Abstract
Recent studies have demonstrated the importance of recipient HLA-DRB1 allele disparity in the development of acute graft-versus-host disease (GVHD) after unrelated donor marrow transplantation. The role of HLA-DQB1 allele disparity in this clinical setting is unknown. To elucidate the biological importance of HLA-DQB1, we conducted a retrospective analysis of 449 HLA-A, -B, and -DR serologically matched unrelated donor transplants. Molecular typing of HLA-DRB1 and HLA-DQB1 alleles revealed 335 DRB1 and DQB1 matched pairs; 41 DRB1 matched and DQB1 mismatched pairs; 48 DRB1 mismatched and DQB1 matched pairs; and 25 DRB1 and DQB1 mismatched pairs. The conditional probabilities of grades III-IV acute GVHD were 0.42, 0.61, 0.55, and 0.71, respectively. The relative risk of acute GVHD associated with a single locus HLA-DQB1 mismatch was 1.8 (1.1, 2.7; P = 0.01), and the risk associated with any HLA-DQB1 and/or HLA-DRB1 mismatch was 1.6 (1.2, 2.2; P = 0.003). These results provide evidence that HLA-DQ is a transplant antigen and suggest that evaluation of both HLA-DQB1 and HLA-DRB1 is necessary in selecting potential donors.
Keywords: HLA-DQB1 alleles, acute graft-versus-host disease, unrelated donor marrow transplantation
The human major histocompatibility complex is encoded on the short arm of chromosome 6 in the distal portion of the 6p21.3 band and consists of the class I (HLA-A, -B, -C) and class II (HLA-DR, -DQ, -DP) genes. Much attention has been paid to this region because of the importance of the class I and class II genes in the immune response. HLA molecules play a fundamental role by presenting antigen to T cells (1). The role of diversity in the HLA system is of considerable interest in clinical transplantation because HLA molecules are able to elicit strong humoral and cellular alloimmune responses (2–8).
In unrelated marrow transplantation, donor selection has been historically based on serological typing for HLA-A, HLA-B, and HLA-DR antigens. Recently, molecular genetic analysis has disclosed extensive polymorphism among the serologically defined HLA-A, HLA-B, and HLA-DR antigens (9–12). Thus individual serologically defined antigens each comprise a family of alleles, and unrelated donors who are HLA-DR serologically matched with the recipient may have HLA-DRB1 allele disparity as detected by DNA-based methods (8).
HLA-DRB1 allele mismatching between the donor and recipient is associated with an increased risk of acute graft-versus-host disease (GVHD) after unrelated marrow transplantation (8). Additional undetected allele mismatching within the class II region can occur at the HLA-DQB1 locus (13) in HLA-DRB1 matched or mismatched unrelated transplant pairs. The clinical importance of the HLA-DQ locus in unrelated transplantation has not been elucidated, in part because investigators have assumed that matching for HLA-DRB1 would obviate the need to match for HLA-DQB1 (14–16).
The HLA-DQ locus maps 110 kb centromeric to the HLA-DR locus and is comprised of a polymorphic HLA-DQB1 gene encoding at least 22 alleles and a HLA-DQA1 gene encoding at least 12 alleles (17). HLA-DQB1 and HLA-DQA1 gene products define the serological antigen families designated DQw1, DQw2, DQw3, and DQw4 (18, 19). Linkage disequilibrium of HLA-DQA1, DQB1, and DRB1 alleles in defined populations produce predictable extended haplotypes. For Caucasian populations, most HLA-A, -B, and -DRB1 matched donor-recipient transplant pairs are also HLA-DQB1 and HLA-DQA1 allele matched (20, 21), and thus the analysis of transplant cases with isolated HLA-DQB1 allele disparity was not possible until a large clinical experience had accumulated. With broadened racial diversity among volunteer donor registries worldwide, unusual HLA-DR/DQ associations are likely to become more frequent among HLA-A, -B, and -DRB1 matched pairs.
We hypothesized that undetected HLA-DQB1 disparity could increase the risk of acute GVHD independently of HLA-DRB1 disparity in patients undergoing HLA-A, -B, and -DR matched unrelated marrow transplantation. The current analysis has dissected the roles of HLA-DQB1 and HLA-DRB1 disparity in contributing to the risk of acute GVHD. HLA-DQB1 disparity was found in a significant number of HLA-A, -B, and -DRB1 matched transplant pairs, and matching for both HLA-DRB1 and HLA-DQB1 alleles was associated with a substantial reduction in the risk of acute GVHD. These results provide evidence that HLA-DQ functions as a transplantation antigen.
MATERIALS AND METHODS
Study Population.
Patients who had hematologic malignancies and received unmanipulated marrow from HLA-A, -B, or -DR serologically matched donors between May 1985 and June 1995 were included in this analysis. In the interest of decreasing heterogeneity with respect to other acute GVHD risk factors, we further restricted the study to patients who received cyclophosphamide with 1200–1440 cGy of total body irradiation (TBI) in the conditioning regimen, and cyclosporine and methotrexate as GVHD prophylaxis (n = 449). All patients were included for analysis, regardless of the duration of posttransplant survival.
Before 1991, donor selection criteria included matching for HLA-A, -B, -DR, and -Dw with allowance for a single HLA-Dw mismatch (22) within serological HLA-DR matches for patients younger than 36 years of age if a HLA-Dw matched donor could not be identified. After 1991, HLA-DRB1 allele matching superseded Dw matching; a single HLA-DRB1 allele mismatch was permitted for patients younger than 36 years of age if a HLA-DRB1 matched donor could not be identified. Beginning in 1992, prospective HLA-DQB1 allele typing was performed for all patients and donors. Where possible, HLA-DQB1 matched donors were selected in preference to HLA-DQB1 mismatched donors.
Histocompatibility Testing.
All patients and donors were serologically typed for HLA-A and HLA-B antigens using the standard two-stage National Institutes of Health complement-dependent microcytotoxicity test. Serological typing for HLA-DR and HLA-DQ was performed using Dynabead-purified (Dynal, Great Neck, NY) B cells in a microcytotoxicity assay (8). Molecular typing of HLA-DRB1 and HLA-DQB1 alleles was performed as described (8, 23). In this analysis, HLA-DRB1 and HLA-DQB1 alleles were determined retrospectively for all individuals lacking allele-level resolution at the time of the original transplant. Mismatching was defined as the presence of patient HLA-DRB1 and HLA-DQB1 alleles not shared by the donor (GVHD vector) (23). Using this definition, 70 of the 73 HLA-DRB1 mismatches were also mismatched for the host-versus-graft (HVG) vector, and 3 were mismatched only for GVHD. One of the HLA-DRB1 matched pairs in group 1 was mismatched for HVG. Of the 66 HLA-DQB1 mismatches, 60 were also mismatched for HVG, and 6 were mismatched only for GVHD. Six HLA-DQB1 matched pairs (groups 1 and 3) were mismatched for HVG.
Transplant Procedure.
Cyclophosphamide (60 mg/kg of recipient body weight) was administered intravenously on each of two successive days followed by TBI administered as 6 fractions of 2.0 Gy, or 11 or 12 fractions of 1.2 Gy from dual opposed 60Co sources. T-cell-replete marrow was infused after the conditioning regimen was completed. GVHD prophylaxis consisted of standard methotrexate and cyclosporine (24). All protocols and consent forms were reviewed and approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center. The severity of acute GVHD was graded according to criteria previously described (25).
Statistical Methods.
The endpoint of this study was the development of clinically significant (grades III-IV) acute GVHD. Conditional probability estimates (26) were used to display the incidence of acute GVHD. These estimates depict the probability of developing acute GVHD by an indicated time after transplant given that recurrent malignancy, graft rejection, or death without acute GVHD have not occurred by that time.
The association between HLA-DQB1 mismatching and the risk of acute GVHD was evaluated in the context of multivariable Cox proportional hazards regression models (27) that stratified for patient age, transplant year, and, where relevant, HLA-DRB1 match/mismatch status. Stratification for patient age was motivated by imbalances for patient age among mismatched groups that resulted from the donor selection criteria. Covariables were included to adjust for potential confounding effects of patient, donor, or treatment characteristics associated with the risk of acute GVHD. Factors considered for inclusion in the base model were patient age, diagnosis and disease status at the time of transplant, transplant year, TBI exposure, marrow cell dose, patient/donor cytomegalovirus serologic tests, patient/donor gender mismatch, history of donor pregnancy, and compliance with methotrexate and cyclosporine GVHD prophylaxis regimens (administration of at least 80% of the recommended dose among patients at risk for acute GVHD). Those factors with the greatest potential for confounding by virtue of their contribution to the model and their imbalance with respect to mismatch groups were retained in the base model. The additional effect of HLA-DQB1 mismatching on the incidence of acute GVHD was then evaluated in the context of the base model.
RESULTS
HLA-DQB1 and HLA-DRB1 Allele Matching.
Four groups of donor–recipient pairs were defined among the 449 pairs evaluated: HLA-DRB1 and DQB1 matched (n = 335, group 1); HLA-DRB1 matched, HLA-DQB1 mismatched (n = 41, group 2); HLA-DRB1 mismatched, HLA-DQB1 matched (n = 48, group 3), and HLA-DRB1 and DQB1 mismatched (n = 25, group 4).
Among the 41 HLA-DQB1 mismatched pairs in group 2, the most prevalent HLA-DQB1 mismatches involved DQB1*0301 versus DQB1*0302 associated with DRB1*0401 (n = 11; 27%) and DQB1*0201 versus DQB1*0303 associated with DRB1*0701 (n = 10; 24%). Only 1 of the 41 pairs was mismatched for both HLA-DQB1 alleles. The double mismatch involved a DQB1*0501, 0604 African American donor and a DQB1*0605, 0605 African American recipient.
In group 3, 25 (52%) of the HLA-DRB1 mismatches involved alleles within the DRB1*11 family, all of which were matched for DQB1*0301; 18 (38%) of the HLA-DRB1 mismatches involved alleles within the DRB1*04 family, all of which were matched for DQB1*0302.
Demographics of the Study Population.
Several potential GVHD risk factors were unevenly distributed among groups 1–4 (Table 1). Because the criteria for donor selection allowed a single HLA-DRB1 mismatch only in patients younger than 36 years, the distribution of patient age in groups 3 and 4 was lower than in groups 1 and 2. A slightly higher proportion of transplants occurred between a male patient and male donor in group 3, and between a male patient and female donor in group 4. There was a higher frequency of female donor parity in group 3 and lower female donor parity in group 4.
Table 1.
Characteristics of the study population
| HLA-DR/DQ match group
|
||||
|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | |
| DRB1 matched | DRB1 matched | DRB1 mismatched | DRB1 mismatched | |
| DQB1 matched | DQB1 mismatched | DQB1 matched | DQB1 mismatched | |
| Number of pairs (%) | 335 (74) | 41 (9) | 48 (11) | 25 (6) |
| Transplant year, no. (%) | ||||
| 1987–1989 | 56 (17) | 9 (22) | 9 (19) | 0 |
| 1990–1992 | 157 (47) | 19 (46) | 22 (46) | 15 (60) |
| 1993–1995 | 121 (36) | 13 (32) | 17 (35) | 10 (40) |
| Mean patient age, no. (range) | 32 (0-55) | 31 (1-49) | 26 (1-50) | 24 (0-43) |
| 0–20 yr | 65 (19) | 8 (20) | 16 (33) | 8 (32) |
| 21–35 yr | 105 (31) | 16 (39) | 21 (44) | 13 (52) |
| >36 yr | 165 (49) | 17 (41) | 11 (23) | 4 (16) |
| Mean donor age, yr (range) | 37 (19-57) | 39 (22-53) | 39 (20-51) | 35 (18-48) |
| Patient/donor gender, no.(%) | ||||
| M/M | 120 (36) | 10 (24) | 20 (42) | 8 (32) |
| F/F | 61 (18) | 10 (24) | 8 (17) | 5 (20) |
| M/F | 70 (21) | 10 (24) | 10 (21) | 8 (32) |
| F/M | 84 (25) | 11 (27) | 10 (21) | 4 (16) |
| Female donor parity, no. (%) | ||||
| Yes | 85 (65) | 12 (60) | 13 (72) | 7 (54) |
| No | 40 (31) | 8 (40) | 4 (22) | 6 (46) |
| Unknown | 6 (5) | 0 | 1 (6) | 0 |
| Diagnosis, no. (%) | ||||
| Acute leukemia | ||||
| Rem | 43 (13) | 3 (7) | 10 (21) | 7 (28) |
| Rel | 60 (18) | 7 (17) | 12 (25) | 3 (12) |
| De novo | 1 (1) | 0 | 0 | 0 |
| Unknown | 1 (1) | 0 | 0 | 0 |
| Chronic leukemia | ||||
| CP | 138 (41) | 19 (46) | 12 (25) | 6 (24) |
| AP | 44 (13) | 5 (12) | 6 (13) | 0 |
| BC | 18 (5) | 3 (7) | 4 (8) | 3 (12) |
| BC/rem | 14 (4) | 1 (2) | 1 (2) | 2 (8) |
| Juv | 1 (1) | 0 | 2 (4) | 0 |
| MDS | 6 (2) | 3 (7) | 0 | 4 (16) |
| Lymphoma | ||||
| Rem | 1 (1) | 0 | 0 | 0 |
| Rel | 8 (2) | 0 | 1 (2) | 0 |
| TBI exposure, no. (%) | ||||
| 12 Gy | 133 (40) | 19 (46) | 9 (19) | 0 |
| 13.2 Gy | 151 (45) | 14 (34) | 26 (54) | 20 (80) |
| >13.2 Gy | 51 (15) | 8 (20) | 13 (27) | 5 (20) |
| Patient/donor CMV serostatus, no. (%) | ||||
| −/− | 127 (38) | 15 (37) | 14 (29) | 4 (16) |
| −/+ | 45 (13) | 6 (15) | 6 (13) | 6 (24) |
| +/− | 100 (30) | 11 (27) | 17 (35) | 9 (36) |
| +/+ | 63 (19) | 9 (22) | 11 (23) | 6 (24) |
| MTX compliance,* no. (%) | ||||
| Day 1 | 320/325 (98) | 39/41 (95) | 42/47 (89) | 23/24 (96) |
| Day 3 | 313/325 (96) | 40/41 (98) | 45/47 (96) | 23/24 (96) |
| Day 6 | 292/315 (93) | 38/40 (95) | 42/47 (89) | 23/24 (96) |
| Day 11 | 224/276 (81) | 27/32 (84) | 29/38 (76) | 17/20 (85) |
| CSP compliance,* no. (%) | ||||
| Week 1 | 277/326 (85) | 35/40 (88) | 41/47 (87) | 23/24 (96) |
| Week 2 | 215/274 (78) | 26/30 (87) | 27/36 (75) | 14/18 (78) |
| Week 3 | 145/218 (67) | 16/21 (76) | 17/30 (57) | 8/10 (80) |
| Week 4 | 130/200 (65) | 12/18 (67) | 18/26 (69) | 8/9 (89) |
| Week 5 | 114/194 (59) | 13/17 (76) | 15/24 (63) | 6/8 (75) |
Rem, remission; Rel, relapse; CP, chronic phase; AP, accelerated phase; BC, blast crisis; BC/rem, remission following initial BC; Juv, juvenile; MDS, myelodysplastic syndrome; MTX, methotrexate; CSP, cyclosporine; CMV, cytomegalovirus.
MTX and CSP compliance denotes the number of patients who received at least 80% of the recommended dose among patients at risk for acute GVHD (alive without acute GVHD, relapse, death, or second transplant).
More patients were transplanted in blast crisis or in remission following an initial blast crisis in group 4 than in any other group. This result likely reflects the urgency of transplantation in cases where malignancy could not be controlled by conventional treatment. Patients with more advanced disease and patients undergoing HLA-DRB1 mismatched transplants were prepared for transplantation by using higher TBI exposures. Consequently, patients in groups 3 and 4 received exposures of 13.2 Gy or higher more frequently than patients in groups 1 or 2.
Acute GVHD.
The conditional probabilities of grades III–IV acute GVHD were 0.42 (group 1), 0.61 (group 2), 0.55 (group 3), and 0.71 (group 4) (Fig. 1). Univariate analysis of the association of a HLA-DQB1 and/or HLA-DRB1 mismatch with the development of grades III-IV acute GVHD yielded a relative risk (RR) of 1.4 (0.91, 2.2; P = 0.12) for a single locus HLA-DRB1 mismatch; a RR of 1.7 (1.1, 2.6; P = 0.02) for a single locus HLA-DQB1 mismatch; and a RR of 1.9 (1.1, 3.3; P = 0.02) for both a HLA-DQB1 and HLA-DRB1 mismatch compared with HLA-DRB1 and HLA-DQB1 matched recipients.
Figure 1.
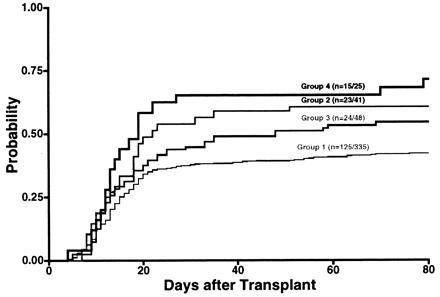
Conditional probability of grades III–IV acute GVHD was 0.42 for group 1 (DRB1 and DQB1 matched, n = 335), 0.61 for group 2 (DRB1 matched and DQB1 mismatched, n = 41), 0.55 for group 3 (DRB1 mismatched and DQB1 matched, n = 48), and 0.71 for group 4 (DRB1 and DQB1 mismatched, n = 25) by day 80 after transplantation. Data in parentheses denote the number of patients who developed grades III–IV acute GVHD.
The effect of HLA-DQB1 mismatching was further examined in the context of multivariable models which were constructed to avoid biases introduced by the possible confounding effects of other GVHD risk factors (Table 1; demographics of the study population). Of the four categories defined by gender mismatch and prior donor pregnancy, only the gender mismatched/parous donor category was distinguishable from the others with respect to acute GVHD risk (RR 1.7, P < 0.01). Therefore an indicator for this category was retained in the model. Likewise, TBI exposure greater than 1200 cGy was included in the models because of its relative strength of association with GVHD risk (RR 1.3; P = 0.1). Stratification for transplant year (1991–1992 versus other years) was motivated by the relatively high incidence of acute GVHD in these 2 years. The 1991–1992 transplant years represent a surrogate marker for as yet unidentified risk factors. Because recipient age was used to define permissible HLA-DRB1 mismatched donors, patient age was incorporated in the stratification (age < 36 years versus 36 or greater).
To determine whether HLA-DQB1 is an independent risk factor for GVHD, a model stratifying for HLA-DRB1 match status was examined (Table 2). The presence of HLA-DQB1 mismatching conferred a RR of 1.6 (1.1, 2.3; P = 0.02) for grades III–IV acute GVHD and a RR of 1.4 (1.0, 1.9; P = 0.02) for grades II–IV acute GVHD when assuming a common relative risk across HLA-DRB1 strata. In the second model, we examined the relative risk for each HLA-DRB1/DQB1 defined group (Table 3). A HLA-DQB1 mismatch conferred a RR of 1.8 (1.1, 2.8; P = 0.01) for grades III–IV acute GVHD and a RR of 1.5 (1.0, 2.1; P = 0.04) for grades II–IV acute GVHD when compared with a HLA-DQB1 and HLA-DRB1 match. The presence of both a HLA-DRB1 and HLA-DQB1 mismatch conferred a RR of 1.9 (1.0, 3.2; P = 0.03) for grades III–IV acute GVHD and a RR of 1.6 (1.0, 2.4; P = 0.05) for grades II–IV acute GVHD. These results indicate that HLA-DQB1 mismatching is an independent risk factor for acute GVHD.
Table 2.
Cox regression model of grades III–IV acute GVHD, stratifying on DRB1 match status
| Factor | RR | 95% CI | P value* |
|---|---|---|---|
| DQB1 mismatch† | 1.6 | 1.1, 2.3 | 0.02 |
| Parous female donor and male patient | 1.7 | 1.1, 2.4 | <0.01 |
| Dose of TBI >1200 cGy | 1.3 | 0.91, 1.7 | 0.17 |
Eight strata were modeled based on patient age (≥36 versus <36 years), transplant year (1991–1992 versus all other), and DRB1 match/mismatch status. CI, confidence interval.
P values correspond to the test of the null hypothesis that the RR = 1.0.
Likelihood ratio test P value for the contribution of DQB1 mismatch on this model is 0.03.
Table 3.
Stratified Cox regression model of grades III–IV acute GVHD
| Factor | RR | 95% CI | P value* |
|---|---|---|---|
| HLA mismatch† | |||
| DQB1 only | 1.8 | 1.1, 2.8 | 0.01 |
| DRB1 only | 1.4 | 0.92, 2.3 | 0.11 |
| DRB1 and DQB1 | 1.9 | 1.0, 3.2 | 0.03 |
| Parous female donor and male patient | 1.7 | 1.1, 2.4 | <0.01 |
| Dose of TBI >1200 cGy | 1.3 | 0.92, 1.8 | 0.15 |
The model contained four strata based on patient age (≥36 versus <36 years) and transplant year (1991–1992 versus all other years). CI, confidence interval.
P values correspond to the test of the null hypothesis that the RR = 1.0.
Likelihood ratio test P value for the contribution of HLA mismatch to this model is 0.02.
The presence of any HLA-DRB1 and/or HLA-DQB1 mismatch (groups 2, 3, and 4 combined) was associated with a higher conditional probability of grades III–IV acute GVHD compared with no mismatch (Fig. 2). In a multivariable model analysis of the same variables included in Table 3, any HLA-DRB1 and/or HLA-DQB1 mismatch conferred a RR of 1.6 (1.2, 2.2; P = 0.003) as compared with complete matches.
Figure 2.
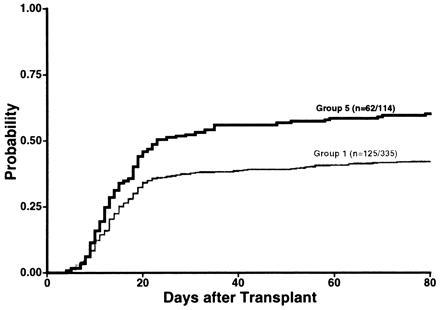
Conditional probability of grades III–IV acute GVHD in transplants mismatched for HLA-DRB1 and/or HLA-DQB1 (groups 2, 3, and 4 combined defined as group 5) was 0.60.
Tests for a difference in GVHD between groups 2 and 3 yielded P = 0.50. Tests for a difference between groups 4 and 2 yielded P = 0.89. Taken together, these results indicate that HLA-DRB1 and HLA-DQB1 mismatching each contribute to increased risk of GVHD, and the effect of HLA-DQB1 mismatching is at least as great as that contributed by HLA-DRB1.
DISCUSSION
In this study, we provide evidence that gene products of HLA-DQB1 function as transplantation antigens by influencing acute GVHD risk. The effect of HLA-DQB1 disparity appears to be at least as strong as that contributed by HLA-DRB1. Because 50% of the HLA-DQB1 allele mismatches were not detected by conventional serological reagents for DQw1-DQw4, the results demonstrate the importance of using molecular methods for accurate definition of HLA-DQB1 alleles. The improved outcome associated with donor–recipient HLA-DQB1 matching might also apply to solid organ transplantation (28–30).
The findings of a previous retrospective study of unrelated marrow transplant recipients demonstrated that mismatching for HLA-DRB1 alleles was associated with a significantly increased risk of grades III–IV acute GVHD (8). In that analysis, it was not possible to ascertain whether the increased risk of GVHD could be associated with cumulative effects of multiple disparities that were present in association with HLA-DRB1 disparity. This question has been answered in part by the current study which became feasible only after a significantly larger transplant experience allowed the HLA-DQB1 effect to be measured independently of HLA-DRB1. The matched group of transplants in the present study is restricted to pairs that are both HLA-DRB1 and HLA-DQB1 identical, and this definition is fundamentally an important distinction to be made from the “matched” group in the former study. When groups 3 and 4 from the current study were recombined (HLA-DRB1 mismatched), the RR estimate for a HLA-DRB1 mismatch was consistent with that obtained in the previous study (RR 1.5; 1.0, 2.1; P = 0.05).
Any comparison of HLA-DRB1 and HLA-DQB1 mismatching in contributing to the risk of GVHD must be interpreted cautiously. By virtue of our donor selection criteria, all donors were matched for serologically defined HLA-DR antigens (DR1-DR18) and only HLA-DRB1 mismatches corresponding to allele differences within antigen families were allowed. As a result, the composition of the HLA-DRB1 allele mismatches in our study was limited to allele pairs having a single amino acid difference (40%, of which the majority were valine/glycine mismatches at codon 86), two differences (42%), three differences (15%), or four differences (4%). In contrast, 50% of the HLA-DQB1 mismatches occurred between serologically definable DQw1-DQw4 antigen groups. The most frequent HLA-DQB1 mismatches involved 4 amino acid differences (DQB1*0301 versus 0302, 35%) or 15 differences (DQB1*0201 versus 0303, 28%). Hence, HLA-DR and HLA-DQ mismatches differed qualitatively for the number, the position and the nature of amino acid differences, making it difficult to compare the relative importance of matching at these loci.
Although most (75%) transplant pairs in this study were matched for both HLA-DRB1 and HLA-DQB1 alleles (group 1), the frequency of single locus mismatches (group 2, 9%; group 3, 11%) was higher than expected, and the frequency of two-locus mismatches (group 4, 6%) was lower than expected. The high frequency of single locus mismatches can be explained by the presence of specific HLA-DR antigens which are known to associate with more than one HLA-DQ antigen. These haplotypes have been extensively studied at both the serological and allele level in many populations and frequently involve DR4 and DR7 (14, 15, 20). In groups 1 and 2, patients who were DR4 or DR7-positive had a higher frequency of HLA-DQB1 mismatching (16%) than those who were DR4 or DR7-negative (6%). Because the proportion of DR4 and DR7-positive pairs in groups 1 and 2 were equivalent (29%) and the DR4 and DR7 haplotypes were similar, there is no a priori reason to believe that prospective attempts to match for HLA-DQB1 alleles in DR4 or DR7-positive individuals would not be successful. In any given patient, the likelihood of identifying a match would depend both on the HLA phenotype of the patient and on the composition of donor registries (31).
Strong positive linkage disequilibrium occurs not only between HLA-DRB1 and HLA-DQB1 but also between HLA-DQA1 and HLA-DQB1 (21), particularly on extended HLA haplotypes. Three HLA-DQB1 alleles are known to associate with more than one HLA-DQA1 allele (DQB1*0201 with DQA1*0201 or 0501; DQB1*0301 with DQA1*0301, 0501, or 0601; DQB1*0303 with DQA1*0301 or 0201). Each HLA-DQB1/DQA1 pairing, however, is strongly associated with a specific HLA-DRB1 haplotype. Because none of the study pairs encoded a major (DR1-DR18) disparity, the probability of HLA-DQA1 disparity among the 48 HLA-DRB1 mismatched, DQB1 matched (group 3) is likely to be quite low. Nevertheless, the HLA-DQα chain contributes epitopes important to the tertiary structure and peptide binding specificity of HLA-DQ molecules (32–36), and additional mismatching at HLA-DQA1 among HLA-DQB1 mismatched recipients could contribute to the overall effect of HLA-DQ on acute GVHD. Retrospective allele typing of HLA-DQA1 is in progress to determine the extent of HLA-DQA1 mismatching among our study pairs.
In conclusion, the results of our study provide evidence supporting the biological importance of HLA-DQB1 gene products in clinical transplantation. Mismatching for HLA-DQB1 alone was associated with a significantly increased risk of acute GVHD. The lowest incidence of acute GVHD was observed in patients with HLA-A, -B, -DRB1, and -DQB1 matched donors. Our data indicate that the results of unrelated donor marrow transplantation for the treatment of hematologic malignancy can be improved through more complete and precise matching of donors and recipients for both HLA-DQB1 and HLA-DRB1 alleles.
Acknowledgments
We thank Amy Mellon for data collection and Alison Sell for preparation of the manuscript. This work was supported by Grants AI33484 and CA18029 from the National Institutes of Health, a grant from the Friends of Allison Atlas Foundation, and a Special Fellowship from the Leukemia Society of America (E.W.P.).
Footnotes
Abbreviations: GVHD, graft-versus-host disease; TBI, total body irradiation; RR, relative risk.
References
- 1.Ploegh H L, Orr H T, Strominger J L. Cell. 1981;24:287–299. doi: 10.1016/0092-8674(81)90318-4. [DOI] [PubMed] [Google Scholar]
- 2.Hows J M, Yin J L, Marsh J, Swirsky D, Jones L, Apperley J F, James D C, Smithers S, Batchelor J R, Goldman J M, Gordon-Smith E C. Blood. 1986;68:1322–1328. [PubMed] [Google Scholar]
- 3.Anasetti C, Amos D, Beatty P G, Appelbaum F R, Bensinger W, et al. N Engl J Med. 1989;320:197–204. doi: 10.1056/NEJM198901263200401. [DOI] [PubMed] [Google Scholar]
- 4.Ash R C, Casper J T, Chitambar C R, Hansen R, Bunin N, Truitt R L, Lawton C, Murray K, Hunter J, Baxter-Lowe L A, Gottschall J L, Oldham K, Anderson T, Camitta B, Menitove J. N Engl J Med. 1990;322:485–494. doi: 10.1056/NEJM199002223220801. [DOI] [PubMed] [Google Scholar]
- 5.Weisdorf D, Hakke R, Blazar B, Miller W, McGlave P, Ramsay N, Kersey J, Filipovich A. Transplantation. 1991;51:1197–1203. doi: 10.1097/00007890-199106000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Beatty P G, Anasetti C, Hansen J A, Longton G M, Sanders J E, et al. Blood. 1993;81:249–253. [PubMed] [Google Scholar]
- 7.Kernan N A, Bartsch G, Ash R C, Beatty P G, Champlin R, Filipovich A, Gajewski J, Hansen J A, Henslee-Downey J, McCullough J, McGlave P, Perkins H A, Philips G L, Sanders J, Stroncek D, Thomas E D, Blume K G. N Engl J Med. 1993;328:593–602. doi: 10.1056/NEJM199303043280901. [DOI] [PubMed] [Google Scholar]
- 8.Petersdorf E W, Longton G M, Anasetti C, Martin P J, Mickelson E M, Smith A G, Hansen J A. Blood. 1995;86:1606–1613. [PubMed] [Google Scholar]
- 9.Saiki R K, Walsh P S, Levenson C H, Erlich H A. Proc Natl Acad Sci USA. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine J E, Yang S Y. Tissue Antigens. 1995;46:368–373. doi: 10.1111/j.1399-0039.1995.tb03128.x. [DOI] [PubMed] [Google Scholar]
- 11.Bunce M, O’Neill C M, Barnardo M C N M, Krausa P, Browning M J, Morris P J, Welsh K I. Tissue Antigens. 1996;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 12.Ng J, Hurley C K, Carter C, Baxter-Lowe L A, Bing D, Chopek M, Hegland J, Lee T D, Li T C, Hsu S, KuKuruga D, Mason J M, Monos D, Noreen H, Rosner G, Schmeckpeper B, Dupont B, Hartzman R J. Tissue Antigens. 1996;47:21–26. doi: 10.1111/j.1399-0039.1996.tb02510.x. [DOI] [PubMed] [Google Scholar]
- 13.Tiercy J M, Morel C, Freidel A C, Zwahlen F, Gebuhrer L, Betuel H, Jeannet M, Mach B. Proc Natl Acad Sci USA. 1991;88:7121–7125. doi: 10.1073/pnas.88.16.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Begovich A B, McClure G R, Suraj V C, Helmuth R C, Fildes N, Bugawan T L, Erlich H A, Klitz W. J Immunol. 1992;148:249–258. [PubMed] [Google Scholar]
- 15.Pera C, Delfino L, Angelini G, Longo A, Ferrara G B. Eur J Immunogenet. 1992;19:373–380. doi: 10.1111/j.1744-313x.1992.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 16.Termijtelen A, D’Amaro J, van Rood J J, Schreuder G M T. Tissue Antigens. 1995;46:387–390. doi: 10.1111/j.1399-0039.1995.tb03131.x. [DOI] [PubMed] [Google Scholar]
- 17.Bodmer J G, Marsh S G E, Albert E D, Bodmer W F, Bontrop R E, Charron D, Dupont B, Erlich H A, Mach B, Mayr W R, Parham P, Sasazuki T, Schreuder G M Th, Strominger J L, Svejgaard A, Terasaki P I. Tissue Antigens. 1995;46:1–18. doi: 10.1111/j.1399-0039.1995.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 18.van Rood J J, van Leeuwen A, Keuning J J, Termijtelen A. Scand J Immunol. 1977;6:373–384. doi: 10.1111/j.1365-3083.1977.tb02092.x. [DOI] [PubMed] [Google Scholar]
- 19.Duquesnoy R J, Marrari M, Annen K. Transplant Proc. 1979;11:1757–1760. [PubMed] [Google Scholar]
- 20.Imanishi T, Akaza T, Kimura A, Tokunaga K, Gojobori T. In: HLA 1991. Tsuji K, Aizawa M, Sasazuki T, editors. Oxford: Oxford Univ. Press; 1992. pp. 1174–1178. [Google Scholar]
- 21.Imanishi T, Akaza T, Kimura A, Tokunaga K, Gojobori T. In: HLA 1991. Tsuji K, Aizawa M, Sasazuki T, editors. Oxford: Oxford Univ. Press; 1992. pp. 1201–1204. [Google Scholar]
- 22.Dupont B, Braun D W, Yunis E J, Carpenter C B. In: Histocompatibility Testing 1980. Terasaki P I, editor. Los Angeles: Univ. of California Press; 1980. pp. 229–267. [Google Scholar]
- 23.Petersdorf E W, Smith A G, Mickelson E M, Longton G M, Anasetti C, Choo S Y, Martin P J, Hansen J A. Blood. 1993;81:1923–1932. [PubMed] [Google Scholar]
- 24.Storb R, Deeg H J, Whitehead J, Appelbaum F, Beatty P, et al. N Engl J Med. 1986;314:729–735. doi: 10.1056/NEJM198603203141201. [DOI] [PubMed] [Google Scholar]
- 25.Thomas E D, Storb R, Clift R, Fefer A, Johnson F L, Neiman P E, Lerner K G, Glucksberg H, Buckner C D. N Engl J Med. 1975;292:895–902. doi: 10.1056/NEJM197504242921706. [DOI] [PubMed] [Google Scholar]
- 26.Pepe M S, Mori M. Stat Med. 1993;12:737–751. doi: 10.1002/sim.4780120803. [DOI] [PubMed] [Google Scholar]
- 27.Kalbfleish J D, Prentice R L. In: The Statistical Analysis of Failure Time Data. Bradley R A, Hunter J S, Kendall D G, Watson G S, editors. New York: Wiley; 1980. pp. 70–118. [Google Scholar]
- 28.Duquesnoy R J, Annen K B, Marrari M M, Kauffman H M., Jr N Engl J Med. 1980;302:821–825. doi: 10.1056/NEJM198004103021501. [DOI] [PubMed] [Google Scholar]
- 29.Opelz G, Mytilineos J, Wujciak T, Schwarz V, Back D. Clin Invest. 1992;70:767–772. doi: 10.1007/BF00180746. [DOI] [PubMed] [Google Scholar]
- 30.Opelz G, Wujciak T. N Engl J Med. 1994;330:816–819. doi: 10.1056/NEJM199403243301203. [DOI] [PubMed] [Google Scholar]
- 31.Beatty P G, Mori M, Milford E. Transplantation. 1995;60:778–783. [PubMed] [Google Scholar]
- 32.Todd J A, Bell J I, McDevitt H O. Nature (London) 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 33.Horn G T, Bugawan T L, Long C M, Erlich H A. Proc Natl Acad Sci USA. 1988;85:6012–6016. doi: 10.1073/pnas.85.16.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sollid L M, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. J Exp Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scharf S J, Freidmann A, Steinman L, Brautbar C, Erlich H A. Proc Natl Acad Sci USA. 1989;86:6215–6219. doi: 10.1073/pnas.86.16.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok W W, Nepom G T, Raymond F C. J Immunol. 1995;155:2468–2476. [PubMed] [Google Scholar]


