Abstract
Lipid transfer proteins are important in membrane vesicle biogenesis and trafficking, signal transduction and immunological presentation processes1–3. The conserved and ubiquitous mammalian glycolipid transfer proteins (GLTPs) serve as potential regulators of cell processes mediated by glycosphingolipids, ranging from differentiation and proliferation to invasive adhesion, neurodegeneration and apoptosis4,5. Here we report crystal structures of apo-GLTP (1.65 Å resolution) and lactosylceramide-bound (1.95 Å) GLTP, in which the bound glycosphingolipid is sandwiched, after adaptive recognition, within a previously unknown two-layer all-α-helical topology. Glycosphingolipid binding specificity is achieved through recognition and anchoring of the sugar-amide headgroup to the GLTP recognition centre by hydrogen bond networks and hydrophobic contacts, and encapsulation of both lipid chains, in a precisely oriented manner within a ‘moulded-to-fit’ hydrophobic tunnel. A cleft-like conformational gating mechanism, involving two interhelical loops and one α-helix of GLTP, could enable the glycolipid chains to enter and leave the tunnel in the membrane-associated state. Mutation and functional analyses of residues in the glycolipid recognition centre and within the hydrophobic tunnel support a framework for understanding how GLTPs acquire and release glycosphingolipids during lipid intermembrane transfer and presentation processes.
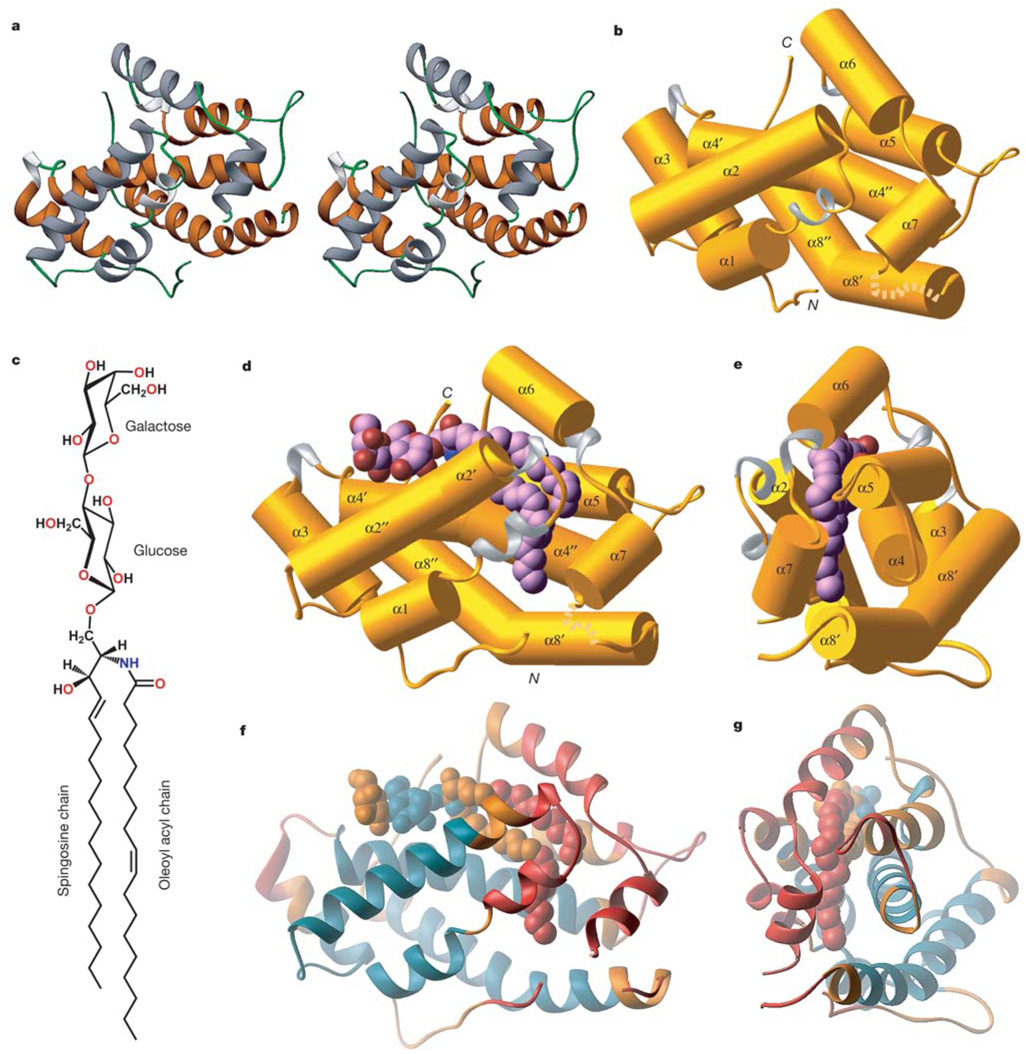
GLTPs are soluble proteins discovered initially in the membrane-free cytosolic extract of bovine spleen, and later in a wide variety of tissues, that selectively accelerate the transfer of glycolipids between membranes6–8. The 23–24-kDa GLTPs display absolute specificity for glycolipids9 and are highly conserved among mammals10, with some orthologues implicated in apoptosis11,12. Although specific folding motifs and lipid-binding domains have been characterized in other lipid transfer/binding proteins13–19, this is not true of GLTP. We have solved the structure of apo-GLTP crystal from human skin fibroblasts at 1.65 Å resolution and the corresponding lactosylceramide–GLTP complex co-crystal at 1.95 Å resolution (see Methods and Supplementary Table S1). The novel protein folding of apo-GLTP (Fig. 1a, b) is organized as a two-layer arrangement of α-helices, with helices 1, 2, 6 and 7 forming the front layer, and helices 3, 8, 4 and 5 belonging to the back layer (Supplementary Fig. S1), with two long α-helices, 8 and 4, coiling around each other to form a superhelix. Such a sandwich topology generates a lactosylceramide-binding site between the α-helical layers in the lactosylceramide–GLTP complex (Fig 1d, e; Supplementary Fig. S2a, b).
Figure 1.
Overall structures of apo-GLTP and the lactosylceramide–GLTP complex. a, Stereo view of the apo-protein in a ribbon representation. Four α-helices on the front face are coloured grey, and four other α-helices on the back face are coloured brown. b, A view of apo-GLTP: α-helices (coloured gold) are shown in a cylinder representation; 310 helices (coloured silver) and loop segments (coloured gold) are shown in a ribbon representation. c, Chemical formula of lactosylceramide. d, e, Two perpendicular views of the lactosylceramide–GLTP complex, with α-helices (coloured gold) in a cylinder representation, 310 helices (coloured silver) and loop segments (coloured gold) in a ribbon representation, and bound lactosylceramide in a space-filling representation. The bound glycolipid atoms are coloured lavender, red and blue for carbon, oxygen and nitrogen atoms, respectively. f, g, Views (corresponding to d and e) of the crystallographic B-factors for the lactosylceramide–GLTP complex. The B-factor scale is colour coded as follows: blue–green, B < 34; orange, 34 < B < 46; red, B < 46.
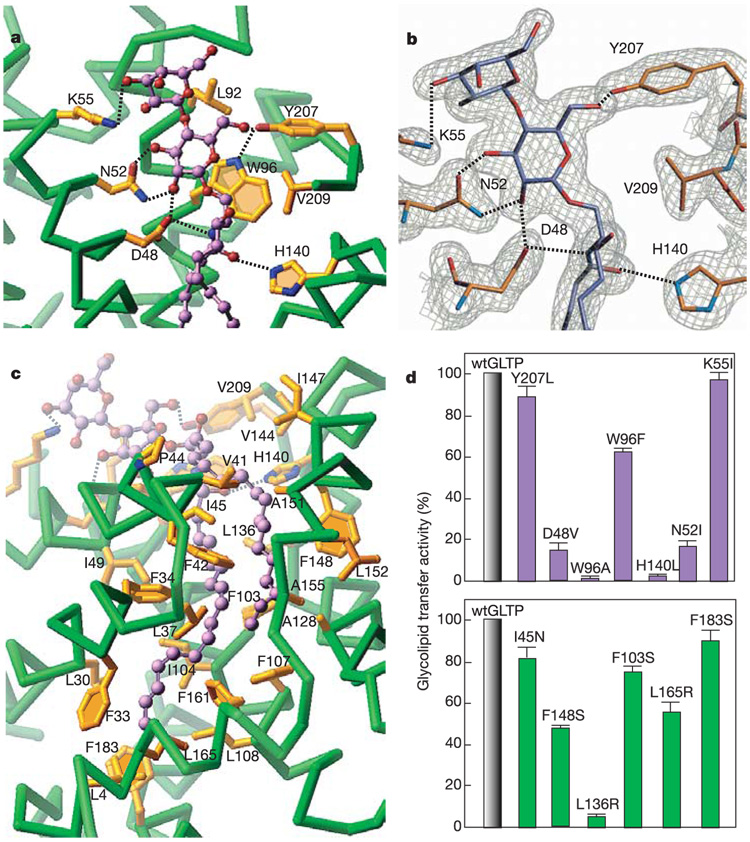
Lactosylceramide (Fig. 1c) binding to GLTP is achieved through recognition and anchoring of the hydrophilic headgroup to the protein surface, and through accommodation of the hydrophobic lipid chains within the interior of the protein. The lactosyl headgroup is anchored within the GLTP recognition centre, located on the protein surface, through a network of hydrogen-bonding (with Asp 48, Asn 52 and Lys 55 of α2, and Tyr 207 towards the carboxy terminus) and hydrophobic (Trp 96 of α4 stacks over the glucose ring, whereas Leu 92 contacts the galactose ring) interactions (Fig. 2a; Supplementary Fig. S3), analogous to carbohydrate–lectin interactions20,21. The ceramide amide group is oriented through a pair of hydrogen bonds (with negatively charged Asp 48 and positively charged His 140), whose alignment is facilitated by hydrophobic contacts (with Val 209) of the initial three-carbon ceramide segment (Fig. 2a; Supplementary Fig. S3). The importance of individual recognition events within the headgroup recognition centre was probed by point mutation studies, as monitored by glycolipid intervesicular transfer assays with radiolabelled glycolipid (Supplementary Fig. S4). The experimental data support the key role of Trp 96, His 140, Asp 48 and Asn 52 for recognition of the glycosyl and ceramide amide moieties in the complex. Mutations W96A and H140L resulted in almost complete inactivation (residual activity 1–3%), whereas mutations D48V and N52I resulted in significant inactivation (residual activity about 15%) (Fig. 2d, top). The extent of inactivation is different for the W96F mutation (residual activity 63%) relative to its W96A counterpart (residual activity 1%), highlighting the importance of aromatic ring stacking over the glucose ring to recognition (Fig. 2d, top). By contrast, Tyr 207 and Lys 55, which form single intermolecular hydrogen bonds, seem to be much less crucial for recognition, because the mutations Y207L and K55I retain practically full (90–97%) activity (Fig. 2d, top). We have verified the integrity of our GLTP headgroup mutants for unanticipated structural alterations. The crystal structure of the D48V mutant shows only local rearrangements (Supplementary Fig. S5), whereas circular dichroism (CD) spectra of other mutants indicate minimal perturbation of a-helical global folds (far-ultraviolet CD; Supplementary Fig. S6, top) and of aromatic amino acid environments (near-ultraviolet CD; Supplementary Fig. S7, top).
Figure 2.
Intermolecular interactions in the lactosylceramide–GLTP complex. a, The headgroup recognition centre residues interacting with the two sugars and the ceramide amide group of bound lactosylceramide. Hydrogen bonds are shown by dashed lines. The bound glycolipid atoms are coloured lavender, red and blue for carbon, oxygen and nitrogen atoms, respectively. The GLTP Cα backbone is coloured green, the side chains are shown in gold, and oxygen and nitrogen atoms are red and blue, respectively. b, Final 2Fo – Fc electron density map for the headgroup recognition centre with bound lactosylceramide. The hydrogen bonds are indicated by dashed lines. c, The hydrophobic tunnel residues surrounding the longer N-acyl and shorter sphingoid-base chains of bound lactosylceramide. d, Glycolipid transfer activities for GLTPs with point mutations within the headgroup recognition centre (top) and within the hydrophobic tunnel (bottom). Error bars show standard deviations. Additional details of radiolabelled glycolipid assay conditions are provided in Methods and in Supplementary Fig. S4.
When GLTP acquires lactosylceramide, both hydrocarbon chains of the ceramide moiety become buried within a single completely hydrophobic tunnel, lined by the side chains of nonpolar phenylalanine, leucine, isoleucine and alanine residues, together with a few valine and proline residues (Fig. 2c). Residual activities ranging from 50% to 90% were observed when individual phenylalanines were replaced by serines at positions 103, 148 and 183 (Fig. 2d, bottom), indicative of a possible additive role in creating the favourable hydrophobic environment for the nonpolar chains. Significant retention of activity (82%) was also found for the I45N mutant (Fig. 2d, bottom), located near the entrance to the channel. Replacement of leucines by arginines affected activity differently depending on their location within the channel. Thus, the L165R, mutant located towards the end of the channel, retained 54% of activity, whereas L136R, located in the interior of the channel, retained only 5% of activity (Fig. 2d, bottom). We have verified the integrity of the GLTP channel mutants by recording the far-ultraviolet and near-ultraviolet CD spectra (Supplementary Fig S6 and Supplementary Fig S7, bottom panels).
Neither the hydrophobic tunnel nor lipid chains (a Δ9,10 cis-double bond occurs halfway down the N-acyl chain) are straight in the complex (Fig 1d, e and Fig 2c). The N-acyl chain extends deeper into the hydrophobic tunnel because it is longer than the adjacent sphingoid-base chain (Fig. 1c, d). The terminal segments of both chains become nearly orthogonal with respect to the long axis of the sphingoid base (Fig 1d and Fig 2c). Our measured solvent-accessible volume of about 315 Å3 for the lipid-binding tunnel in the complex is much smaller than reported values for other lipid-binding pockets19, indicative of a very tight fit between the inserted portion of the lipid chains, with an estimated volume of about 330 Å3, and the walls of the tunnel. The measured solvent-accessible volume is further reduced to 170 Å3 in apo-GLTP, so that this residual space can no longer accommodate lipid chains.
Our structure of the lactosylceramide–GLTP complex (1.95 Å) can be compared with the structures of the ganglioside GM2–CD1b (2.8 Å)16 and sulphated-galactosylceramide–CD1a (2.15 Å)17 complexes. The recognition specificity associated with the network of hydrogen-bonding interactions involving the glycosyl and ceramide moieties distinguishes the GLTP complex (Fig. 2a) from the corresponding CD1a complex, which involved far fewer hydrogen-bonding recognition contacts17. Further, whereas the dual-chain lipid inserts into a common hydrophobic tunnel in the GLTP complex (Fig. 2c), the sphingoid-base and N-acyl chains insert into separate interconnecting hydrophobic pockets in the CD1a complex17. It therefore seems that unique glycolipid–protein interactions are associated with distinct functional events, namely glycolipid transfer in the GLTP complex in contrast to glycolipid presentation in the CD1b16 and CD1a17 major histocompatibility complexes.
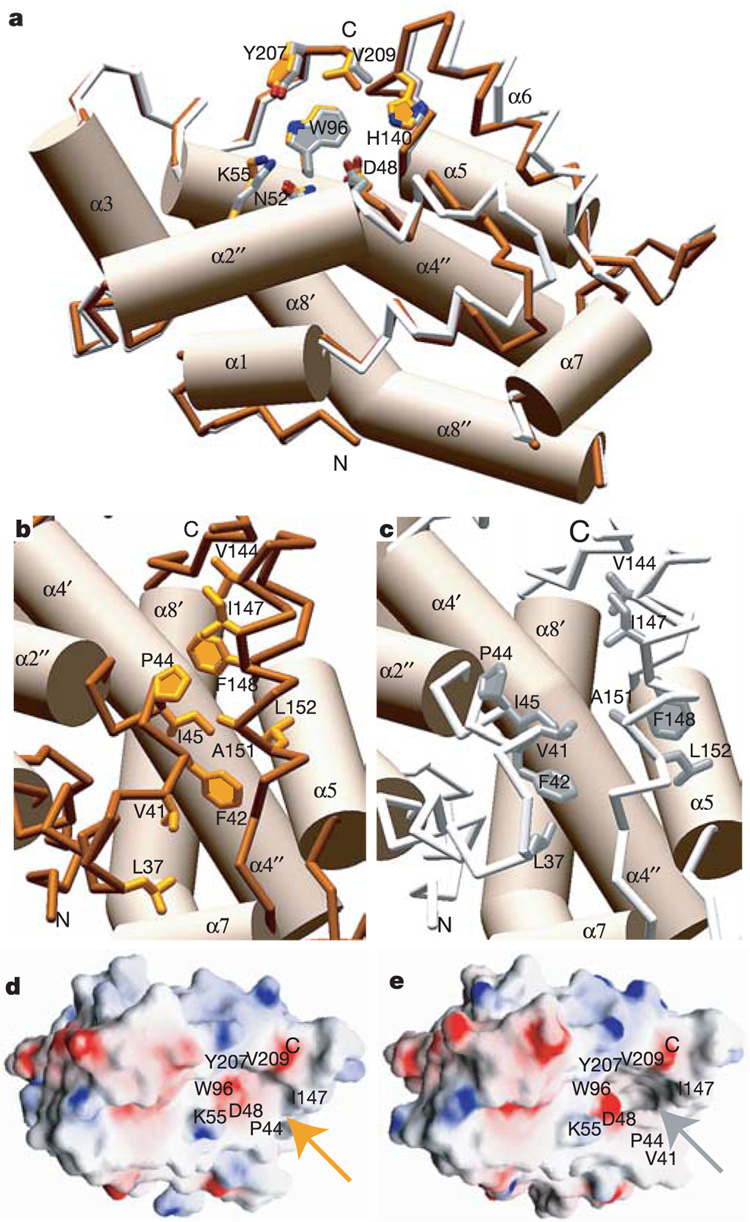
Superposition of the apo-GLTP and lactosylceramide-bound GLTP structures reveals essentially no difference in the headgroup recognition centre but provides clear evidence of localized conformational differences related to the hydrophobic channel (Fig. 3a). The differences, positioned along the front layer of GLTP, are associated with interhelical loops α1–α2 and α6–α7, helix α6 and the amino terminus of helix α2 (Fig. 3b, c). The conformational consequences of glycolipid acquisition are: first, bending of the α2-helix; second, rearrangement of the α1–α2 loop, accompanied by the appearance of a new 310 helix near the N terminus of the α2 helix; third, outward displacement of the α6 helix by 2.7 Å; and fourth, shortening of the C terminus of the α6 helix that is compensated for by formation of a new 310 helix (Fig 1b, d and Fig 3a; Supplementary Fig S1, Supplementary Fig S2c and Supplementary Fig S8c). Acquisition of glycolipid by GLTP results in amino acids Pro 44, Ile 45, Val 41, Phe 42, Leu 37 and Ile 147, Phe 148, Ala 151, Leu 152, associated with separate regions, moving farther apart with respect to each other (Fig. 3b, c), while the side chains of Phe 148 and Leu 152 rotate around their Cα–Cβ bonds to move from an inside-facing orientation in apo-GLTP (Fig. 3b) to a swung-out orientation in lactosylceramide-bound GLTP (Fig. 3c). The net result of all movements and rearrangements is an opening and expansion of a hydrophobic tunnel (Fig. 3d, e) when GLTP acquires the lactosylceramide molecule from the membrane.
Figure 3.
Comparison of apo-GLTP and lactosylceramide-bound GLTP. a, Superposition of apo-GLTP and lactosylceramide-bound GLTP structures, with similarly positioned α-helices shown in a cylinder representation, while 310 helices and loop segments are coloured orange for free GLTP and white for lactosylceramide-bound GLTP. Note the similarity of the glycosyl recognition centre conformations (top centre) and differences in the structure of the α1–α2 loop, and in mutual positioning of the α6-helix and the N terminus of the α2-helix. b, c, Detailed views of the positioning of the backbone and side chains between the α1–α2 loop and the α6-helix in apo-GLTP (b) and in the lactosylceramide–GLTP complex (c). Note the conformational rotation of Phe 148 (F148) and Leu 152 (L152) from an inwardly pointing orientation in b to a swung-out orientation in c. d, e, Electrostatics surface of apo-GLTP (d) and lactosylceramide-bound GLTP (e). The channel (indicated by a grey arrow) leading from the glycolipid recognition centre is open in the lactosylceramide–GLTP complex (e), but is closed (indicated by a gold arrow) in apo-GLTP (d). Amino acids lining the entrance to the channel are labelled.
It is difficult to imagine penetration of the lipid chains into the protein interior via an ‘end-on digging or tunnelling mechanism’ (pushing the hydrophobic walls apart to get the terminal methyl groups buried deeper and deeper). Rather, the GLTP structure most probably provides a cleft-like gate, which could open and close to let the lipid chains in and out. Indeed, the crystallographic B-factor distribution reveals local conformational mobility within the lactosylceramide-bound GLTP (Fig. 1f, g; highest B-factors in red). A comparative analysis for apo-GLTP and lactosylceramide-bound GLTP structures suggests that the loops α1–α2 and α5–α6, helix 6 and possibly helix 7 could form the lipid ‘gate’ in GLTP. The proper orientation of the glycolipid recognition centre and the ‘gate’ region with respect to the membrane surface might be aided by Trp 142 as well as Tyr 81, Tyr 153, Tyr 157 and Tyr 207, nearby residues known to interact favourably with membrane interfaces22,23. The GLTP-membrane interface could potentially provide a favourable environment for protein conformational changes that enhance the binding and desorption of glycolipid amphiphiles from the membrane into GLTP.
To our knowledge, the results on apo-GLTP and lactosylceramide-bound GLTP are the first demonstration of a two-layered α-helical motif functioning to accommodate a glycolipid by the creation and expansion of a ‘moulded-to-fit’ hydrophobic tunnel (Fig. 3e) that does not pre-exist in the free protein (Fig. 3d). This architecture differs from all previously reported motifs of lipid-binding and lipid-transfer proteins, which are either α-helical with extensive disulphide crosslinking or contain extensive β-structure. Thus, GLTP represents a new emerging family of sphingolipid-binding/transfer proteins. Our results provide a potential framework for understanding how GLTPs, with their structurally conservative and conformationally flexible segments, acquire and release membrane glycolipids during lipid transport and presentation processes.
Methods
Plasmid construction and mutagenesis
The open reading frame encoding human GLTP (NCBI GenBank accession no. AF209704) was subcloned into the pET-30 Xa/LIC expression vector (Novagen) by ligation-independent cloning, resulting in N-terminal His tags and S-tags and a factor Xa cleavage site. Site-directed mutations were obtained with a QuickChange site-directed mutagenesis kit (Stratagene).
Protein expression and purification
Transformed BL21 (DE3) cells (Novagen) were grown in Luria–Bertani medium at 37 °C, induced with 0.1 mM isopropyl β-d-thiogalactoside and grown for a further 16–20 h at 15 °C. Purification of recombinant GLTP (rGLTP) from soluble lysate protein was accomplished by Ni-affinity chromatography. The His-tag and S-tag sequences were removed from rGLTP by using factor Xa. rGLTP was then purified by fast protein liquid chromatography size-exclusion chromatography. Selenomethionine (Se-Met)-labelled GLTP, incorporated by inhibiting the methionine biosynthesis pathway24, retained full activity. Point mutants of the liganding site (Fig. 2d) had similar high yields and solubilities to those of the wild-type protein, consistent with folding to native-like conformations. In contrast, point mutations of essential residues in connecting regions between helices and far removed from the liganding site (for example W85-cis-P86 (Supplementary Fig. S8b), W85R), resulted in inactive protein, characterized by poor yield and very low solubility during expression and consistent with global folding changes.
Activity of rGLTP
The glycolipid transfer activity of GLTP was measured by using established radiolabelled assays. Negatively charged donor vesicles containing tritiated galactosylceramide (2 mol%) were incubated with GLTP (1 µg) and a tenfold excess of neutral acceptor vesicles at 37 °C. Quantification of glycolipid transfer was achieved by liquid scintillation counting of acceptor vesicles recovered from DEAE minicolumns8,12.
Crystallization and X-ray data collection
Crystals of GLTP were grown by the hanging-drop vapour-diffusion method. Protein samples (7–15 mg ml−1) were mixed with equal amount of well solution (15–20% (w/v) PEG 3350, 50 mM KH2PO4 pH 4.5). For Se-Met–GLTP, 10 mM dithiothreitol was included in crystallization reagents. Single crystals appeared in 2–5 days at room temperature. GLTP–glycolipid crystal complexes were obtained by co-crystallization of protein with lactosylceramide (0.4 mM in 50% ethanol). The crystals appeared in 2–3 months. Lactosylceramide, which contained homogeneous oleoyl (C18:1) acyl chains, was synthesized and purified as described previously25. Crystals were transferred into well solution including 20% glycerol as cryoprotectant, then mounted in a fibre loop and flash-frozen in a cold nitrogen stream for X-ray data collection.
Structure determination and refinement
Crystallization of apo-GLTP and its selenomethionine derivative was achieved in space group P21 with similar unit cell parameters and with one protein molecule in the asymmetric unit (Supplementary Table S1). The crystal structure of GLTP was determined by the multiwavelength anomalous dispersion method26. Five of the six selenium sites were identified and refined by the SOLVE/RESOLVE package27. The resulting electron density map was calculated after automatic maximum-likelihood density modification in SOLVE/RESOLVE. On the basis of this map, most of the protein chain was traced with the ARP/wARP program28. The protein model was finally built through several cycles of manual model building with TURBO. The model was refined in REFMAC29 at 1.65 Å resolution against the native data set to a final R-factor/R-free of 0.171/0.224 (Supplementary Table S1). The automatic procedure ARP/wARP was used to add solvent molecules to a final model. Two examples of the electron density map (Supplementary Fig. S8a, b) indicate the high quality of the final model. The distribution of secondary structure elements along the GLTP amino-acid sequence is shown in Supplementary Fig. S8c. The final model contains two disordered regions (1–7 and 168–171). Database searches with the SCOP (http://scop.mrclmb.cam.ac.uk/scop) and DALI (http://www.ebi.ac.uk/dali) programs revealed no structural similarity between the helical sandwich topology of apo-GLTP and deposited protein topologies.
Co-crystallization of GLTP with lactosylceramide was achieved in space group C2 with one molecule in the asymmetric unit (Supplementary Table S1). The apo-GLTP structure was used as a search model in the molecular replacement method with AMoRe30. A molecular replacement electron density map calculated at 1.95 Å resolution indicated the location of protein regions involved in conformational rearrangements. The amino-acid residues that belonged to these regions were removed from the initial model and retraced de novo with ARP/wARP. The rebuilt model was then refined with REFMAC. The difference electron density map calculated on the basis of the refined protein model clearly exhibited extra density, which was manually interpreted as a bound lactosylceramide molecule. Solvent molecules were added to the model by the automatic procedure ARP/wARP. The model was refined in REFMAC at 1.95 Å resolution to a final R-factor/R-free of 0.192/0.244. Figure 2b shows the headgroup and its interactions for a bound lactosylceramide molecule in the final 2Fo – Fc electron density map, and Supplementary Fig. S9 displays the omit 2Fo – Fc map for a ligand. The distribution of the secondary structure elements along the amino acid sequence (Supplementary Fig. S2c) shows distinctions from those observed for apo-GLTP (Supplementary Fig. S8c). The final model contains two disordered regions (1–3 and 168). The apo-GLTP and lactosylceramide-bound GLTP crystals contain a small piece of unknown hydrocarbon, acquired during the expression of protein in Escherichia coli, an unsurprising situation for glycosphingolipid-binding proteins19. The CASTp program (http://cast.engr.uic.edu) was used to estimate solvent-accessible volumes of cavities in apo-GLTP and lactosylceramide-bound GLTP.
Supplementary Material
Acknowledgements
We thank the personnel at SBC beamline 19BM of the Advanced Photon Source beamline staff for assistance with data collection from multiwavelength anomalous dispersion; A. Serganov for technical support; X. Lin, T. Chung and H. Pike for their contributions to the cloning and expression of the recombinant human GLTP; X.-M. Li for synthesizing and purifying N-18:1 lactosylceramide; A. J. Windebank for help with DNA sequencing at the Mayo Molecular Biology Core Facility; T. Burghardt for help with recording near-ultraviolet CD spectra; and S. Venyaminov in the Franklyn Prendergast laboratory for recording the far-ultraviolet CD spectra. This research was supported by NIH and the Hormel Foundation. Use of the ANL SBC beamlines at the APS was supported by the US Department of Energy, Office of Energy Research.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
Coordinates have been deposited in the Protein Data Bank under accession codes 1SWX for apo-GLTP and 1SX6 for lactosylceramide-bound GLTP.
References
- 1.Rogers DP, Bankaitis VA. Phospholipid transfer proteins and physiological functions. Int. Rev. Cytol. 2000;197:35–81. doi: 10.1016/s0074-7696(00)97002-5. [DOI] [PubMed] [Google Scholar]
- 2.Wirtz KWA. Phospholipid transfer proteins revisited. Biochem. J. 1997;324:353–360. doi: 10.1042/bj3240353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakomori S. The glycosynapse. Proc. Natl Acad. Sci. USA. 2002;99:225–232. doi: 10.1073/pnas.012540899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwek RA, Butters TD, Platt FM, Zitzmann N. Targeting glycosylation as a therapeutic approach. Nature Rev. Drug Discov. 2002;1:65–75. doi: 10.1038/nrd708. [DOI] [PubMed] [Google Scholar]
- 6.Metz RJ, Radin NS. Purification and properties of a cerebroside transfer protein. J. Biol. Chem. 1982;257:12901–12907. [PubMed] [Google Scholar]
- 7.Abe A, Sasaki T. Purification and some properties of the glycolipid transfer protein from pig brain. J. Biol. Chem. 1985;260:11231–11239. [PubMed] [Google Scholar]
- 8.Brown RE, Jarvis KL, Hyland KJ. Purification and characterization of glycolipid transfer protein from bovine brain. Biochim. Biophys. Acta. 1990;1044:77–83. doi: 10.1016/0005-2760(90)90221-i. [DOI] [PubMed] [Google Scholar]
- 9.Yamada K, Abe A, Sasaki T. Specificity of the glycolipid transfer protein from pig brain. J. Biol. Chem. 1985;260:4615–4621. [PubMed] [Google Scholar]
- 10.Lin X, Mattjus P, Pike HM, Windebank AJ, Brown RE. Cloning and expression of glycolipid transfer protein from bovine and porcine brain. J. Biol. Chem. 2000;275:5104–5110. doi: 10.1074/jbc.275.7.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodersen P, et al. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattjus P, Turcq B, Pike HM, Molotkovsky JG, Brown RE. Glycolipid intermembrane transfer is accelerated by HET-C2, a filamentous fungus gene product involved in the cell–cell incompatibility response. Biochemistry. 2003;42:535–542. doi: 10.1021/bi026896x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nature Struct. Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 14.Roderick SL, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nature Struct. Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 15.Min KC, Kovall RA, Hendrickson WA. Crystal structure of α-tocopherol transfer protein bound to it ligand: implications for ataxia with vitamin E deficiency. Proc. Natl Acad. Sci. USA. 2003;100:14713–14718. doi: 10.1073/pnas.2136684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 Å, a maze of alkyl chains. Nature Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 17.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nature Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 18.Mahfoud R, et al. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and HIV-1 proteins. J. Biol. Chem. 2002;277:11292–11296. doi: 10.1074/jbc.M111679200. [DOI] [PubMed] [Google Scholar]
- 19.Schubert Wright C, Zhao Q, Rastinejad F. Structural analysis of lipid complexes of GM2-activator protein. J. Mol. Biol. 2003;331:951–964. doi: 10.1016/s0022-2836(03)00794-0. [DOI] [PubMed] [Google Scholar]
- 20.Weis WI, Drickamer K. Structural basis of lectin–carbohydrate interactions. Annu. Rev. Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg H, Mitchell DA, Drickamer K, Weis WI. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 22.Wimley WW, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 23.Killian JA, von Heijne G. How proteins adapt to a membrane–water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 24.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 25.Li X-M, Momsen MM, Brockman HL, Brown RE. Lactosylceramide: effect of acyl chain structure on phase behavior and molecular packing. Biophys. J. 2002;83:1535–1546. doi: 10.1016/S0006-3495(02)73923-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. D. 1993;49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1997;50:157–163. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.