, Invitrogen) in a total volume of 20µl. After RT reaction, cDNA was diluted to 100µl with diethyl pyrocarbonate-treated water. Two microliters of first-strand cDNA were amplified using the following PCR primer sets: hMnSOD sense (5′-AGGTTGTTCACGTAGGCCGC-3′ and antisense (5′-AGCATGTTGAGCCGGGCAAGT-3′), wild type p53 sense (5′-ATGGAGGAGCCGCAGTCAGATCC-3′ and antisense (5′-TTCTGTCTTCCCGGACTGAGTCTGACT-3′), Bax sense (5′-AGGATGATTGCTGACGTGGACACG-3′) and antisense (5′-GGTGTACCGTCTGTCACTGGTAGAA-3), and β-actin sense (5′-TGTTACCAACTGGGACGACA-3′) and antisense (5′-CTGGGTCATCATCTTTTCACGGT-3′). For each PCR reaction, 2µl of diluted cDNA were amplified using specific primers (50 pmol) in a final volume of 50µl. For hMnSOD, PCR was performed for 30 cycles at 94 °C (30 s), 65 °C (15 s), and 72 °C (20 s) followed by a final 72 °C step (7 min). For p53, PCR was performed for 30 cycles at 94 °C (30 s), 60 °C (30 s), and 72 °C (45 s) followed by a final 72 °C step (7 min). For Bax, PCR was performed for 30 cycles at 94 °C (30 s), 58 °C (30 s), and 72 °C (45 s) followed by a final 72 °C step (7 min). For β-actin, PCR was performed for 30 cycles at 94 °C (30 s), 60 °C (30 s), and 72 °C (40 s) followed by a final 72 °C step (7 min). PCR products were separated on 1% agarose gel and visualized by ethidium bromide staining.
Northern Blotting
For RNA analysis by Northern blotting, 30 µg of total RNA were separated on 1.1% formaldehyde-agarose gel (1.1% agarose and 4.6% formaldehyde in 1× MOPS, sodium acetate running buffer (pH 7.0)), transferred onto a nylon membrane, and hybridized with γ-32P-labeled hMnSOD cDNA and β-actin cDNA.
Statistical Analysis
Data were analyzed using two-way analysis of variance for comparison. Bonferroni’s post-test multiple comparisons procedure was used to determine the statistical significance. Data shown represent the mean ± S.D.
RESULTS
Identification of Transcription Factors Involved in TPA-induced MnSOD Expression
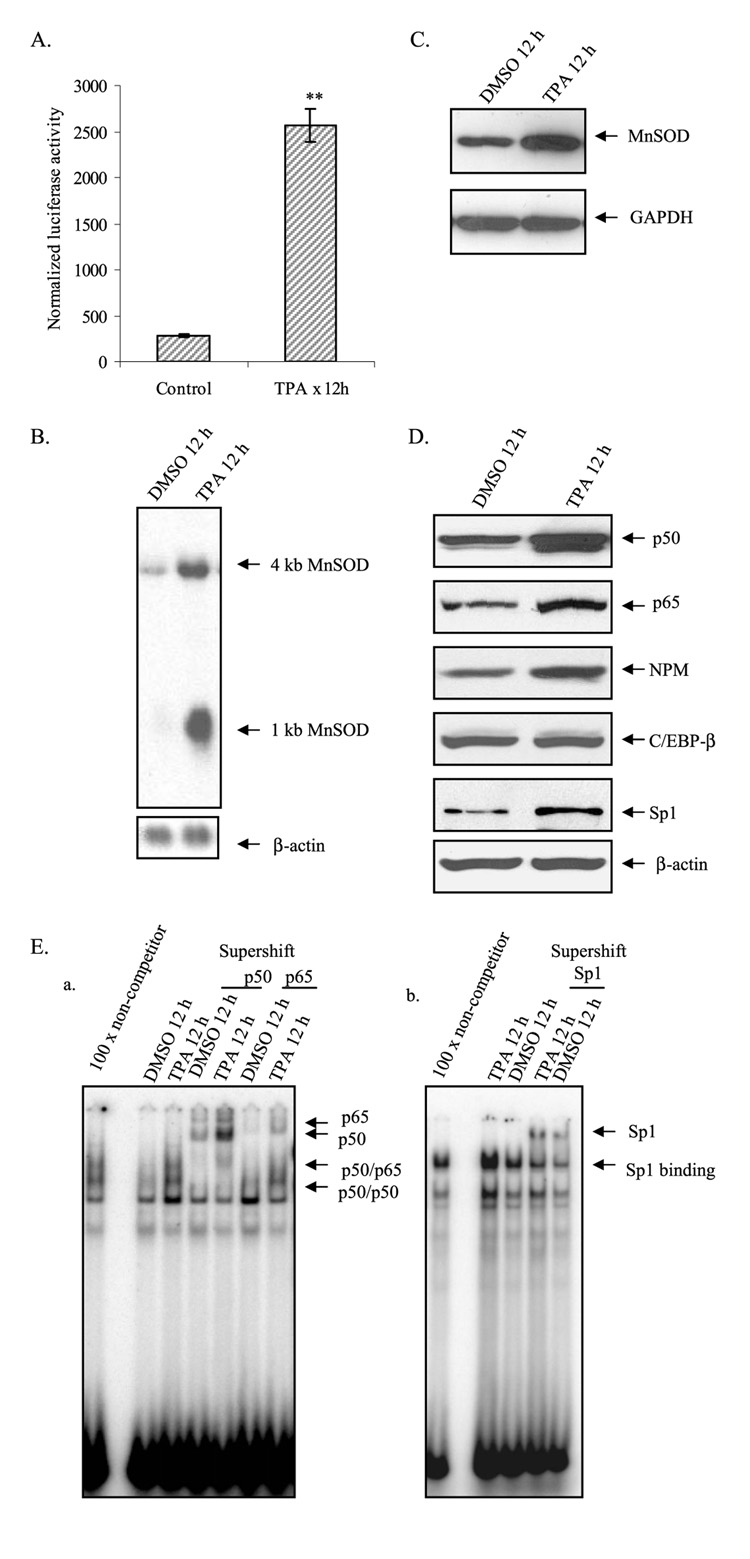
We have previously shown that treatment with TPA increases the levels of MnSOD in HepG2 cells. To verify the effect of TPA on the MnSOD gene, we transiently transfected MnSOD promoter containing the I2E driving the expression of luciferase reporter gene (−555 to +24/I2E/pGL3) into HepG2 cells and measured the reporter gene activity. Consistent with our previous findings, treatment of HepG2 cells with TPA increased the reporter gene activity (Fig. 1A), significantly increased both 1- and 4-kilobase MnSOD transcripts (Fig. 1B), and also significantly increased MnSOD protein levels (Fig. 1C).
FIGURE 1. TPA induces MnSOD gene transcription.
A, MnSOD gene transcription was measured as luciferase activities from MnSOD promoter- and enhancer-driven pGL3 reporter vector. B, Northern analysis was performed using a human MnSOD cDNA probe. Arrows point to the 1- and 4-kilobase human MnSOD transcripts. The same membrane was stripped and re-probed with β-actin cDNA probe as an internal control (bottom arrow).DMSO,Me2SO. C, Western analysis of MnSOD in whole cell extracts after TPA treatment. The membranes were stripped and re-probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as an internal control. D, Western analysis of p50, p65, NPM, C/EBP-β, and Sp1 was performed for nuclear extracts. The same membrane was re-probed with β-actin antibody for the loading control. E, EMSA was carried out as described under “Experimental Procedures.” For supershift experiments EMSA reaction mixtures were incubated with 1 µg of antibodies specific to p50, p65, and Sp1. The arrows point to protein-DNA complexes and supershift complexes. Panel a, complexes of NF-κB and DNA were supershifted by antibodies specific to p50 and p65;panel b,Sp1-DNA complexes were super shifted by the antibody specific to Sp1. Each data point represents the mean of three independent experiments %S.D. Significant difference compared with the control: **, p<0.01.
The increase in the level of MnSOD gene transcription caused by TPA is associated with increased levels of Sp1, NPM, and NF-κB family members. As shown in Fig. 1D, when both control and treated nuclear extracts were subjected to 10% SDS-polyacrylamide gel electrophoresis and Western blotting using antibodies specific to each protein, the results show enhanced accumulation of p50, p65, Sp1, and NPM but not C/EBP-β or β-actin in the nucleus after treatment with TPA. To determine whether the increase in NF-κB and Sp1 levels is accompanied by an increase in DNA binding activity, we examined NF-κB and Sp1 DNA binding activity using EMSA. As expected, TPA-treated nuclear extracts from HepG2 cells showed enhanced DNA binding complexes of both NF-κB and Sp1 (Fig. 1E, panels a and b).
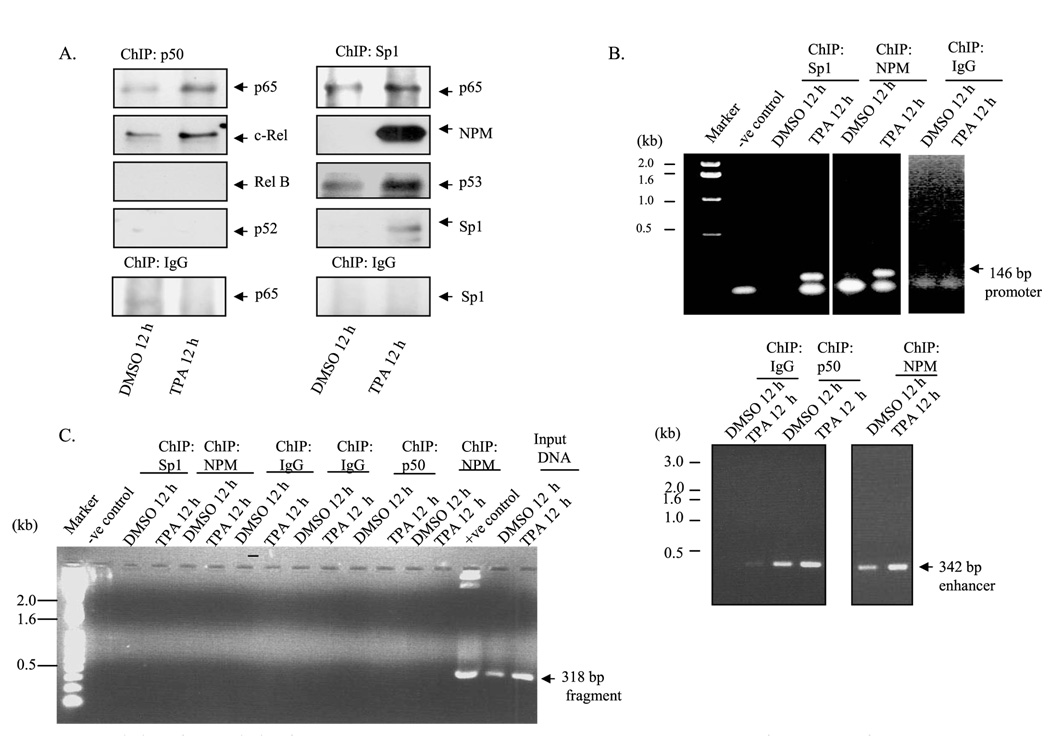
To verify the results of the interactions between Sp1 and NF-κB on sod2, we cross-linked the cellular protein with cellular DNA in vivo by using formaldehyde. The cross-linked DNA-protein complexes were subjected to chromatin immunoprecipitation using antibody specific to p50, a member of the NF-κB family of transcription factors present in HepG2 cells. The product of chromatin immunoprecipitation was analyzed by PCR amplification and Western blotting. For Western analysis, chromatin immunocomplexes were subjected to 10% SDS-polyacrylamide gel, transferred onto nitrocellulose membrane, and probed with an antibody specific for each member of the NF-κB family. As shown in Fig. 2A, an antibody to p50 was able to immunoprecipitate p65 and c-Rel but not p52 and Rel B, and the interaction with p50, p65, or c-Rel increased after TPA treatment. Similarly, chromatin immunoprecipitation using Sp1 antibody was able to immunoprecipitate p65 and NPM. The levels of p65 orNPM increased after TPA treatment. Unexpectedly, we also found p53 protein co-immunoprecipitated with Sp1 antibody (Fig. 2A), suggesting that p53 may play a role in MnSOD transcription. For PCR identification of the MnSOD promoter/enhancer where these proteins bind to drives transcription, we de-cross-linked DNA protein complexes and purified DNA for PCR amplification. Using the purified DNA as the template, both the promoter and intronic enhancer (I2E) fragments were detected (Fig. 2B). Template DNA that was obtained from parallel chromatin immunoprecipitation using non-immune IgG did not yield any PCR product (Fig. 2B).
FIGURE 2. Association of transcription factors to MnSOD promoter and enhancer element.
The binding of transcription factors to the promoter or enhancer region of MnSOD gene was evaluated by ChIP assay in HepG2 cells as described under “Experimental Procedures.” A, chromatins were immunoprecipitated by Sp1 and p50 antibodies or IgG. The pull-down proteins in the immunoprecipitates were detected by Western analysis. DMSO, Me2SO. B, immunoprecipitated DNA was amplified by PCR using primers targeted to the enhancer element (1742–2083) and the promoter elements (−154 to −6) of the MnSOD gene. kb, kilobases. C, the immunoprecipitated DNA was amplified by PCR using primer specific to an off-target region on MnSOD gene (+521 to +839). Total genomic DNA obtained from HepG2 cells were parallel-amplified by using the same off-target primer as input DNA control.
Because we had previously identified NPM as a co-activator of NF-κB in the induction of MnSOD by cytokine and TPA, we also performed ChIP analysis with NPM to amplify the promoter and enhancer elements of the MnSOD gene. ChIP assay results confirmed that NPM is associated with both the promoter and enhancer elements of the MnSOD gene. To further verify the specificity of transcription factors binding to the promoter and enhancer elements, we amplified the DNA obtained from the same ChIP DNA using off target primers, which amplified the fragment between +521 to +839 of the MnSOD gene. As shown in Fig. 2C, we were unable to amplify 318-bp MnSOD off-target fragments using ChIP DNA but were able to amplify appropriate segments of the gene using the same amount of genomic DNA that was used as input control.
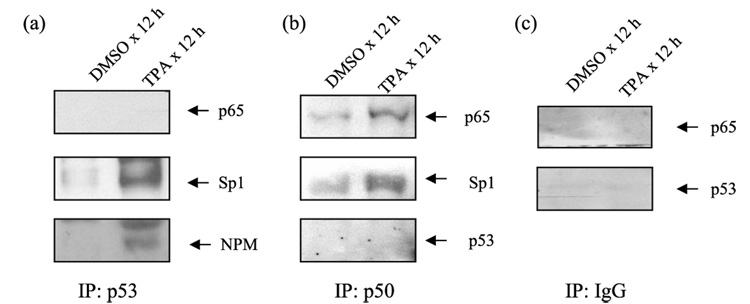
p53 Physically Interacts with Sp1
Because p53 was identified in complex with Sp1, we tested the possibility that p53 may be a component of the transcription complex regulating the expression of the sod2. Using antibodies against p53, we found that p53 antibody was able to immunoprecipitate Sp1 and NPM but not p65 (Fig. 3a), suggesting that p53 did not directly interact with NF-κB family members. Consistent with this possibility, p50 antibodies immunoprecipitated p65 and Sp1 but not p53 (Fig. 3b), and non-immune serum was not able to precipitate either p65 or p53 (Fig. 3c). The interaction between p50 and p65, p50, and Sp1 increased after TPA treatment (Fig. 3, a and b). These results indicate that Sp1 forms complexes with p53 in vivo, and TPA treatment enhances the interaction among proteins within the promoter and enhancer sites.
FIGURE 3. Physical interaction of p53 with Sp1.
In vivo association of p53 with Sp1 and NPM but not with p65 was revealed by co-immunoprecipitation (IP) analysis in isolated nuclear extract (panel a). Both p65 and Sp1 were co-immunoprecipitated with p50 antibody, but p53 was not (panel b). Co-immunoprecipitation with normal IgG was shown as control (panel c). DMSO, Me2SO.
p53 Represses Transcription of the MnSOD Gene
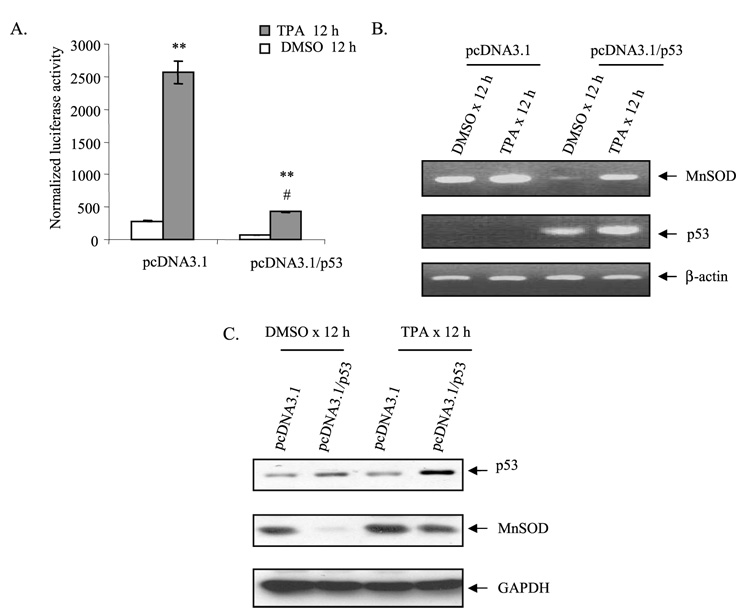
To investigate whether p53 plays a role in MnSOD gene transcription, we overexpressed p53 in HepG2 cells using the pcDNA3.1 vector expressing p53. HepG2 cells were co-transfected with a MnSOD promoter construct (+555 to +24) and p53 expression vector. Overexpression of p53 led to an increase of p53 protein compared with the corresponding empty vector (pcDNA/3.1)-transfected cells. Overexpression of p53 led to a decrease in luciferase activity, whereas transfection with the corresponding empty vector had no effect on reporter gene activity (Fig. 4A). The transcription activity of the MnSOD gene was also suppressed after TPA treatment of HepG2 cells for 12 h compared with the corresponding empty vector-transfected cells.
FIGURE 4. p53 overexpression suppresses MnSOD gene transcription.
HepG2 cells were co-transfected with either the p53 expression vector (pcDNA3.1/p53) or empty vector (pcDNA3.1) along with the MnSOD promoter (−555 to +24)-driven luciferase reporter vector. After 24 h of co-transfection, cells were split into separate dishes and grown for another 24 h before being treated with TPA (100 nm).DMSO,Me2SO. A, 12 h later, cells were collected for luciferase activity as a measure of MnSOD gene transcription. B, RT-PCR of RNA isolated from control and TPA-treated cells was carried out using primers for each specific gene as described under “Experimental Procedures.” C, Western analysis of MnSOD and p53. The membrane was stripped and re-probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as the loading control. Statistical analyses were performed in three independent experiments **, p<0.01 compared with the control; #, p<0.01 compared with the corresponding treatment.
To verify the role of p53 on the endogenous hMnSOD gene, we overexpressed wild-type p53 in HepG2 cells, and the hMnSOD mRNA levels were measured by RT-PCR. As shown in Fig. 4B, p53 over-expression increased p53 mRNA levels and correspondingly decreased hMnSOD mRNA levels. Consistent with the effect on MnSOD mRNA, wild-type p53 overexpression led to decreased hMnSOD protein levels compared with no decrease in the corresponding vector control (Fig. 4C). Both mRNA and protein levels of hMnSOD increased after TPA treatment, but the induction of MnSOD by TPA was reduced in p53 transfected cells compared with empty vector transfected cells similarly treated (Fig. 4, B and C).
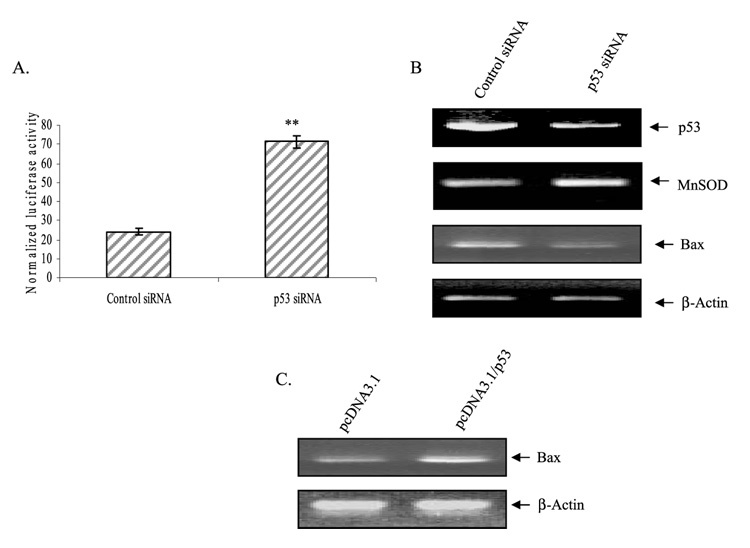
p53 siRNA Enhances hMnSOD Transcription
To further verify the role of p53 in the expression of MnSOD, we used the siRNA approach. HepG2 cells were transfected with the control siRNA or p53 siRNA, and then the luciferase activity and endogenous mRNA levels were evaluated. Suppression of endogenous p53 by expression of p53 siRNA had a significant effect on MnSOD gene transcription as measured by MnSOD-driven reporter gene activity and MnSOD mRNA levels (Fig. 5, A and B). Expression of p53 siRNA significantly decreased the p53 mRNA but not the non-target gene β-actin. p53 siRNA expression also decreased the mRNA levels of Bax, a well recognized p53 target gene, whereas β-actin mRNA level remain unchanged (Fig. 5B). To further verify the p53 effect on its known target genes, we overexpressed p53 expression vector in HepG2 cells and examined the p53 target gene expression. p53 overexpression increased Bax mRNA expression (Fig. 5C). These results further confirm that p53 target genes are transcriptionally regulated by p53 in HepG2 cells.
FIGURE 5. p53 modulates transcription of MnSOD as well as other target genes.
HepG2 cells were transfected with control siRNA or p53 siRNA along with MnSOD promoter (−555 to+24)-driven luciferase reporter vectors. After 24 h of co-transfection, cells were split into separate dishes and grown for another 24 h. A, cells were collected for luciferase activity as a measure of MnSOD gene transcription. B, total RNA was isolated from control siRNA- or p53 siRNA-transfected HepG2 cells. RT-PCR was carried out using primers for each specific gene as described under “Experimental Procedures.” C, HepG2 cells were transfected with p53 expression vector (pcDNA3.1/p53, 4µg) or empty-vector (pcDNA3.1, equivalent amount), and total RNA was purified. The mRNA level of Bax gene under p53 overexpression conditions was determined by RT-PCR. A transcriptional off-target, the β-actin mRNA, was examined as an internal control. Statistical analyses were performed in three independent experiments **, p < 0.01 compared with the control.
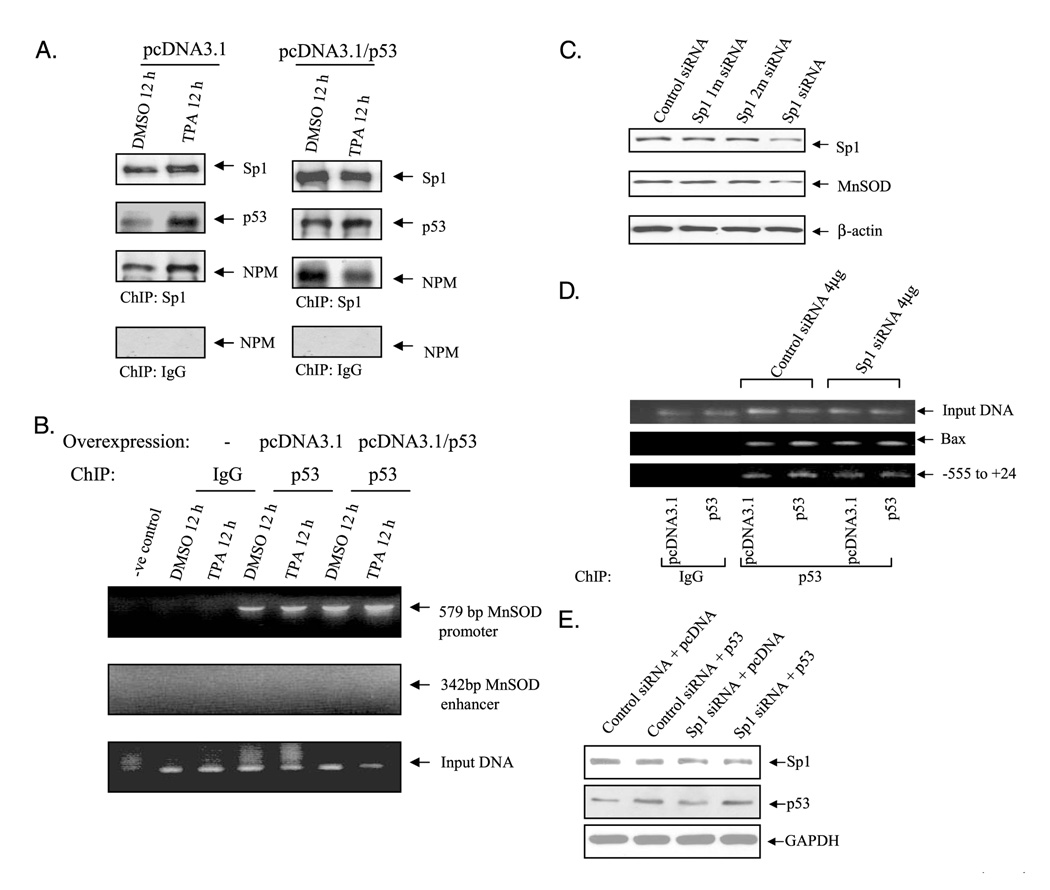
p53 Is Associated with Sp1 and NPM at Promoter
Although we could not determine the direct interaction of p53 with NF-κB members, Sp1 and NPM both interact with NF-κB to enhance MnSOD expression. To investigate whether p53 interferes with MnSOD expression at the promoter or enhancer level, we overexpressed p53 in HepG2 cells and performed a ChIP assay to evaluate the recruitment of NPM and Sp1 to the promoter/enhancer of the MnSOD gene. ChIP assay, conducted with Sp1 antibody, showed that Sp1 and p53 association increased in p53-transfected cells in both untreated and TPA treated conditions. Both Sp1/p53 and Sp1/NPM association increased after TPA treatment compared with Me2SO control in empty vector-transfected cells. In contrast, Sp1/NPM association decreased after TPA treatment compared with control in p53-transfected cells (Fig. 6A). In p53-overexpressed cells, p53 recruited more Sp1 and NPM in the nucleus of Me2SO control, whereas under TPA stimulated condition, association of NPM with Sp1 or p53 decreased. These results suggest that in stimulated condition, p53 may preferentially form complexes with Sp1 when NPM is linked to an enhancer-binding protein such as NF-κB family members. To identify whether p53 interaction is at the promoter or enhancer level, we performed a ChIP assay with p53 antibody in either vector controls or p53-overexpressed HepG2 cells. Chromatin immunoprecipitation with p53 antibody and subsequent PCR amplification of 579-bp promoter fragments (−555 to+24) were detected, as shown in Fig. 6B. However, although we could not amplify the intronic enhancer element using the same DNA as template (Fig. 6B, middle panel), we did secure positive results from input DNA. This result suggests that p53 interaction with Sp1 occurs in the promoter region but not at the enhancer region. To determine whether p53 interaction in the MnSOD promoter is Sp1 dependent, we co-transfected Sp1 siRNA with p53 expression vector and performed a ChIP assay using a p53 antibody. The results shown in Fig. 6D indicate that Sp1 siRNA decreases p53 loading onto the promoter (lower panel), whereas Sp1 siRNA does not have any effect on the p53 target gene such as Bax promoter (middle panel). This result suggests that the effect of p53 on MnSOD transcription is Sp1-dependent.
FIGURE 6. Interaction of p53 with MnSOD promoter.
HepG2 cells were transfected with either p53 expression vector (pcDNA3.1/p53, 4µg) or empty-vector (pcDNA3.1, equivalent amount) and then stimulated with TPA for 12 h. Similarly, cells were co-transfected with p53 expression vector or empty vector with Sp1 siRNA or control siRNA. ChIP assay was carried out as described under “Experimental Procedures.” A, chromatin-immunoprecipitated proteins were detected by Western analysis. DMSO, Me2SO. B, in vivo interactions of p53 with the promoter or the enhancer region of MnSOD gene were determined by ChIP assay. Precipitated DNA was analyzed by PCR using primers specific to MnSOD promoter (−555 to +24) and enhancer (1742 to 2083). Purified DNA from the ChIP assay before immunoprecipitation was amplified with β-actin primer and used as an input loading control. C, cells were transfected with single or double nucleotide mismatches Sp1 siRNA. Sp1 and MnSOD protein levels were determined to verify the control siRNA and target Sp1 siRNA effects. D, in vivo interactions of p53 with MnSOD promoter or Bax promoter upon suppression of Sp1 protein by Sp1 siRNA was determined by ChIP assay. E, Western blotting was used to determine that overexpression of Sp1 siRNA suppresses Sp1 protein and overexpression of p53 increases p53 protein levels. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
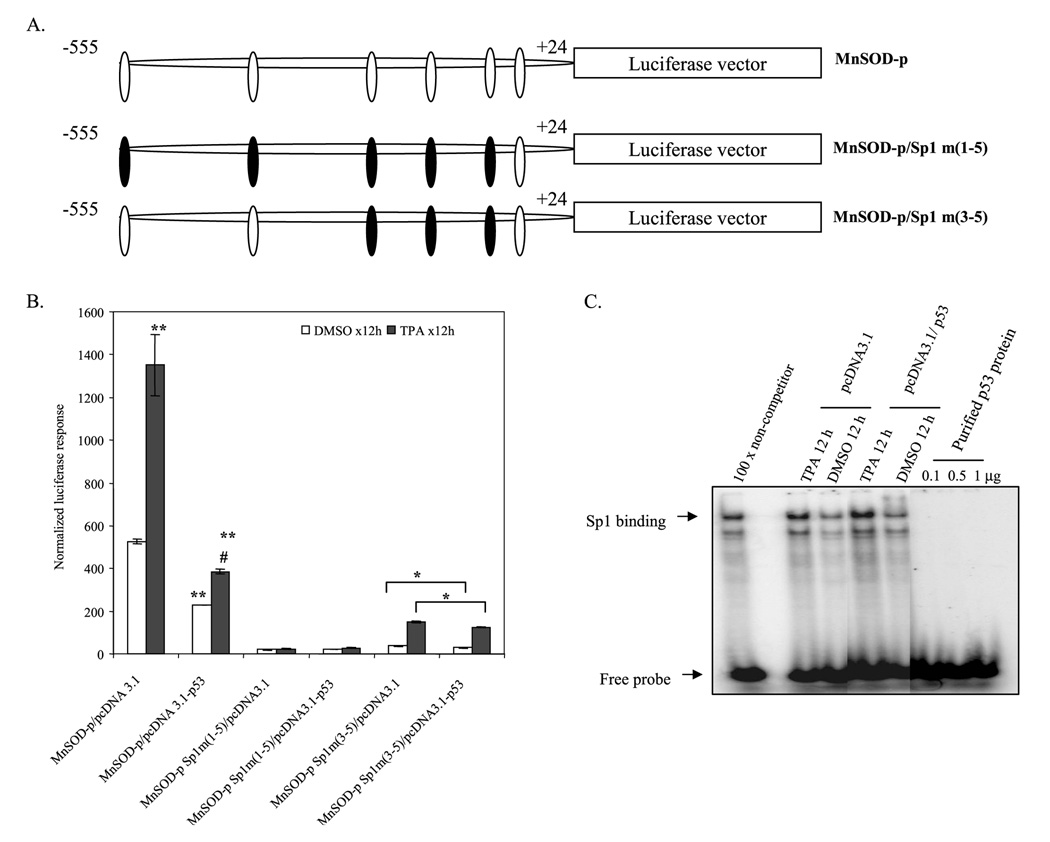
Inefficient Suppression of MnSOD by p53 in Sp1-mutated Promoter
In a site-directed mutagenesis study, we previously demonstrated that Sp1 is essential and sufficient for the basal promoter activity of MnSOD (9). To further confirm the role of p53 on MnSOD promoter activation, we used Sp1-mutated MnSOD promoter constructs and cotransfected with p53 expression vector (Fig. 7A). Consistent with our previous findings, mutations of five Sp1-binding sites in the −555 to +24 promoter construct caused a complete suppression of basal and induced levels of reporter gene activity. Mutation of three Sp1 sites in MnSOD promoter restores a low level of transcriptional activity. Under this condition, p53-induced suppression of MnSOD transcriptional activity is significantly reduced compared with non-mutated MnSOD promoter (Fig. 7B). This result indicates that the suppression effect of p53 on MnSOD transcription is dependent on Sp1 sites in the promoter. To determine whether p53 can directly bind to the Sp1 site and compete for the binding site with Sp1, we performed EMSA with nuclear extract from HepG2 cells. Overexpression of p53 did not alter the Sp1 binding activity under basal or treatment conditions. EMSA carried out with purified p53 (EMSA grade) proteins using consensus Sp1 probe confirmed that p53 was unable to directly bind to the Sp1 site (Fig. 7C).
FIGURE 7. Mutation of Sp1 binding sites abrogates p53 effects on MnSOD transcription.
A, schematic representation of the MnSOD basal promoter and specific mutations of its Sp1 sites in the construct were shown as black boxes. B, either p53 expression-vector (4 µg) or empty vector (equivalent amount) along with MnSOD promoter-driven luciferase reporter vector were transfected in HepG2 cells. Parallel transfection experiments were performed using MnSOD promoter-driven luciferase reporter vectors, where three or five Sp1 binding sites in the promoter region were mutated. After 24 h of co-transfection, cells were washed and grown for another 24 h, and then cells were treated with TPA (100 nm) for 12 h. Cell lysates were collected, and luciferase activity was measured as a determinant of MnSOD gene transcription. DMSO, Me2SO. C, EMSA was performed to check whether there was interference due to the direct binding of p53 to Sp1 binding sites. No binding of purified p53 protein (EMSA grade) to the Sp1 consensus sequence was detected. Each data point represents the mean ± S.D. of three experiments. **, p < 0.01 compared with the control; #, p < 0.01 compared with corresponding treatment; *, p < 0.05.
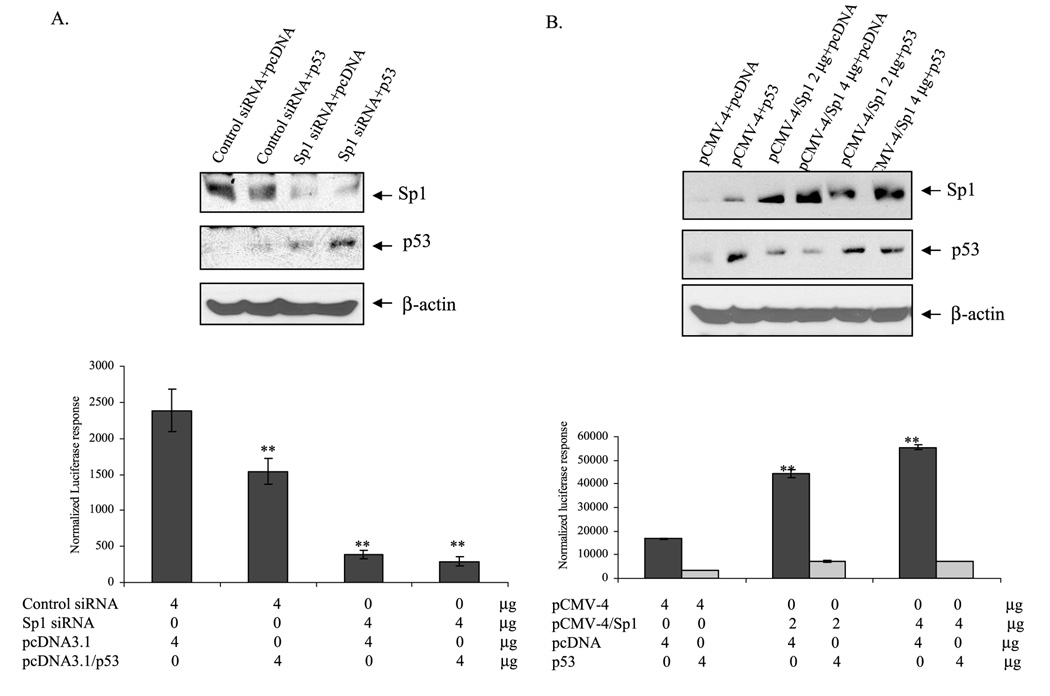
Suppression of MnSOD Promoter Activity by p53 Is Sp1-dependent
To further confirm that p53 leading to the suppression of MnSOD gene transcription is dependent on the availability of Sp1, we overexpressed p53 in cells with reduced levels of Sp1 using Sp1 siRNA. Sp1 siRNA decreased Sp1 protein, and p53 overexpression increased p53 protein levels (Fig. 8A). Co-transfection of wild-type p53 with control siRNA suppressed MnSOD transcription, and co-transfection of wild-type p53 with Sp1 siRNA suppressed both the basal and siRNA-mediated effects on MnSOD promoter. To further confirm that suppression of MnSOD transcription by p53 is Sp1-dependent, we overexpressed wild-type p53 in cells with increased levels of Sp1 using Sp1 expression vector. The expression of Sp1 and p53 proteins was confirmed by Western analysis of nuclear extracts (Fig. 8B, top). Overexpression of Sp1 increased MnSOD transcription in a dose-dependent manner (Fig. 8B). Overexpression of p53 reduced both the basal and Sp1-mediated MnSOD transcription level. In addition, p53 overexpression led to a sustained suppression of Sp1-mediated MnSOD transcription (Fig. 8B).
FIGURE 8. p53 suppresses Sp1-mediated MnSOD gene transcription.
HepG2 cells were co-transfected with Sp1 siRNA and p53 expression (4µg) vector (A) or Sp1 expression vector and p53 expression vector (4µg) (B) along with the MnSOD promoter-driven luciferase reporter vector. After 24 h of co-transfection, cells were washed and grown for another 24 h and harvested, and then the reporter gene activity was determined. The corresponding nuclear extract was subjected to 10% SDS-polyacrylamide gel electrophoresis, and proteins were determined by Western blotting. Statistical analyses were performed in three independent experiments. **, p < 0.01 compared with the control.
DISCUSSION
Induction of MnSOD in response to oxidative stress-inducing conditions is well documented. However, the mechanism by which transcription of the MnSOD gene is suppressed is not fully understood. hMnSOD gene transcriptional regulation is highly dependent on key transcription factors such as NF-κB and Sp1. We and others have demonstrated that NF-κB is essential for the induction of sod2 in response to TPA, tumor necrosis factor-α, and interleukin-1β (17, 24, 25). NF-κB family transcription factors consist of five important members: p50, p52, p65 (Rel A), c-Rel, and Rel B. Cell type and relative abundance determine their differential participation in the transcription of the human MnSOD gene. For example, p50/p50 homodimer is a negative regulator of MnSOD gene transcription (26), whereas p50/p65 and p50/Rel B are potent positive regulators (27, 28). In the present investigation, using ChIP and co-immunoprecipitation assays we confirmed that p50/p65, Sp1, and NPM are key components of the transcription factor complex during TPA-induced MnSOD expression at the chromatin level. We also identified p53 as a component of this complex that can antagonize the positive effect of Sp1 on MnSOD gene transcription. The presence of p53 in the MnSOD promoter complex was verified by the corresponding changes in the level of PCR products representing promoter/enhancer elements, suggesting that p53 interacts with these transcription factors at the chromosomal level.
We have also previously identified NPM as a co-activator of NF-κB in the induction of MnSOD by cytokines (29). It has recently been shown that NPM binds to p53 and prevents p53 from MDM2-mediated degradation (30). In this investigation, we observed that NPM physically interacted with p53 at the chromosomal level, as detected by co-immunoprecipitation and ChIP assays (Fig. 3 and Fig 6) and that the level of p53 increased after TPA treatment (data not shown). Thus, it is tempting to speculate that p53 interacting with NPM may stabilize p53 and facilitate p53 interaction with Sp1. Although the p53 binding sites in the MnSOD gene are not clearly identified, a computer-based F-match search of the MnSOD gene suggested that a p53 binding region is located at 328 bp upstream of MnSOD promoter with a sequence 5′-agGCAGGgcc-3′. The predicted p53-binding site is located at close proximity to Sp1 binding sites. Thus, it seemed possible that p53 binds to the site and hinders the ability of Sp1 to bind with DNA by blocking the Sp1 consensus site. However, this possibility was ruled out because we did not observe p53 binding to the promoter regions containing this consensus site (data not shown). Furthermore, when we added purified p53 protein in the reaction mixture of EMSA using Sp1 consensus element, we found that p53 did not bind to the consensus Sp1 site nor did it block Sp1 binding to DNA. Hussain et al. (31) reported that the p53 transcription factor binding site is present at 2009 bp upstream of the transcription start site in the human MnSOD gene. However, the promoter used in our experiments only contains 555 base pairs of the 5′ flanking region; thus, the possibility of p53 binding to the MnSOD distal site is also excluded. Together, these results indicate that it is not likely that the suppressive effect of p53 on the MnSOD gene is due to p53 binding to MnSOD promoter but, rather, due to p53 interaction with Sp1 protein.
The mechanism by which p53 modulates MnSOD transcription is not known. Various mechanisms have been proposed for p53-mediated transcriptional suppression, including interference with transcription initiation (32), binding of p53 response element (33), and complex formation with other proteins (34). An additional mechanism predicted by the recruitment inhibition model involves p53 interacting with activators at the promoter level and then interfering with the function of the activator by formation of a complex leading to the suppression of gene transcription (35). For instance, the formation of Sp1/p53 complex was proposed to be the mechanism of WRN promoter (36). Others have shown that p53-dependent suppression involves p53 interfering with basal transcription machinery outside gene-specific activators and the binding site (37). It has also been reported that p53- dependent suppression involves the alteration of chromatin structure and recruitment of proteins to the promoter (33, 38).
Our results, which indicate that Sp1 binding sites are necessary for both constitutive and TPA-induced expression of MnSOD, suggest that p53 regulates both basal and induced levels of MnSOD gene transcription by interfering with the basal transcriptional machinery. Thus, complex formation by p53 with Sp1 at the promoter region is likely to have a major impact on MnSOD induction in response to oxidative stress-inducing agents. This possibility is supported by the following findings; 1) Sp1 is essential for both basal- and TPA-induced expression of the MnSOD gene; 2) p53 has no effect on MnSOD expression when all Sp1 sites in the basal promoter were mutated; 3) overexpression of Sp1 enhanced, but reduced expression of Sp1 lessened the effect of p53 on MnSOD expression. These findings are very interesting because p53 acts as a pro-apoptotic factor in cellular response to stress, whereas MnSOD is a pro-survival protein whose functions are involved in protecting cells from oxidative damage. Thus, it is possible that the tumor suppressor function of p53 may be in part mediated by its transcriptional repression ability to suppress the expression of cyto-protective genes such as MnSOD.
Acknowledgments
We thank Dr. Yi Sun, University of Michigan, for the generous gift of p53 expression vector and Dr. Jonathan M. Horowitz, North Carolina State University, for Sp1 expression vector.
Footnotes
This work was supported by National Institutes of Health Grants CA 49797, CA 94853, CA73599, and AG 05119 (to D. K. St. C.).
The abbreviations used are: MnSOD, manganese superoxide dismutase (SOD);hMnSOD, human MnSOD;NPM, nucleophosmin; NF-κB, nuclear factor κB; Sp1, specificity protein 1; TPA, 12-O-tetradecanoylphorbol-13-acetate; PBS, phosphate-buffered saline; EMSA, electrophoretic mobility shift assay; RT, reverse transcriptase; MOPS, 4-morpholinepropanesulfonic acid; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; I2E, intronic enhancer fragment.
REFERENCES
- 1.Dougall WC, Nick HS. Endocrinology. 1991;129:2379–2384. doi: 10.1210/endo-129-5-2376. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Tatsumi H, Satoh S, Sendra T, Naskata T, Fuji J, Takiguchi N. Am. J. Physiol. 1993;265:H1173–H1178. doi: 10.1152/ajpheart.1993.265.4.H1173. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Huang TT, Carison EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 4.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, Mirra SS, Crapo JD, Wallace DC. Nat. Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 5.Wispe JR, Warner BB, Clark JC, Dey CR, Neuman J, Glasser SW, Crapo JD, Chang L, Whitselt JA. J. Biol. Chem. 1992;267:23937–23941. [PubMed] [Google Scholar]
- 6.Baker K, Marcus CB, Huffman K, Kruk H, Malfroy B, Doctrow SR. J. Pharmacol. Exp. Ther. 1998;284:215–221. [PubMed] [Google Scholar]
- 7.Keller JN, Kindy MS, Holtsberg FW, St. Clair DK, Yen H-C, Germeyer AM, Steiner SM, Bruce-Keller AJ, Hutchins JB, Mattson MP. J. Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan XS, Devalaraja MN, St. Clair DK. DNA Cell Biol. 1994;13:1127–1136. doi: 10.1089/dna.1994.13.1127. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Porntadavity S, St. Clair DK. Biochem. J. 2002;362:401–412. doi: 10.1042/0264-6021:3620401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murata Y, Kim HG, Rogers KT, Udvadia AJ, Horowitz JM. J. Biol. Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]
- 11.Coursey AJ, Tjian R. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 12.Pascal E, Tjian R. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 13.Su W, Jackson S, Tjian R, Echols H. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 14.Emili A, Greenblatt J, Ingeles CJ. Mol. Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YH, Williams SC, Baer M, Sterneck E, Gonzalaz FJ, Johnson PF. Mol. Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seto E, Lewis B, Shenk T. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones PL, Ping D, Boss JM. Mol. Cell Biol. 1997;17:6970–6981. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Kiningham KK, Devalaraja MN, Yeh C-C, Majima H, Kasarskis EJ, St. Clair DK. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 19.Grilli M, Chiu JJ, Lenardo MJ. Int. Rev. Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 20.Zabel U, Schreck R, Baeuerle PA. J. Biol. Chem. 1991;266:252–260. [PubMed] [Google Scholar]
- 21.Porntadavity S, Xu Y, Kiningham KK, Rangnekar VM, Prachayasitikul V, St. Clair DK. DNA Cell Biol. 2001;20:473–481. doi: 10.1089/104454901316976109. [DOI] [PubMed] [Google Scholar]
- 22.Katayama K, Ohtsuka K, Takai H, Nakayama H, Doi K. Histol. Histopathol. 2002;17:715–720. doi: 10.14670/HH-17.715. [DOI] [PubMed] [Google Scholar]
- 23.Pascal D, Anne B, Veronique B, Evelyne M. Oncogene. 2001;20:430–439. [Google Scholar]
- 24.Kiningham KK, Xu Y, Daosukho C, Popova B, St. Clair DK. Biochem. J. 2001;353:147–156. [PMC free article] [PubMed] [Google Scholar]
- 25.St. Clair DK, Porntadavity S, Xu Y, Kiningham KK. Methods Enzymol. 2002;349:306–312. doi: 10.1016/s0076-6879(02)49345-7. [DOI] [PubMed] [Google Scholar]
- 26.Tong X, Yin L, Washington R, Rosenberg DW, Giardina C. Mol. Cell. Biochem. 2004;265:171–183. doi: 10.1023/b:mcbi.0000044394.66951.4d. [DOI] [PubMed] [Google Scholar]
- 27.Baeuerle PA, Henkel T. Annu. Rev. Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 28.Josson S, Xu Y, Fang F, Dhar SK, St. Clair DK, St. Clair WH. Oncogene. 2006;25:1554–1559. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhar SK, Lynn BC, Daosukho C, St. Clair DK. J. Biol.Chem. 1994;279:28209–28219. doi: 10.1074/jbc.M403553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chernov MV, Ramana CV, Adler VV, Stark GR. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2284–2289. doi: 10.1073/pnas.95.5.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain SP, Amstad P, He P, Robles A, Lupoid S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 32.Farmer G, Friedlander P, Colgan J, Manley JL, Prives C. Nucleic Acids Res. 1996;24:4281–4288. doi: 10.1093/nar/24.21.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KC, Crowe AJ, Barton MC. Mol. Cell Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy M, Ahn J, Walker KK, Hoffman WH, Evens RM, Levine AJ, George DL. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho J, Benchimol S. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 36.Yamabe Y, Shimamoto A, Goto M, Yokota J, Sugawara M, Furuichi Y. Mol. Cell Biol. 1998;18:6191–6200. doi: 10.1128/mcb.18.11.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mossner J, Engeland K. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirshmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]