Abstract
Purpose
The variant allele of CCND1 G870A encodes a splice variant of the Cyclin D1 protein which possesses an increased half-life. To confirm the phenotypic effect of the variant allele, we examined the immunohistochemical staining pattern of the protein in tumors from a case population of head and neck squamous cell carcinoma (HNSCC) and compared it to the genotype of these individuals. We also examined how this genotype was associated with the risk of HNSCC, and if this genotype-phenotype association was related to patient outcome
Experimental Design
In a population-based case-control study of 698 cases and 777 controls we both genotyped all participants for the CCND1 gene and performed immunohistochemical staining of the Cyclin D1 protein in the HNSCC tumors.
Results
The variant AA genotype was significantly associated with positive IHC staining (P<0.02), and this variant genotype was associated with a significantly elevated OR of 1.5 (95% CI 1.1, 2.0) for HNSCC overall, with risk greatest in oral and laryngeal sites. Positive IHC staining was inversely related to HPV16 DNA present in the tumor (P<0.03). The AA genotype, and super-positive IHC staining for Cyclin D1 also had independent and significant effects on patient survival.
Conclusions
These results strongly suggest that this splice variant, when present in two copies, is a significant predictor of both the occurrence of HNSCC, as well as patient survival after treatment. This data further indicates that this variant protein is an important determinant of individual response to therapy for this disease.
Keywords: HNSCC, Cyclin D1, genotype-phenotype, immunohistochemistry
Introduction
In 2007, almost 46,000 new cases of head and neck squamous cell carcinoma (HNSCC) will be diagnosed, with greater than 11,000 deaths resulting from this disease(1). Tobacco and alcohol use are independently as well as synergistically associated with the incidence of this disease(2, 3), as is human papillomavirus (HPV) infection(4–6). The contribution of genetic variation to HNSCC etiology is an area of intense study, as only a small fraction of those exposed to these agents develop the disease. The precise genes and polymorphisms conferring risk, or modifying the action of alcohol, tobacco or HPV are still being elucidated(7, 8).
Promotion of cell division through dysregulation of the cell cycle is considered a fundamental hallmark of cancer(9) and at the center of this cell cycle regulation are the cyclins and cyclin-dependent kinases(10). Cyclin D1 (encoded by CCND1) interacts with CDK 4/6, and in response to mitogenic signals, leads to phosphorylation of pRB, allowing passage through the restriction point and progression through the G1 phase(11). Cyclin D1 is an established oncogene(12), with overexpression observed in a number of human cancers(13). Overexpression of Cyclin D1 has been observed in HNSCC, and use of neutralizing antibodies to Cyclin D1 in a HNSCC cell line inhibited cell cycle progression(14). In CCND1 a polymorphism (G870A, rs603965) has been identified that leads to a splicing variation and may alter sequences in the protein responsible for protein turnover, thereby rendering this oncogenic protein a greater half-life(15). The variant A allele is associated with an increased risk for HNSCC in a hospital-based case control study in the United States(16), as well as with increased risk for oral premalignant lesions(17). Another European group has described the G allele as variant, and reported this allele to be associated with risk for HNSCC(18), as well as with poorer patient survival(19).
The focus of this study was to examine, in a population-based case control study of HNSCC, the role of CCND1 G870A polymorphism on this disease. We examined the association of the polymorphic alleles of this gene with the expression of the protein in individual tumors using immunohistochemical methods and with the occurrence of HNSCC. Finally, we investigated the association of these biomarkers with patient outcome, defined as overall patient survival.
Materials and Methods
Study population
The study population has been previously described(20, 21). Briefly, incident cases of HNSCC were identified from nine medical facilities in the Boston, MA metropolitan area, with histological classification of malignancy reported by pathology of the participating hospitals, and confirmed by independent study pathologist. Population-based controls were drawn from the same greater Boston population, and were matched to cases by gender, age (±3 years), and town of residence using the Massachusetts town lists. All cases and controls enrolled in the study provided written, informed consent as approved by the IRBs of the participating institutions. Archived pathology specimens were used to construct tissue microarrays for the immunohistochemical analyses. A total of 226 tumors had adequate samples available for tumor array production, and subsequent molecular analyses. Survival time was determined for cases using publicly available databases. Data on HPV16 serology in this case-control study(6), as well as HPV16 DNA in case tumor samples has been previously reported (Furniss, et al Int J Cancer 2007 in press).
Genotyping
DNA was obtained from whole blood or where blood was unavailable, exfoliated buccal cells using the QiAmp DNA extraction system (Qiagen, Valencia, CA). Examination of the CCND1 G870A polymorphism was performed using a commercially available primer/probe set (Applied Biosystems, Foster City, CA) detected using an ABI 3500 Sequence Detection System (Applied Biosystems). Genotyping was performed in a blinded fashion, appropriate controls were included in each run, and approximately 10% of samples were duplicated in a coded fashion as quality assurance with >95% concordance observed between replicates. We have labeled the G allele as the wild-type, and A as variant, as the G allele is considered the “ancestral” allele in the the dbSNP database.
Immunohistochemistry
Each tumor was arrayed in duplicate on the tissue microarray. Array slides were prepared, stained and scored as previously described(22–24). Briefly, antigen retrieval was performed in a pressure cooker for 40 minutes total (19 PSI/127°C) using a citrate buffer (Biogenex, San Ramos,CA). The tissue was incubated with 1:200 dilution of anti-cyclin D1 antibody against the c-terminus of human cyclin D1 (Biocare Meical, Concord CA) for 30 minutes. Primary antibody was detected with peroxidase-conjugated strepavidin and dimethylbenzadene chromagen (Biogenex). Scoring was performed in blinded fashion by the study pathologist (CCB), and staining was scored as insufficient or lacking tumor on slide, negative (no staining), equivocal (1–5% cells staining), positive (>5–90% cells staining), super-positive (>90% cells staining with opaque chromagen). In analyses, samples scoring as negative or equivocal for 1 or both tumor samples were grouped together, samples with 1 or both considered positive were counted as positive, and samples where both samples scored as super-positive were scored as super-positive.
Statistical Analysis
Data were analyzed using the SAS software, and all P values represent two-sided statistical tests. Tests for Hardy-Weinberg equilibrium were conducted. Unconditional logistic regression was used to evaluate the independent effect of the variant CCND1 G870A polymorphism on HNSCC risk, controlling for the matching factors of age and gender, as well as confounders known to be associated with HNSCC risk, including tobacco use (packyears smoked in quartiles based on distribution in controls), alcohol use (lifetime average drinks per week, in quartiles based on distribution in controls), and HPV16 serology (positive or negative based on titer). For analyses by tumor location, cases were grouped according to ICD-9 code, with oral cancer encompassing ICD9 141–145, pharyngeal cancers 146–149, and laryngeal cancers 161. As homozygous GG and heterozygous G/A genotypes had similar ORs, these groups were combined as the referent, and the OR for the association of the homozygous variant (A/A) genotype with disease was determined in all analyses. To evaluate synergistic effects between genotype and these exposures, interaction terms were included and the significance of the interaction evaluated using the likelihood ratio test.
To examine the relationship between Cyclin D IHC score, and CCND1 genotype, as well as tumor HPV16 DNA status, 2 × 2 tables were constructed, and differences in the prevalence of genotype or HPV16 DNA status by Cyclin D IHC staining examined using a Fisher’s exact test. Due to reduced sample size and cells in the table containing no observations, logistic regression analysis could not be performed.
Patient survival was first examined using Kaplan-Meier survival probability curves, and differences between strata tested using the log-rank test. To control for additional variables related to patient survival, Cox proportional hazards modeling was employed. These survival probability models included variables representing the Cyclin D1 IHC superpositivity and CCND1 genotype and were controlled for patient age (in decades), and tumor stage (1/2 vs. 3/4).
Results
823 eligible cases were invited to participate; of these, 57 refused to participate, 44 did not complete the questionnaire, and 24 did not provide DNA samples for genotyping studies. Therefore, 698 cases are included in the analysis. Of these cases, pathological tumor stage data was available on 433 individuals of which 60 (14%) are stage 1, 68 (16%) stage 2, 90 (21%) stage 3, and 215 (50%) stage 4. Similarly, 1,623 subjects were identified as potential controls, and a total of 777 consented to participation, completed the questionnaire, and provided a DNA sample, and are thus included in the analysis.
Table 1 describes the characteristics of the study population. As expected, the mean age and distribution of gender was similar between cases and controls. As previously described(20, 21), there was an increasing relative risk of HNSCC with increasing pack-years smoked, with individuals in the highest quartile having an OR of 3.7 (95% CI 2.7, 5.1). The estimated magnitude of cancer risk for tobacco use was similar for oral and pharyngeal cancers, but was greatly elevated (OR 12.6 95% CI 6.2, 25.5) for laryngeal cancer. There was a non-linear dose-response for HNSCC risk with increasing lifetime average drinks per week. Compared to subjects drinking <2.5 drinks per week on average, those with low average weekly alcohol consumption (2.5 to <6 drinks per week) had a significantly reduced relative risk of HNSCC overall (OR 0.6, 95% CI 0.4, 0.9). This was observed in all subsites, but was statistically significant only amongst the oral cancers (OR 0.6, 95% CI 0.4, 0.9). At the same time, subjects whose lifetime average alcohol consumption was >14 drinks per week had a significantly elevated overall relative risk for HNSCC (OR 2.2, 95% CI 1.5, 3.1), with similar ORs across the subsites. Also as previously reported(6), seropositivity for HPV16 was associated with significantly increased HNSCC risk (OR 4.4 95% CI 3.1, 6.3), with the greatest OR for tumors of the pharynx (OR 8.3 95% CI 5.1, 13.7).
Table 1.
Variant CCDN1 Genotype is Associated with Oral and Laryngeal Cancers.
| All HNSCC |
Oral Cancer (N=355) |
Pharyngeal Cancer (N=202) |
Laryngeal Cancer (N=139) |
|||
|---|---|---|---|---|---|---|
| Characteristic | Controls (n=777) |
Cases (n=698) |
OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) |
| Age, year, mean (SD) | 61.0 (11.3) | 59.9 (11.8) | ||||
| Gender, n (%) | ||||||
| Male | 568 (71.9) | 530 (72.6) | ||||
| Female | 222 (28.1) | 200 (27.4) | ||||
| Race, n (%) | ||||||
| Non-white | 69 (8.7) | 81 (11.1) | ||||
| White | 721 (91.3) | 649 (88.9) | ||||
| Tobacco, Lifetime pack-years, n (%) | ||||||
| ≤ 1 | 302 (38.2) | 147 (20.1) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| > 1 to ≤ 9 | 94 (11.9) | 60 (8.2) | 1.4 (0.9, 2.1) | 1.3 (0.8, 2.1) | 1.1 (0.5, 2.2) | 2.5 (0.9, 6.8) |
| > 9 to ≤ 34 | 199 (25.2) | 166 (22.7) | 1.6 (1.2, 2.2) | 1.3 (0.9, 2.0) | 1.5 (0.9, 2.6) | 4.0 (1.9, 8.4) |
| > 34 | 195 (24.7) | 357 (49.0) | 3.7 (2.7, 5.1) | 2.4 (1.6, 3.5) | 4.3 (2.6, 7.0) | 12.6 (6.2, 25.5) |
| Alcohol Use, Lifetime Ave. | ||||||
| Drinks Weekly, n (%) | ||||||
| < 2.5 | 178 (22.5) | 127 (17.4) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| 2.5 to < 6 | 218 (27.6) | 98 (13.4) | 0.6 (0.4, 0.9) | 0.6 (0.4, 0.9) | 0.7 (0.4, 1.2) | 0.7 (0.3, 1.3) |
| 6 to < 14 | 194 (24.6) | 129 (17.7) | 0.9 (0.6, 1.3) | 1.1 (0.7, 1.8) | 0.7 (0.4, 1.3) | 0.6 (0.3, 1.2) |
| ≥ 14 | 200 (25.3) | 375 (51.5) | 2.2 (1.5, 3.1) | 2.6 (1.7, 4.0) | 2.0 (1.1, 3.3) | 1.7 (0.9, 3.1) |
| HPV16 Seropositivity, n (%)* | ||||||
| No | 478 (89.2) | 308 (68.0) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| Yes | 58 (10.8) | 145 (32.0) | 4.4 (3.1, 6.3) | 3.1 (2.0, 4.6) | 8.3 (5.1, 13.7) | 2.2 (1.1, 4.3) |
| CCND1 Genotype | ||||||
| Wt/Wt + Wt/Var | 634 (81.6) | 524 (75.1) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| Var/Var | 143 (18.4) | 174 (24.9) | 1.5 (1.1, 2.0) | 1.6 (1.2, 2.2) | 1.1 (0.7, 1.7) | 1.8 (1.1, 2.8) |
Note: Models are controlled for al variables in table. Two cases did not have ICD-9 coding and thus were not included in location-specific models.
Serum HPV antibody status was available for 989 subjects. Remaining subjects were coded as missing and included in the model.
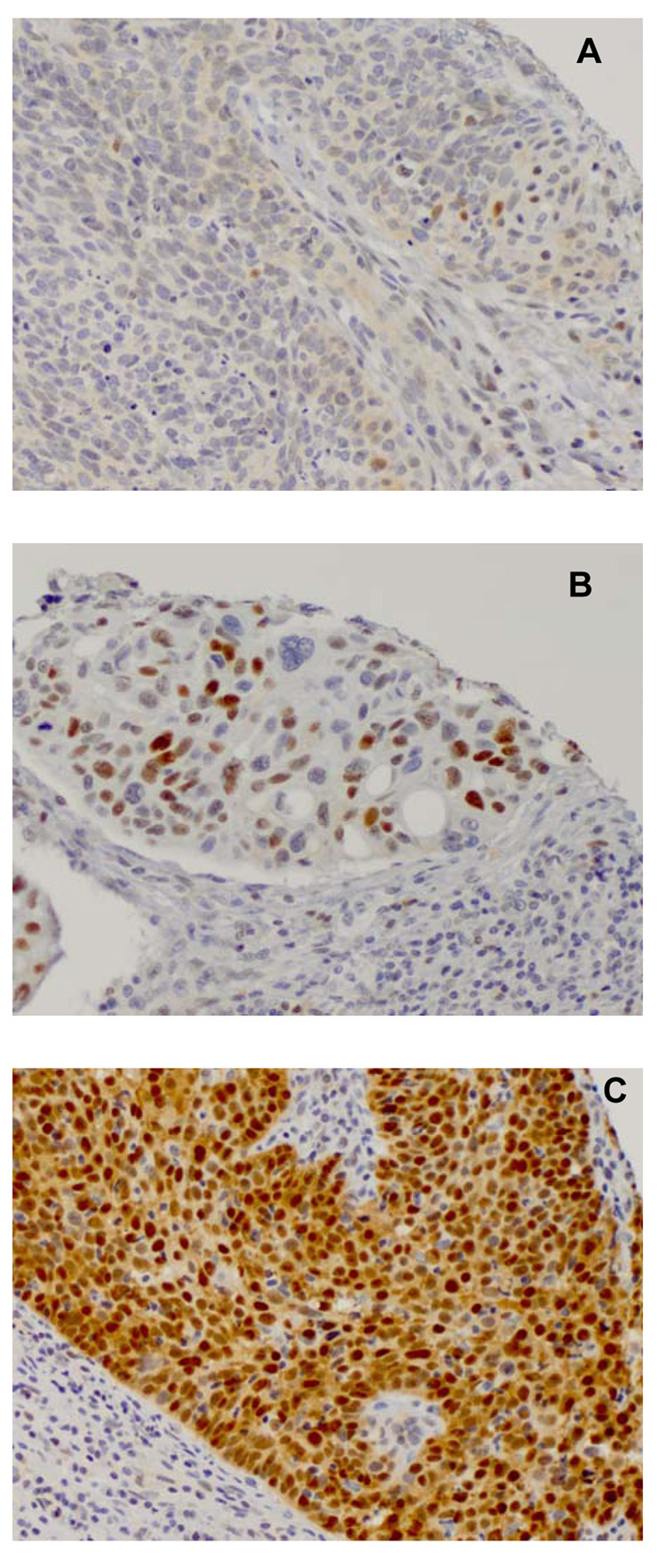
We examined the impact of the variant CCND1 genotype on a phenotypic marker in tumors from patients harboring this genotype, as this variant has previously been associated with HNSCC risk. In an effort to ascribe a definite phenotype to this polymorphism, we investigated the relationship between CCND1 (G870A) genotype and Cyclin D1 IHC staining. Figure 1 shows representative photomicrographs of the staining patterns of Cyclin D1. Table 2 shows that tumors arising in homozygous variant individuals had a significantly greater prevalence of positive or super-positive Cyclin D1 IHC staining compared to tumors arising in homozygous wildtype (GG) or heterozygous (GA) individuals (P<0.02).
Figure 1.
Representative array photomicrographs of Cyclin D1 IHC staining pattern in HNSCC, where brown nuclear color represents antibody positivity. All photomicrographs are at 20X magnification. Tumor samples considered (A) equivocal, (B) positive, and (C) super-positive are shown.
Table 2.
Altered Cyclin D immunohistochemical staining is associated with variant genotype and inversely associated with tumor HPV positivity in HNSCC cases.
| Cyclin D1 IHC Staining | |||
|---|---|---|---|
| Tumor Characteristic | Negative or Equivocal (n=17) |
Positive or Super- positive (n=98) |
P* |
| CCND1 genotype | 0.02 | ||
| Wt/Wt + Wt/Var | 17 (100) | 74 (75.5) | |
| Var/Var | 0 (0) | 24 (24.5) | |
| Tumor HPV16 DNA Status† | 0.03 | ||
| Absent | 7 (46.7) | 71 (74.7) | |
| Present | 8 (53.3) | 24 (25.2) | |
Fisher’s exact test.
Tumor HPV16 status available on 110 tumors.
As our data strongly indicated that the protein expression of CCND1 was altered by this polymorphism, we sought to determine if it was associated with HNSCC in our population. The prevalence of the homozygous wildtype CCND1 (GG) genotype was 30.6% (238/777) in controls and 30.1% (210/698) in cases. The heterozygous (GA) genotype was found in 51.0% (396/777) of controls and 45.0% (314/698) of cases, while the homozygous variant (AA) genotype was found in 18.4% (143/777) controls and 24.9% (174/698) cases. These allele frequencies in controls did not deviate from Hardy-Weinberg equilibrium. The AA genotype was associated with a significantly increased relative risk for HNSCC overall (OR 1.5 95% CI 1.1, 2.0) in models controlled for confounders. This effect was most prominent in oral (OR 1.6 95% CI 1.2, 2.2) and laryngeal cancers (OR 1.8 95% CI 1.1, 2.8), and was not significant in pharyngeal cancers. There was no significant interaction of this genotype with tobacco smoking, alcohol use, or HPV16 seropositivity (data not shown). As the homozygous variant genotype was significantly associated with disease in the tumor subsites that show a lesser association with HPV16 serology, we examined whether Cyclin D1 overexpression differed in tumors based on the concomitant presence of HPV16 detectable in tumor DNA. Table 2 shows that tumors positive for HPV16 DNA were significantly less likely to have positive or super-positive staining compared to those negative for HPV16 DNA (P<0.03).
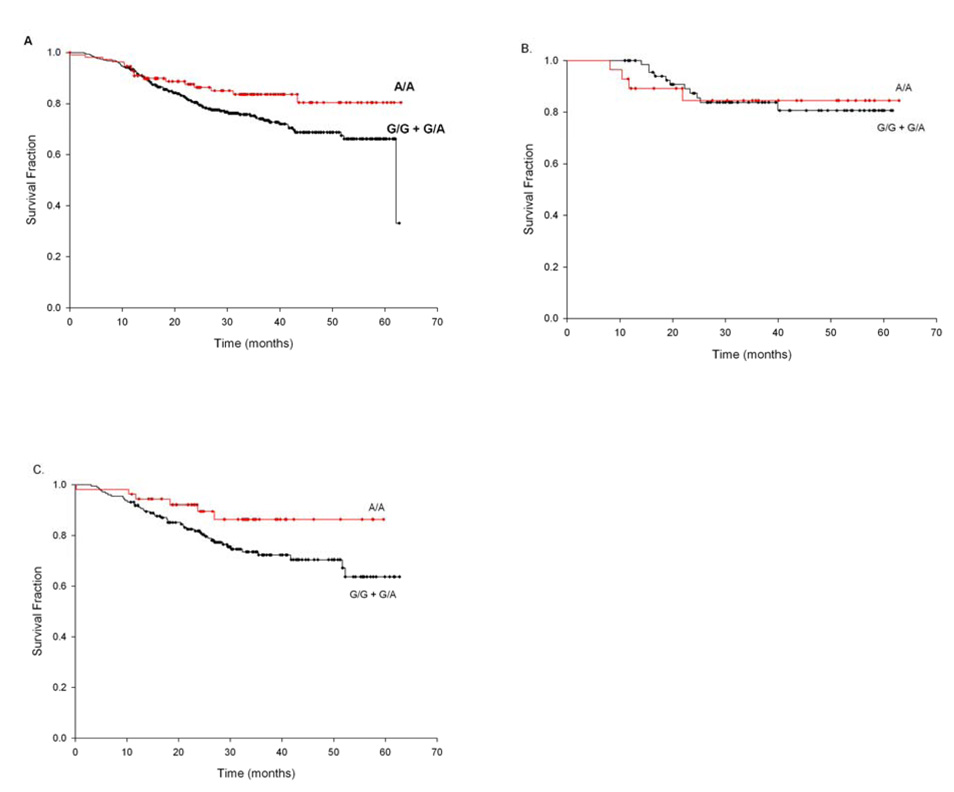
The inverse relationships of HPV status (both seropositivity and tumor DNA) with Cyclin D staining led us to then examine if CCND1 genotype or Cyclin D1 IHC staining was associated with patient outcome, as we and others have previously demonstrated that HPV16-associated HNSCC has better overall survival than those not associated with HPV(6, 25). Results of Kaplan-Meier analysis of CCND1 G870A genotype on overall patient survival are presented in Fig. 2. Amongst cases, those with the CCND1 AA genotype had significantly better overall survival (Fig. 2A, log-rank P<0.05), compared to those with the GG or GA genotypes. Stratifying by tumor stage, this effect appears most strikingly amongst the higher stage tumors but is no longer statistically significant (Fig 2B and 2C). Stratifying by HPV16 seropositivity, we find that the association of CCND1 genotype with survival is present and strongly significant (log-rank P<0.01, data not shown) only amongst HPV16 seronegative cases.
Figure 2.
Kaplan-Maier survival probability curves comparing CCND1 G870A genotypes in patients with HNSCC both overall and stratified by tumor stage. The difference between the groups was tested by use of the log-rank method. (A) CCND1 G870A genotype (AA vs. GG/GA) in all patients (log-rank P<0.05); (B) CCND1 G870A genotype (AA vs. GG/GA) in low stage (1/2) patients (log-rank P<1.0); (C) CCND1 G870A genotype (AA vs. GG/GA) in high stage (3/4) patients (log-rank P<0.06). Survival time is defined as the time from initial diagnosis to patient’s death. Tick marks represent censored values.
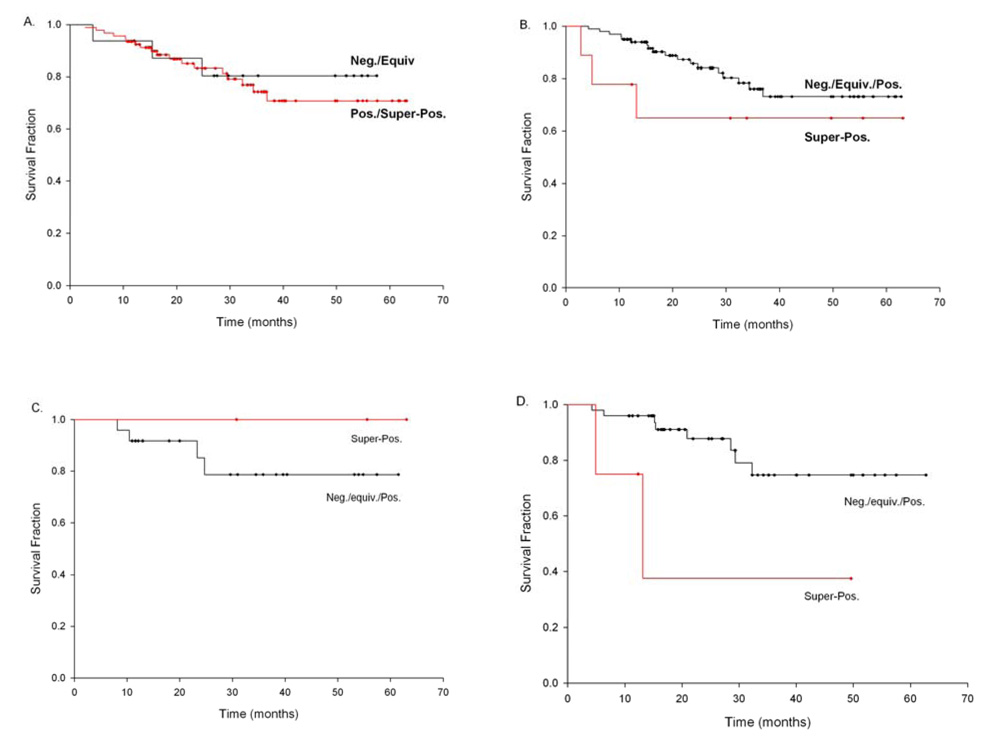
Dichotomizing Cyclin D1 IHC as in Table 2 showed no significant difference in overall patient survival (Fig 3A). On the other hand, comparing super-positive staining tumors to all others, there was a trend towards poorer overall survival in the super-positive tumors (Fig 3C). This effect was highly significant amongst the stage 3/4 tumors (log rank P<0.03, Fig 3D) but was not apparent in the stage 1/2 cases (Fig. 3C). Stratified by CCND1 genotype, the association between Cyclin D1 superpositivity is only amongst the CCND1 GG or GA genotype individuals (log rank P<0.07, data not shown), while there no association between Cyclin D1 IHC superpositivity and survival amongst the variant AA genotype cases. In order to examine these two measures simultaneously, as well as to control for confounders of survival, we employed a multivariate Cox’s proportional hazards model of survival (Table 3), controlling for patient age and tumor stage. We have chosen to control for tumor stage in these models, instead of specific treatment regiments, as there is significant co-linearity between stage and treatment plans, with low stage cases being treated with either surgery (majority) or radiation alone, while higher stage (3/4) disease is treated with both surgery and radiation, as well as chemotherapeutic approaches(26). This model suggests a significantly reduced hazards ratio (P<0.05) for CCND1 AA genotype (HR 0.6 95% CI 0.3, 1.0) compared to AG and GG genotypes, and a significantly increased HR (P<0.05) for super-positive staining (HR 3.6 95% CI 1.0, 13.0).
Figure 3.
Kaplan-Maier survival probability curves comparing Cyclin D1 IHC staining measures in patients with HNSCC both overall and stratified by tumor stage. The difference between the groups was tested by use of the log-rank method. (A) Cyclin D1 IHC staining positive/super-positive vs. negative/equivocal amongst all patients (log-rank P<0.67); (B) Cyclin D1 IHC staining super-positive vs. positive/equivocal/negative amongst all patients (log-rank P<0.26); (C) Cyclin D1 IHC staining super-positive vs. positive/equivocal/negative amongst low stage (1/2) patients (log-rank P<0.4); and (D) Cyclin D1 IHC staining super-positive vs. positive/equivocal/negative amongst high stage (3/4) patients (log-rank P<0.03). Survival time is defined as the time from initial diagnosis to patient’s death. Tick marks represent censored values.
Table 3.
CCND1 genotype and Cyclin D1 immunohistochemical staining super-positivity are associated with better survival in a model controlled for tumor stage, and patient age.
| Cyclin D1 Co-variate | n | Hazard Ratio (95% CI) |
|---|---|---|
| CCND1 genotype | ||
| Wt/Wt + Wt/Var | 344 | 1.0 (referent) |
| Var/Var | 110 | 0.6 (0.4, 1.0) |
| Cyclin D1 Immunohistochemistry | ||
| Negative, Equivocal, Weak Positive | 100 | 1.0 (referent) |
| Super-positive | 9 | 3.6 (1.0, 13.0) |
Note: Model controlled for age, stage, and all variables in table. Subjects missing IHC or stage data were coded as missing and included in the model.
Discussion
Our initial work defined an additional phenotypic consequence of the CCND1 polymorphism, showing that cases possessing the homozygous variant genotype had an increased prevalence of positive or super-positive IHC staining for the Cyclin D1 protein (Table 2). This is consistent with the molecular evidence that the variant A allele codes for an alternate transcript, whose protein product exhibits a greater half-life compared to that encoded by the G allele(15). These findings, taken together, provide an excellent grounding for further study of this variant in a case-control studies of cancer risk.
We also found, in one of the largest studies to date, that the variant AA CCND1 genotype was associated with an increased relative risk of HNSCC. The magnitude of the risk estimate for this variant AA genotype of 1.5, is similar to what has been previously reported in a smaller, hospital-based study of HNSCC(16), but lower than that reported for the association between this genotype and oral premalignant lesions(17). At the same time, our results are opposite to those reported by Holley et al (18) in a German population, who list the G allele as variant, and find significantly elevated risk (OR 3.37, 95% CI 1.61, 9.80) in subjects possessing the GG genotype for oral squamous cell carcionomas(18). Interestingly, Matthias et al (19) observed no association of this genotype with cancer risk in a population of predominantly laryngeal cancers(19). These studies, however, controlled only for age and gender in their logistic regression analyses, and thus the differences observed between their results and ours may be due to a large level of residual confounding in their model that did not control for smoking, alcohol consumption or HPV16.
Our observation that the risk associated with the CCND1 variant allele is greatest in oral and laryngeal, but not pharyngeal, is also of interest, particularly as this is opposite what we have observed with HPV16 seropositivity. HPV acts as a carcinogen through the expression of the E6/E7 proteins which inactivate the tumor suppressor proteins p53 and pRB, amongst other functions(27). Patients possessing the variant Cyclin D1 protein may already be pre-disposed to abrogation of the pRB protein product through increased or longer-lived expression of the Cyclin D1 protein, which, through interaction with CDK4/6 leads to the phosphorylation and subsequent degradation of pRB. The inverse relationship between Cyclin D1 positive IHC staining and HPV16 DNA we observed in the primary tumor samples adds additional strong evidence for this hypothesis.
We also observed that cases possessing the AA genotype had significantly better overall survival, compared to those having the GA or GG genotypes (Fig 2A) and this effect was evident amongst higher stage disease. This result is consistent with the report of Matthias, et al. who observed poorer survival with the GG genotype(19). We have previously demonstrated that HPV16 seropositive cases have significantly better survival rates, and, consistent with other work, these cases are more often pharyngeal(6). Interestingly, if our population of cases is stratified by HPV16 seropositivity, we find that the association of CCND1 genotype with survival is present and strongly significant only amongst HPV16 seronegative cases. This suggests that possibly the same survival advantage, be it less aggressive disease, or better treatment response, associated with HPV may also be present in the individuals with variant genotype.
At the same time, Cyclin D1 protein IHC super-positivity was related to poorer patient survival, achieving statistical significance in the high stage disease. While this seems at odds with the observation that variant CCND1 genotype is significantly associated with positive staining, if the survival analysis is stratified by CCND1 genotype, the association between Cyclin D1 superpositivity is only amongst the CCND1 GG or GA genotype individuals (log rank P<0.07, data not shown). There is no appreciable relationship amongst the variant AA genotype cases. These results suggest that the super-positive staining may be resulting from different molecular events than those related to more moderate IHC positivity. This is borne out in the multivariate proportional hazards model which demonstrates independent effects of CCND1 genotype and Cyclin D1 IHC super-positivity on survival (Table 3).
We have observed a genotype-phenotype correlation suggesting that the AA genotype of CCND1 G870A polymorphism is associated with positive IHC staining for the Cyclin D1 protein in HNSCC tumors. This motivated our examination of the association between CCND1 genotype and HNSCC, where we have observed a 50% increased relative risk of HNSCC in individuals harboring the CCND1 AA genotype independent of the risks associated with tobacco smoking, alcohol use, and HPV16 infection. Further, cases with the homozygous variant genotype appear to have better overall survival, suggesting that these individuals may be similar to those with HPV-related HNSCC, as they also experience enhanced survival from this disease. Finally, we have noted that super-positive IHC staining for Cyclin D1 protein may be a potential prognostic marker of poorer survival in HNSCC, but additional, larger studies must be undertaken in order to confirm these findings.
Acknowledgements
The authors would like to thank the collaborating clinicians and research staff involved in this study.
Grant support: NIH (R01CA078609 and R01CA100679) and the Flight Attendants Medical Research Institute (CJM).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ, McLaughlin JK, Winn DM, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer research. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 3.Rothman K, Keller A. The effect of joint exposure to alcohol and tobacco on risk of cancer of the mouth and pharynx. J Chronic Dis. 1972;25:711–716. doi: 10.1016/0021-9681(72)90006-9. [DOI] [PubMed] [Google Scholar]
- 4.Gillison ML, Shah KV. Human papillomavirus-associated head and neck squamous cell carcinoma: mounting evidence for an etiologic role for human papillomavirus in a subset of head and neck cancers. Curr Opin Oncol. 2001;13:183–188. doi: 10.1097/00001622-200105000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Loning T, Ikenberg H, Becker J, Gissmann L, Hoepfer I, zur Hausen H. Analysis of oral papillomas, leukoplakias, and invasive carcinomas for human papillomavirus type related DNA. J Invest Dermatol. 1985;84:417–420. doi: 10.1111/1523-1747.ep12265517. [DOI] [PubMed] [Google Scholar]
- 6.Furniss CS, McClean MD, Smith JF. Human papillomavirus 16 and head and neck squamous cell carcinoma. International journal of cancer. 2007;120:2386–2392. doi: 10.1002/ijc.22633. [DOI] [PubMed] [Google Scholar]
- 7.Hung RJ, van der Hel O, Tavtigian SV, Brennan P, Boffetta P, Hashibe M. Perspectives on the molecular epidemiology of aerodigestive tract cancers. Mutation research. 2005;592:102–118. doi: 10.1016/j.mrfmmm.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Sturgis EM, Wei Q. Genetic susceptibility--molecular epidemiology of head and neck cancer. Curr Opin Oncol. 2002;14:310–317. doi: 10.1097/00001622-200205000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Lundberg AS, Weinberg RA. Control of the cell cycle and apoptosis. Eur J Cancer. 1999;35:531–539. [PubMed] [Google Scholar]
- 11.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 12.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd R, Hinds PW, Munger K, et al. Cell cycle dysregulation in oral cancer. Crit Rev Oral Biol Med. 2002;13:51–61. doi: 10.1177/154411130201300106. [DOI] [PubMed] [Google Scholar]
- 14.Bartkova J, Lukas J, Muller H, Strauss M, Gusterson B, Bartek J. Abnormal patterns of D-type cyclin expression and G1 regulation in human head and neck cancer. Cancer research. 1995;55:949–956. [PubMed] [Google Scholar]
- 15.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- 16.Zheng Y, Shen H, Sturgis EM, et al. Cyclin D1 polymorphism and risk for squamous cell carcinoma of the head and neck: a case-control study. Carcinogenesis. 2001;22:1195–1199. doi: 10.1093/carcin/22.8.1195. [DOI] [PubMed] [Google Scholar]
- 17.Huang M, Spitz MR, Gu J, et al. Cyclin D1 gene polymorphism as a risk factor for oral premalignant lesions. Carcinogenesis. 2006;27:2034–2037. doi: 10.1093/carcin/bgl048. [DOI] [PubMed] [Google Scholar]
- 18.Holley SL, Matthias C, Jahnke V, Fryer AA, Strange RC, Hoban PR. Association of cyclin D1 polymorphism with increased susceptibility to oral squamous cell carcinoma. Oral oncology. 2005;41:156–160. doi: 10.1016/j.oraloncology.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Matthias C, Branigan K, Jahnke V, et al. Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998;4:2411–2418. [PubMed] [Google Scholar]
- 20.Peters ES, McClean MD, Marsit CJ, Luckett B, Kelsey KT. Glutathione Stransferase polymorphisms and the synergy of alcohol and tobacco in oral, pharyngeal, and laryngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:2196–2202. doi: 10.1158/1055-9965.EPI-06-0503. [DOI] [PubMed] [Google Scholar]
- 21.Ting Hsiung D, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 22.Brambilla E, Moro D, Gazzeri S, Brambilla C. Alterations of expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. The Journal of pathology. 1999;188:351–360. doi: 10.1002/(SICI)1096-9896(199908)188:4<351::AID-PATH385>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Frierson HF, Jr., Gaffey MJ, Zukerberg LR, Arnold A, Williams ME. Immunohistochemical detection and gene amplification of cyclin D1 in mammary infiltrating ductal carcinoma. Mod Pathol. 1996;9:725–730. [PubMed] [Google Scholar]
- 24.Lee CC, Yamamoto S, Morimura K. Significance of cyclin D1 overexpression in transitional cell carcinomas of the urinary bladder and its correlation with histopathologic features. Cancer. 1997;79:780–789. doi: 10.1002/(sici)1097-0142(19970215)79:4<780::aid-cncr15>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 25.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24:2606–2611. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland JF. Cancer medicine. 4th ed. Baltimore: Williams & Wilkins; 1997. pp. 1655–1660. [Google Scholar]
- 27.Munger K. The role of human papillomaviruses in human cancers. Front Biosci. 2002;7:d641–d649. doi: 10.2741/a800. [DOI] [PubMed] [Google Scholar]