Abstract
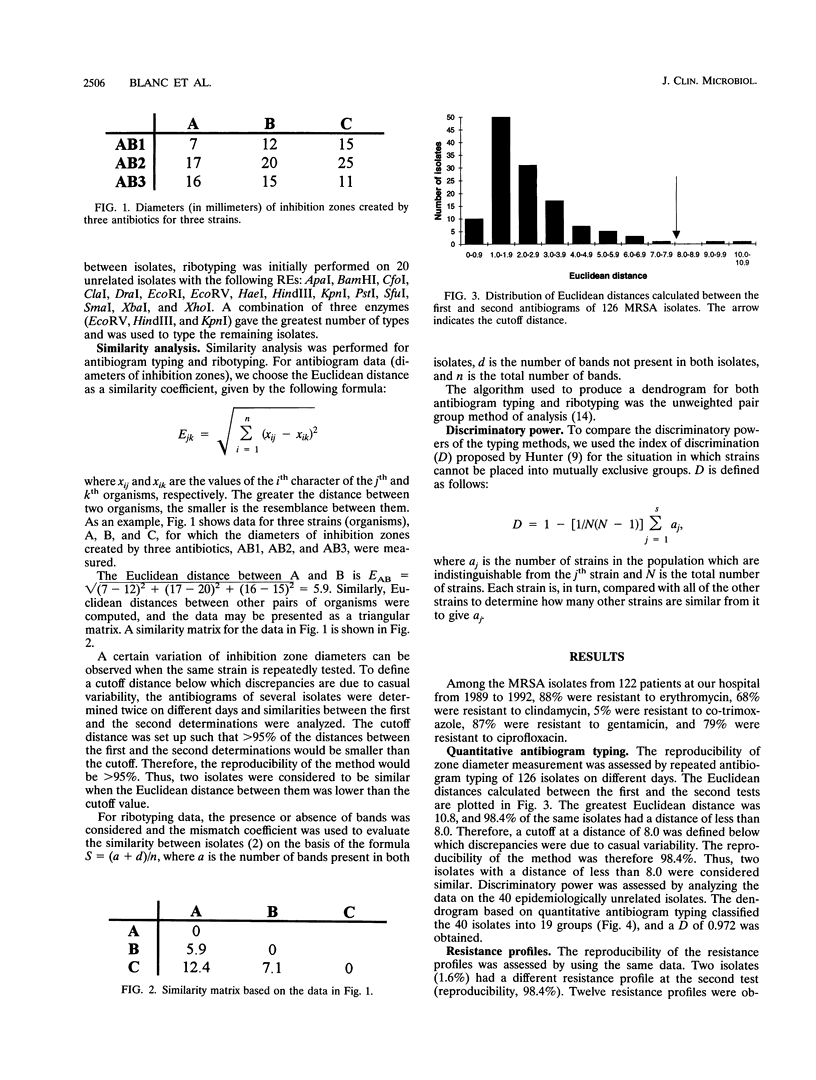
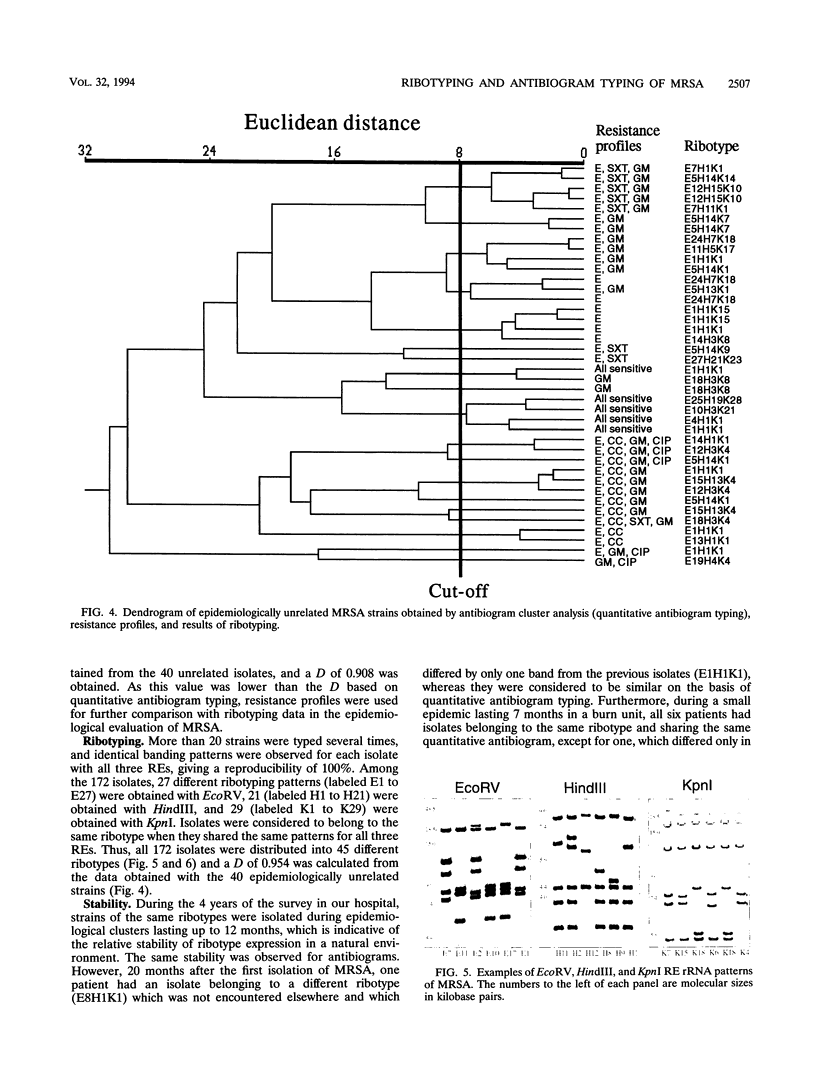
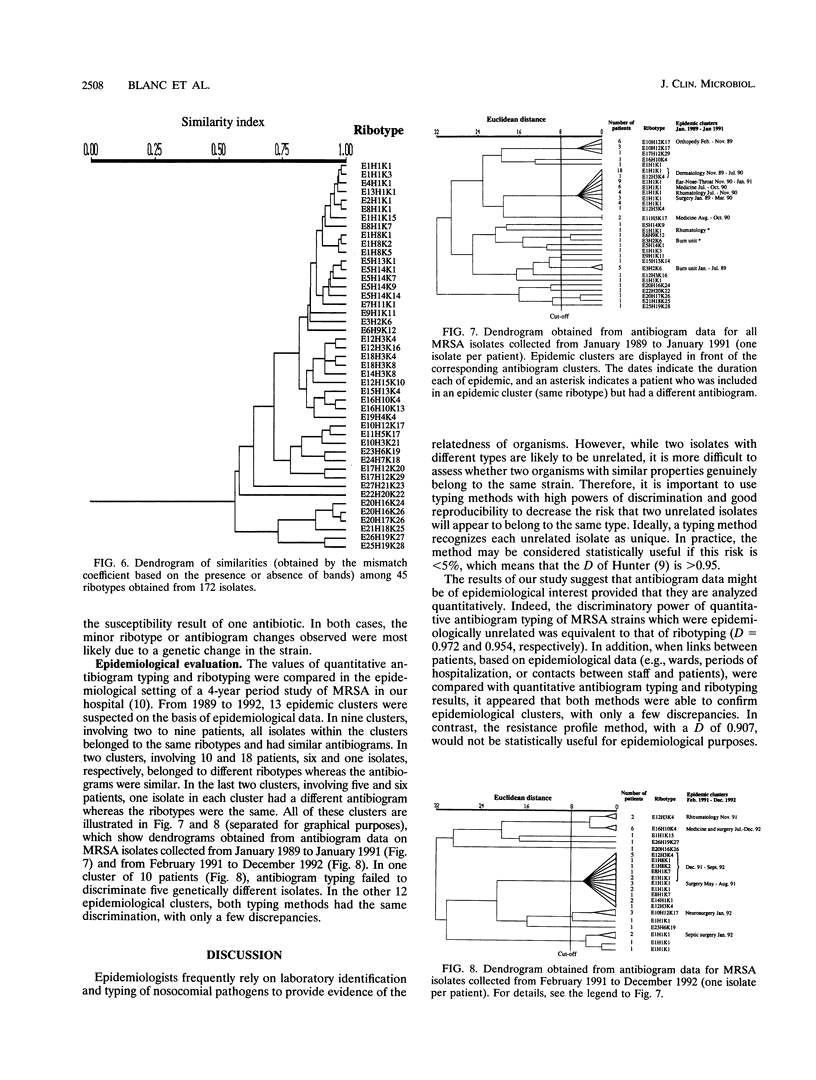
Antibiogram typing of methicillin-resistant Staphylococcus aureus with selected antibiotics was evaluated as a primary epidemiological typing tool and compared with ribotyping. Antibiograms were derived with the Kirby-Bauer disk diffusion method by using erythromycin, clindamycin, cotrimoxazole, gentamicin, and ciprofloxacin. For typing, antibiogram data were analyzed by similarity analysis of disk zone diameters (quantitative antibiogram typing). One hundred seventy-two isolates were typed. Reproducibility reached 98% for the quantitative antibiogram and 100% for ribotyping. With three selected restriction enzymes (EcoRV, HindIII, and KpnI), 40 epidemiologically unrelated isolates could be classified into 21 ribotypes, whereas quantitative antibiogram typing classified these isolates into 19 groups. To evaluate the discriminatory power of the methods, we calculated an index of discrimination from data obtained with these 40 isolates. This index takes into consideration both the number of types defined by the typing method and their relative frequencies. With both ribotyping and quantitative antibiogram typing, high discrimination indices (0.972 and 0.954, respectively) were obtained. When epidemiological links between patients (ward, period of hospitalization, and contacts between staff and patients) were compared with the results of ribotyping or the quantitative antibiogram typing method, it appeared that both methods were able to discriminate epidemiological clusters, with only a few discrepancies. In conclusion, quantitative antibiogram typing, although not necessarily based on genomic markers, is a simple method which enables a reliable workup of methicillin-resistant S. aureus epidemic when sophisticated molecular typing methods are not available.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altwegg M., Mayer L. W. Bacterial molecular epidemiology based on a nonradioactive probe complementary to ribosomal RNA. Res Microbiol. 1989 May-Jun;140(4-5):325–333. doi: 10.1016/0923-2508(89)90024-7. [DOI] [PubMed] [Google Scholar]
- Blanc D. S., Siegrist H. H., Sahli R., Francioli P. Ribotyping of Pseudomonas aeruginosa: discriminatory power and usefulness as a tool for epidemiological studies. J Clin Microbiol. 1993 Jan;31(1):71–77. doi: 10.1128/jcm.31.1.71-77.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H. M., Rimland D., Kiehlbauch J. A., Terry P. M., Wachsmuth I. K. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J Clin Microbiol. 1992 Feb;30(2):362–369. doi: 10.1128/jcm.30.2.362-369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Costas M., Cookson B. D., Talsania H. G., Owen R. J. Numerical analysis of electrophoretic protein patterns of methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1989 Nov;27(11):2574–2581. doi: 10.1128/jcm.27.11.2574-2581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M., Menzo S., Trojan S., Monti-Bragadin C. Cluster analysis of antibiotic susceptibility patterns of clinical isolates as a tool in nosocomial infection surveillance. Eur J Epidemiol. 1987 Jun;3(2):155–163. doi: 10.1007/BF00239753. [DOI] [PubMed] [Google Scholar]
- Grimont F., Chevrier D., Grimont P. A., Lefevre M., Guesdon J. L. Acetylaminofluorene-labelled ribosomal RNA for use in molecular epidemiology and taxonomy. Res Microbiol. 1989 Sep;140(7):447–454. doi: 10.1016/0923-2508(89)90065-x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Hunter P. R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990 Sep;28(9):1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolle L. E., Bialkowska-Hobrzanska H., Romance L., Harry V. S., Parker S. Clonal diversity of methicillin-resistant Staphylococcus aureus in an acute-care institution. Infect Control Hosp Epidemiol. 1992 Jan;13(1):33–37. doi: 10.1086/646420. [DOI] [PubMed] [Google Scholar]
- Preheim L., Pitcher D., Owen R., Cookson B. Typing of methicillin resistant and susceptible Staphylococcus aureus strains by ribosomal RNA gene restriction patterns using a biotinylated probe. Eur J Clin Microbiol Infect Dis. 1991 May;10(5):428–436. doi: 10.1007/BF01968023. [DOI] [PubMed] [Google Scholar]
- Trilla A., Nettleman M. D., Hollis R. J., Fredrickson M., Wenzel R. P., Pfaller M. A. Restriction endonuclease analysis of plasmid DNA from methicillin-resistant Staphylococcus aureus: clinical application over a three-year period. Infect Control Hosp Epidemiol. 1993 Jan;14(1):29–35. doi: 10.1086/646627. [DOI] [PubMed] [Google Scholar]
- Wei M. Q., Wang F. U., Grubb W. B. Use of contour-clamped homogeneous electric field (CHEF) electrophoresis to type methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1992 Mar;36(3):172–176. doi: 10.1099/00222615-36-3-172. [DOI] [PubMed] [Google Scholar]



