Abstract
The Epstein-Barr virus (EBV) is associated with lymphoid and epithelial cancers. Initial EBV infection alters lymphocyte gene expression, inducing cellular proliferation and differentiation as the virus transitions through consecutive latency transcription programs. Cellular microRNAs (miRNAs) are important regulators of signaling pathways and are implicated in carcinogenesis. The extent to which EBV exploits cellular miRNAs is unknown. Using micro-array analysis and quantitative PCR, we demonstrate differential expression of cellular miRNAs in type III versus type I EBV latency including elevated expression of miR-21, miR-23a, miR-24, miR-27a, miR-34a, miR-146a and b, and miR-155. In contrast, miR-28 expression was found to be lower in type III latency. The EBV-mediated regulation of cellular miRNAs may contribute to EBV signaling and associated cancers.
Keywords: Epstein-Barr virus, EBV, miRNA, microrna, latency, miR-21, miR-23a cluster, miR-23a, miR-24, miR-27a, miR-28, miR-34a, miR-146, miR-155
Introduction
The Epstein-Barr virus (EBV) has been implicated in a variety of lymphoid and epithelial cancers, including Burkitt’s lymphoma, Hodgkin’s disease, post-transplant lymphoproliferative disease, AIDS-associated immunoblastic lymphoma, and nasopharyngeal carcinoma. This ubiquitous human herpesvirus persists for the life of its host by establishing latent infection predominantly in resting memory B lymphocytes (Babcock et al., 1998; Miyashita et al., 1997). The process by which EBV establishes latency in memory B cells is not fully understood. In the model proposed by Thorley-Lawson and colleagues (Thorley-Lawson, 2005), EBV establishes infection in naïve B cells, and through successive viral transcription programs, drives the proliferation and differentiation of the cell into a memory B cell. During this differentiation process, EBV expresses proteins that mimic antigen activation of B cells and effectively bypasses the normal host signals that drive B cell differentiation. In a newly-infected naïve B cell, EBV initiates the growth program (also referred to as latency type III), and expresses a total of nine protein-coding genes (Kieff and Rickinson, 2007). These proteins – Epstein-Barr nuclear antigen (EBNA)-1, -2, -3a, -3b, -3c, and -LP, and latent membrane protein (LMP) -1, -2a, and -2b, – contribute the surrogate proliferation, migration, and survival signals that antigen-activated naïve B cells receive. Escape of growth program-associated lymphoblasts from normal immune surveillance in post-transplant and HIV-associated immune-suppressed individuals leads to lymphoproliferative diseases, demonstrating the oncogenic potential of this EBV transcriptional program. The oncogenic potential of the EBV growth/latency III program is also demonstrated by its expression in primary B cells transformed in vitro by infection with EBV, which generates long-lived replicating cell lines referred to as lymphoblastoid cell lines (LCLs).
According to Thorley-Lawson’s model, as the EBV-infected cell matures into a memory B cell, both the virus and the cell enters a quiescent state. The virus becomes transcriptionally silent, allowing it to escape immune recognition and preventing the subsequent immune-mediated destruction of the host cell (Hochberg and Thorley-Lawson, 2005). Homeostatic maintenance of the memory cell population drives expression of EBNA-1 alone (latency program, or latency type I) during cell replication in order to concurrently replicate the viral genome and appropriately segregate the viral episomes into the daughter cells (Hochberg et al., 2004; Yates, Warren, and Sugden, 1985).
Both antigen-driven B cell activation and EBV-driven activation are tightly regulated. Recently a new class of cellular regulatory elements, the small non-coding microRNAs (miRNAs), has been shown to play critical roles in a variety of cell signaling pathways. Through incorporation of the ~22 nucleotide single-stranded mature miRNA into the RNA-induced silencing complex (RISC) and subsequent imperfect base pairing within the 3′ untranslated region (UTR) of target messenger RNA transcripts, miRNAs suppress translation, thereby regulating protein levels (He and Hannon, 2004). This mechanism enables miRNAs to regulate processes such as growth, differentiation and apoptosis. Expression profiling of human miRNAs in different cellular contexts has demonstrated that distinct cell phenotypes have unique miRNA signatures. For instance, B lymphocyte populations can be distinguished based on miRNA expression profile: miR-7, miR-9, and miR-155 are upregulated in activated B cells, while miR-224 is upregulated and miR-181a is downregulated in memory B cells (Lawrie et al., 2008). Distinct miRNA expression patterns have also been shown to distinguish normal cells from tumor cells, and the constitutive upregulation of oncogenic miRNAs (oncomirs) contribute to the maintenance of the tumor phenotype (Calin and Croce, 2006). One such oncomir is miR-155, which causes tumors in miR-155-transgenic mice (Costinean et al., 2006) and is highly expressed in multiple types of B cell lymphoma (Eis et al., 2005; Kluiver et al., 2005; Metzler et al., 2004).
Viruses often exploit cellular pathways in order to promote the viral life cycle. We hypothesized that EBV actively regulates the expression of cellular miRNAs to promote a favorable host environment for the virus. Previous studies have shown a correlation between Epstein-Barr virus type III latency and expression of miR-155 (Jiang, Lee, and Schmittgen, 2006; Kluiver et al., 2006), and we have recently shown that EBV latency gene expression drives expression of miR-155 (Yin et al., 2008a). In addition, we have recently demonstrated that miR-146a is also induced by EBV type III latency at least in part through the latency-associated oncoprotein LMP-1 (Cameron et al., 2008). Finally, work published by Mrazek et. al suggests that EBV may regulate other cellular miRNAs in addition to miR-155 and miR-146a (Mrazek et al., 2007). To further investigate the potential contribution of EBV in regulating cellular microRNAs, array profiling was performed in a panel of transformed B cell lines displaying EBV type III and type I latency gene expression as well as EBV negative derivatives of the type I cell line, Mutu I.
Results
EBV latency gene expression pattern correlates with distinct cellular and viral microRNA expression profiles
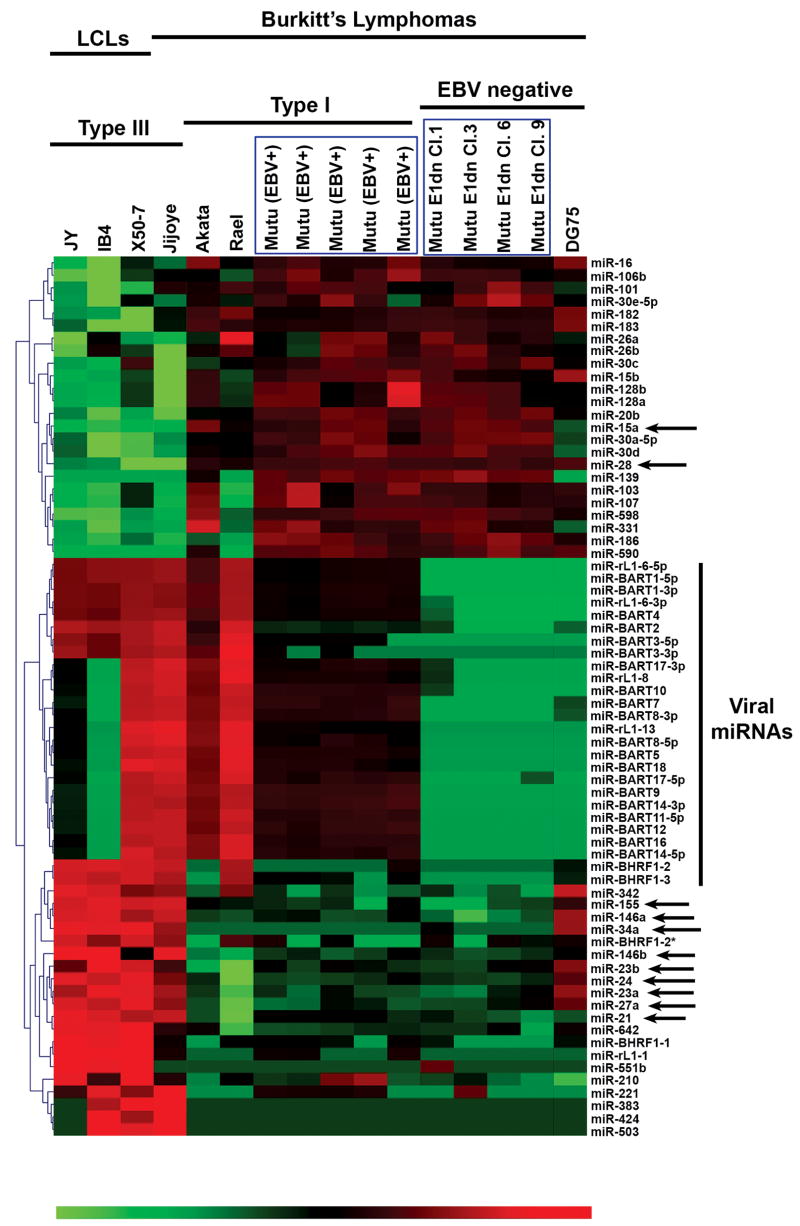
Micro-array chips containing probes for 470 unique human miRNAs and 108 unique viral miRNAs were used to obtain miRNA expression profiles for a panel of lymphoma cell lines grown to similar densities. Analysis of variance (ANOVA) was performed on the data set across three groups: EBV type III latency (Jijoye, JY, IB4, and X50-7), EBV type I latency (Akata, Rael, Mutu I), and EBV-negative cell lines [Burkitt’s lymphoma DG75 and EBV-negative derivatives of Mutu I, EBNA-1 dominant-negative clones 1, 3, 6, and 9 (Cameron et al., 2008)]. Cluster analysis of microRNAs displaying differential expression (p values of < 0.01) is shown in figure 1. As expected, expression of viral miRNAs was detected in EBV positive but not EBV negative cell lines. A subset of viral miRNA genes were not detected in the latency III cell line IB4, a lymphoblastoid cell line (LCL) derived from the B95-8 virus, which carries a deletion in the region of the EBV genome that encodes these viral BART genes (Cai et al., 2006; Parker et al., 1990). Probes designed to detect expression of the rhesus lymphocryptovirus (RhLCV) miRNAs (eg, rlcv-miR-rL1-13) scoring positive in EBV-infected cells represent genetic homologues of EBV (Cai et al., 2006).
Fig. 1.
EBV latency gene expression pattern correlates with distinct cellular and viral miRNA expression profiles. Micro-array analysis of miRNA expression was performed on RNA isolated from EBV infected, immortalized cell lines expressing viral latency patterns I (Akata, Rael, Mutu) or III (JY, IB4, Jijoye), and EBV negative cell lines (Mutu EBNA-1 dominant negative clones, DG75). Eight separate total RNA pairs were hybridized to miRNA arrays, and the data was grouped for analysis. Cluster analysis of miRNAs differentially expressed (ANOVA, p<0.01) among EBV negative, EBV positive/latency I and EBV positive/latency III cell lines is represented by heat map, with red elements representing higher expression and green elements representing lower expression.
Comparison of the array profiles of EBV-negative Mutu and EBV-positive Mutu I cells indicated no marked changes in cellular miRNA expression. In addition, inclusion of the latency I Burkitt’s lymphoma cell lines Akata and Rael in the EBV-positive group and Burkitt’s lymphoma DG75 in the EBV-negative group also failed to identify consistent cellular miRNA expression differences between EBV-positive/latency I and EBV-negative cells. These results suggest that neither expression of EBNA-1 plus non-coding viral RNAs expressed during type I latency, nor the retention of EBV episomes influences expression of cellular miRNAs.
In contrast to EBV-positive versus EBV-negative Mutu I comparisons, cluster analysis indicated striking differences in cellular microRNA expression between cells exhibiting type III and type I latency (figure 1). As expected, miR-155 and miR-146a were among those miRNAs that were expressed at substantially higher levels in latency III cells (1053-fold and 267–fold, respectively). Of the 41 cellular microRNAs altered by EBV latency phenotype, a subset (miR-15a, miR-21, miR-28, miR-34a, members of the miR-23a cluster [miR-23a, miR-24, and miR-27a], and miR-146b) was selected for further investigation based largely on the magnitude of the difference detected.
Quantitative (real-time) PCR validation of differential cellular miRNA expression
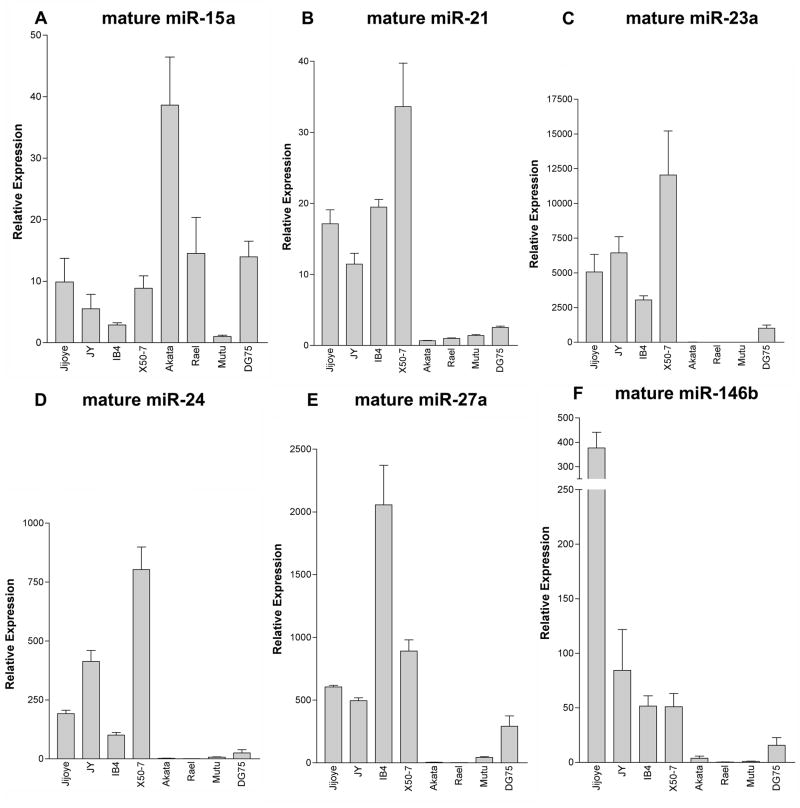
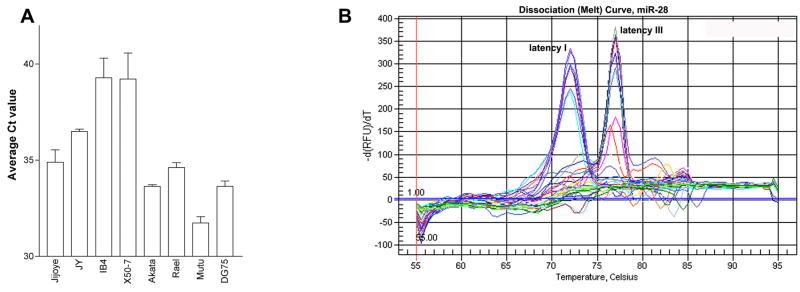
Mammalian miRNAs are transcribed by RNA polymerase II into long polyadenlyated primary transcripts (pri-miRNA), which are then cleaved by the enzyme drosha to form a ~70 base-pair hairpin loop structure (Kim, 2005). This precursor transcript (pre-miRNA) is then exported from the nucleus via exportin-5, and is further cleaved by the enzyme dicer into a small (21–25 nucleotide) duplex RNA molecule containing the mature miRNA and its complement, denoted miRNA*. To validate differential expression of selected miRNAs identified in the miRNA arrays, qRT-PCR for mature miRNAs was performed. Latency III-associated expression of miR-155 and miR-146a has previously been reported by our group (Cameron et al., 2008; Yin et al., 2008a) and others (Jiang, Lee, and Schmittgen, 2006; Kluiver et al., 2006) so validation of type III associated expression of these two microRNAs are not reported herein. As shown in figure 2, expression of mature miR-21, members of the miR-23a cluster (miR-23a, miR-24 and miR-27a), and miR-146b is higher in the type III cell lines Jijoye, JY, IB4 and X50-7 relative to the type I cell lines, Akata, Rael, and Mutu I. In contrast to the array data, mature miR-15a expression in qPCR assays did not correlate with EBV latency type. For quantification of miR-34a expression, analysis of the mature form was considered unreliable due to poor PCR efficiency (data not shown). In addition, although lower Ct values (higher expression) were obtained for mature miR-28 analysis in the type I cell lines versus the type III cell lines (figure 3A), melt curve analysis demonstrated the detection of two different PCR products (figure 3B). Interestingly, the PCR products with the melting temperatures of 72°C were exclusively detected in the latency I cell lines, while PCR products with melting temperatures of 77°C were exclusively detected in the latency III cell lines. Cloning and subsequent sequencing of the PCR products identified mature miR-28 amplicon in the latency I reactions, and non-specific amplification of a segment of the β-actin gene in the latency III reactions. Since the signal detected in the latency type III reactions does not represent miR-28, the differential expression of mature miR-28 in type I versus type III latency cells may be even greater than that represented by qPCR analysis.
Fig. 2.
EBV latency-associated differential expression of mature cellular miRNA demonstrated by quantitative (real-time) polymerase chain reaction (PCR). Expression of mature cellular miRNA differentially regulated during EBV latency was quantified in a panel of immortalized B cell lines by real-time PCR. Total RNA from the following immortalized B cell lines was isolated, poly-adenylated and transcribed into cDNA: a. EBV-positive, latency III cell lines Jijoye, JY, IB4, and X50-7; b. EBV-positive, latency I cell lines Akata, Rael and Mutu; c. EBV negative Burkitt’s lymphoma cell line DG75. The cDNAs were tested for mature cellular miRNA species by real-time, quantitative PCR in triplicate, and results were normalized to U6 small nuclear RNA expression. A–F. mature miRNAs miR-15a, miR-21, miR-23a, miR-24, miR-27a, and miR-146b expression, respectively.
Fig. 3.
A. Ct values for qPCR analysis of mature miR-28 in type I versus type III latency cells. B. Dissociation (melt) curve analysis of mature miR-28 amplification products. Two peaks represent two distinct amplification products, one at 72°C and one at 77°C.
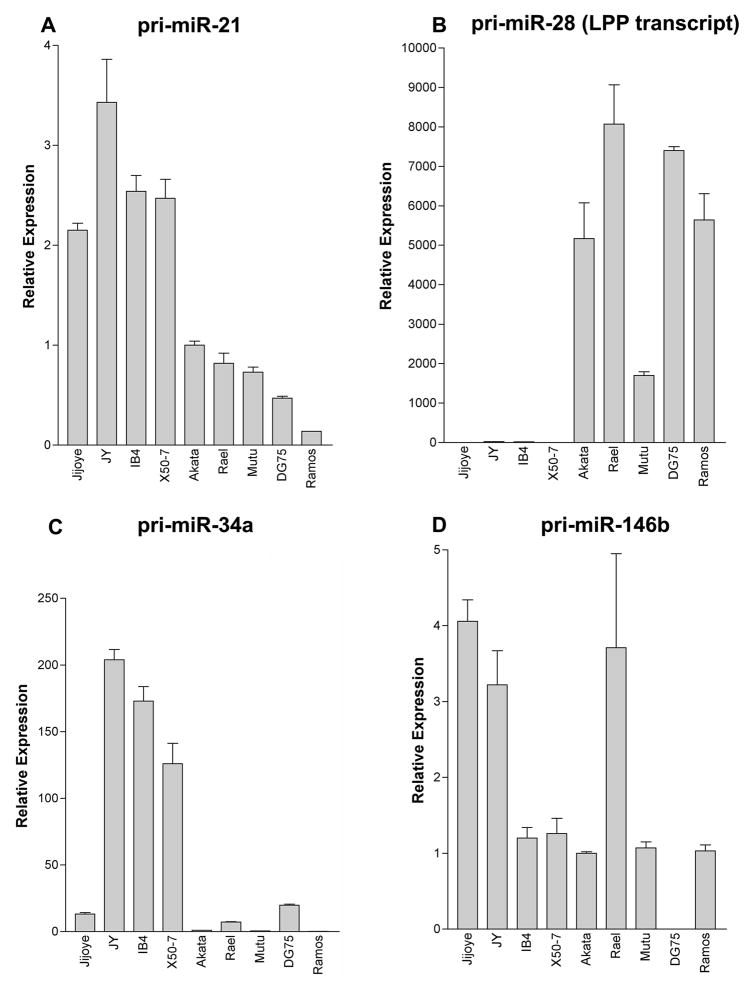
Due to the limitations encountered with qPCR detection of the mature form of miR-28 and miR-34a, as well as the potential for cross-reactivity between the closely-related miR-146a and miR-146b mature miRNAs, qRT-PCR assays were designed to specifically amplify the primary transcripts of these cellular miRNAs. Higher expression of pri-miR-34a was observed in EBV latency III-expressing cell lines (figure 4). MicroRNA-28 is derived from intronic sequences of the protein coding lipoma preferred partner (LPP) transcript. Substantial latency III-associated suppression of the miR-28 primary LPP transcript was seen (figure 4) supporting the conclusions drawn from the qPCR data for mature miR-28.
Fig. 4.
Cellular miRNA primary transcripts differentially regulated during EBV latency. Transcript-specific PCR primers were designed to amplify the primary transcript of cellular miRNAs differentially regulated during EBV latency. Total RNA from the following immortalized B cell lines was isolated and transcribed into cDNA: a. EBV-positive, latency III cell lines Jijoye, JY, IB4, and X50-7; b. EBV-positive, latency I cell lines Akata, Rael and Mutu; c. EBV negative Burkitt’s lymphoma cell lines DG75 and Ramos. The cDNAs were tested for mature cellular miRNA species by real-time, quantitative PCR in triplicate, and results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. A–D. pri-miR-21, pri-miR-28 (LPP transcript), pri-miR-34a and pri-miR-146b, respectively.
Expression of pri-miR-146b was not tightly latency-type associated, despite latency III-associated increases in mature miR-146b. In addition, the latency I Burkitt’s lymphoma cell line Rael, while having no detectable mature miR-146b, expressed readily detectable levels of pri-miR-146b. This suggests a possible defect in miR-146b processing in type I latency cells. However, at this time, we cannot rule out the possibility that detection of mature miR-146b in type III latency cells is due to cross reactivity with mature miR-146a. Nevertheless, we can conclude that the transcription rate of the miR-146b gene and/or stability of the miR-146b transcript is not substantially different in type I vs type III latency cells.
Quantitative RT-PCR assays were also designed to assess the primary transcript levels of miR-21 and the miR-23a cluster to investigate whether differences in the levels of the mature microRNAs is due to altered transcription rates or differences in post-transcriptional processing. Three different primer pairs were designed to amplify the primary miR-23a cluster transcript but all of these failed to amplify a product using RNAs from type I or type III cell lines. We believe that the inability to detect the miR-23a cluster primary transcript may be attributable to efficient processing of the primary transcript resulting from the combined effect of processing at the miR-23a, the miR-24, and the miR-27a loci. This idea is supported by the separate finding that cloning the miR-23a cluster immediately down stream from the GFP reading frame in a CMV expression vector causes abrogation of GFP detection in transfected cells (data not shown). Analysis of miR-21 primary transcript levels demonstrated measurable expression in latency I cells and a relatively modest 2-fold higher expression in latency III cells (figure 4) despite the more substantial increase in mature miR-21 detection (10- to 35-fold, figure 2). This raises the possibility that higher expression of mature miR-21 in type III latency cells may be elicited in part through enhanced post-transcriptional microRNA processing mechanisms.
Cellular miRNAs are regulated during EBV-induced transformation of normal B lymphocytes
To further address the link between cellular miRNA expression changes and EBV, expression of the panel of miRNAs was examined in peripheral B lymphocytes isolated from a pool of normal, healthy donors and three low-passage LCL lines (type III latency) generated by infection of normal B cells with EBV. Lymphocytes (CD19-positive) isolated from normal, healthy donors did not express detectable levels of pri-miR-34a, pri-miR-146a or pri-miR-146b (Figure 5, C-E). These cells also failed to express the mature forms of all three members of the miR-23a cluster (Figure 6A). Predicted levels of miR-92 and miR-150 were detected in the primary B cell sample (Lawrie et al., 2008), indicating that the lack of detectable mature miR-23 cluster members was not due to a lack of small RNAs in the sample (Figure 6B). All three low-passage LCLs demonstrated considerable levels of the primary transcripts of miR-34a, miR-146a, miR-146b (Figure 5C-E) and mature miRNAs miR-23a, miR-24, and miR-27a (Figure 6A). In contrast, expression of the LPP transcript/miR-28 primary transcript was markedly reduced (~90% reduction) in LCLs compared to the normal lymphocytes (Figure 5B). The primary transcript of miR-21, which was expressed in all cell lines, was moderately increased (~2-fold) in LCLs (Figure 5A), while mature miR-21 expression was detected at 3–4 log higher levels in LCLs. Despite detection of primary miR-21 transcripts in peripheral B cells, mature miR-21 was not detected in these cells. Like the primary and mature miR-21 analysis in type III versus type I latency (above), this data also supports the notion that differential expression of miR-21 is facilitated by a post-transcriptional processing mechanism.
Fig. 5.
EBV infection of primary B lymphocytes alters cellular miRNA primary transcript expression. Expression of pri-miRNA in primary B lymphocytes was compared to expression in three low-passage LCL cell lines (Dana, Boston, Alewife). Total RNA was reverse transcribed and the resulting cDNA was assessed for pri-miRNA expression by real-time PCR. A–E. pri-miR21, pri-miR28 (LPP transcript), pri-miR34a, pri-miR146a, and pri-miR146b expression, respectively. Data is normalized to 18S rRNA expression. All samples were tested in triplicate.
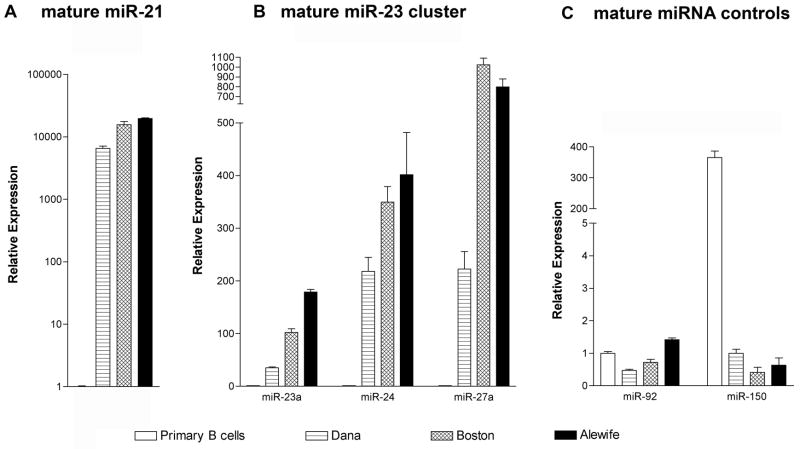
Fig. 6.
Mature miR23 cluster expression is induced by EBV infection of primary B lymphocytes. Peripheral B lymphocytes and low-passage lymphoblastoid cell lines (see figure 5 legend) were tested for mature miRNA expression by NCode real-time PCR. Samples were tested in triplicate, and data is normalized to U6 small nuclear RNA expression. A. mature miR-21 expression. B. mature miR23a cluster expression. C. Mature miR-92 (reportedly expressed similarly in all B cell subsets) and mature miR-150 expression (reportedly upregulated in normal, healthy lymphocytes, see Lawrie, et. al, 2008), demonstrating adequacy of peripheral B cell RNA for mature miRNA expression analysis.
Discussion
In this study, we demonstrate that expression of the EBV latency III transcription program modulates the expression of cellular miRNAs. We had previously described the regulation of cellular miR-155 and miR-146a by EBV latency III transcriptional activity (Cameron et al., 2008; Yin et al., 2008a), and we now demonstrate the regulation of miR-21, miR-23a, miR-24, miR-27a, miR-28, miR-34a, and miR-146b. Some of these results are consistent with a report published by Mrazek et al., which sought to validate a novel subtractive hybridization technique for identifying differentially-expressed noncoding RNAs, using EBV-transformed and EBV-negative lymphoma cells as a model (Mrazek et al., 2007). Despite the fact that Mrazek et al only compared one pair of cells of dissimilar background (EBV-transformed cord blood cells were compared to the EBV-negative Burkitt’s lymphoma cell line BL41), they observed higher expression of the cellular miRNAs miR-21, miR-155, miR-146a, miR-23a, miR-27a, and miR-34a in EBV-transformed cord blood lymphocytes. This study did not detect more subtle changes in miRNA expression, such as the upregulation of miR-146b, and the approach was not designed to detect miRNAs downregulated by EBV expression such as miR-28. The results reported herein expand the findings of Mrazek et al and compliments their observations by demonstrating consistency in EBV-mediated miRNA expression across a variety of cell backgrounds using multiple, highly sensitive assays.
The differential expression of mature miR-21 was consistently found to be greater than predicted by the more subtle differences in pri-miR-21 expression in latency III phenotype cells versus type I latency cells or uninfected B-cells. This discrepancy suggests that miR-21 is processed more efficiently in type III latency cells. Importantly, analysis of known viral and cellular miRNAs did not reveal any miRNAs with sufficient homology to miR-21 to be detected in our assays (data not shown). Nevertheless, the possibility that EBV induces the expression of another as yet unidentified cellular or viral miRNA that shares significant nucleotide homology with mature miR-21 cannot be ruled out.
A broad range of cellular mRNA transcripts may be targeted by a single miRNA; likewise, a single mRNA transcript may be simultaneously regulated by multiple miRNAs. Regulation of cellular pathways involves multiple miRNAs working in concert, and research has only begun to identify some of the transcript targets of individual miRNAs. Nevertheless, previously identified targets of the miRNAs induced by EBV may play a role in the interplay between the virus and the host cell. For instance, miR-24 has been demonstrated to promote cell growth through repression of p16INK4a, consistent with the growth phenotype of cells expressing EBV latency III transcripts (Lal et al., 2008). The reported role of miR-34a in promoting p53-mediated apoptosis is intriguing, given the complex regulatory role of EBV in apoptotic pathways (Chang et al., 2007; He et al., 2007; Raver-Shapira et al., 2007; Tarasov et al., 2007; Tazawa et al., 2007; Welch, Chen, and Stallings, 2007).
The overexpression of miR-21 during EBV latency III has significant implications for both the EBV life cycle and EBV-mediated oncogenesis. Upregulation of miR-21 has been demonstrated in metastatic breast, colorectal, hepatocellular, cervical, pancreatic and glioblastoma cancers (Asangani et al., 2008; Chan, Krichevsky, and Kosik, 2005; Frankel et al., 2008; Lui et al., 2007; Meng et al., 2007; Volinia et al., 2006). In vitro, ectopic expression of miR-21 promotes cell proliferation, migration and invasion, and inhibition of miR-21 suppresses growth, migration, and invasion, and increases apoptosis (Asangani et al., 2008; Chan, Krichevsky, and Kosik, 2005; Frankel et al., 2008; Loffler et al., 2007; Lu et al., 2008; Meng et al., 2007; Si et al., 2007; Zhu et al., 2007; Zhu et al., 2008). Several reports confirmed that miR-21inhibits expression of the tumor suppressors programmed cell death 4 (PDCD4) and tropomyosin 1 (TPM1) (Asangani et al., 2008; Frankel et al., 2008; Lu et al., 2008; Zhu et al., 2007; Zhu et al., 2008). Whether miR-21 inhibits PDCD4 and/or TPM1 translation in the setting of EBV infected B lymphocytes requires investigation.
Since the EBV-driven growth program of an infected naïve B cell mimics the cellular program initiated upon antigen-driven activation, it would be predicted that the miRNA profiles for these two events would also be similar. In a study by Lawrie et al, the miRNA profile of B cells activated by either IL-2 or IgM treatment differed from that of other B cell subsets (eg, naïve, germinal center, memory) by increased expression of eleven miRNAs, including miR-7, miR-9, miR-132, miR-155 and members of the 17–92 cluster (miR-17, miR-20b and miR-106a) and decreased expression of miR-638 (Lawrie et al., 2008). With the exception of miR-155, known to be induced during B cell activation through the activity of the transcription factor AP-1 (Yin et al., 2008b), this profile does not reflect that of EBV latency III-regulated miRNAs identified in our study. This suggests that although EBV- and antigen-activated B cell phenotypes are similar, the regulatory events of the two pathways may be distinct. No direct comparison of miRNA expression in antigen-activated and EBV-activated B cells has been reported.
In summary, we have demonstrated that the EBV latency III transcription (growth) program associated with initial infection in vivo and lymphocyte transformation in vitro is associated with changes in cellular miRNA expression. Two of these miRNAs, miR-155 and miR-21, are putative oncogenes. The role of EBV-mediated cellular miRNAs in EBV pathogenesis is currently under investigation.
Materials and Methods
Maintenance of cell lines
Lymphocyte cell lines (latency III lymphoblastoid cell lines [LCLs] JY, IB4, and X50-7; latency III Burkitt’s lymphoma line Jijoye; latency I Burkitt’s lymphoma cell lines Akata, Rael, and Mutu; EBV-negative Burkitt’s lymphoma cell lines DG75, Ramos, and BL30; EBV-infected Burkitt’s lymphoma BL30/B95-8, and low-passage latency III LCL lines Alewife, Boston, and Dana) were maintained in RPMI media plus 10% fetal calf serum and 0.5% penicillin-streptomycin. Retrovirally transduced EBNA-1 dominant negative (EBV negative) Mutu cells were maintained in RPMI media plus 10% fetal calf serum, 0.5% penicillin-streptomycin, 1μg/ml puromycin, and 1mg/ml gentamicin. Generation of the EBNA-1 dominant negative expression plasmid and EBNA-1 dominant negative Mutu cell lines has been detailed elsewhere (Cameron et al., 2008).
RNA isolation
RNAs used for miRNA array analysis and for quantifying mature miRNAs were generated by a modified TRIzol method. Briefly, 107 cells were suspended in TRIzol reagent (Invitrogen) and processed as per manufacturer’s protocol up through the addition of isopropanol. Samples were allowed to precipitate in isopropanol overnight at −20°C. Samples were then centrifuged at 12,000 × g for 30 min. at 4°C. Isopropanol was decanted and nucleic acid pellets were resuspended in 200 μl nuclease-free water. RNA was then precipitated again by adding 20 μl 3M sodium acetate and 0.5 ml of 100% ethanol, and incubating overnight at −20°C. Samples were centrifuged at 12,000 × g for 30 min. at 4°C, ethanol was decanted, and samples were washed once with 75% ethanol and allowed to air dry for no more than 10 minutes. RNA pellets were resuspended in nuclease-free water, quantified by UV spectrophotometry, aliquoted, and stored at −80°C.
Total RNA used for assessing the levels of primary miRNA transcripts was isolated from 107 cells using a RNEasy kit (Qiagen) according to the manufacturer’s protocol. The RNA was eluted from the column using 50 μl nuclease-free water, quantified by UV spectrophotometry, and stored at −80°C.
Primary B cell RNA used in comparison with EBV-infected primary B lymphocytes was purchased from Miltenyi Biotec (Germany). This sample consisted of total RNA isolated from CD19+ cells purified by positive magnetic selection from buffy coat preparations from a pool of healthy blood donors. Informed consent was obtained from participating blood donors and the procedure adhered to the guidelines set forth by the German Medical Association, as well as pertinent German and EU laws.
Quantitative (Real-Time) Reverse Transcription PCR (qRT-PCR) for mature miRNA and primary transcript (pri-miRNA)
To quantify mature miRNA expression, total RNA isolated by a modified TRIzol protocol (see RNA isolation) was poly-adenylated and reverse-transcribed using the NCode miRNA First Strand Synthesis kit (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA was subjected to quantitative (real-time) PCR using the NCode universal reverse primer in conjunction with a sequence-specific forward primer (forward primers for NCode miRNA detection are the exact sequence of the mature miRNA). Similarly, U6 small nuclear RNA was quantified using the NCode universal reverse primer and a U6-specific primer (5’-CTCGCTTCGGCAGCACA - 3’), and all mature miRNA data was normalized to the U6 data. A master mix was prepared for each PCR run which included Platinum SYBRgreen SuperMix plus UDG (Invitrogen), 50 nM fluorescein-NIST traceable dye, 100 nM forward and reverse primers, and nuclease-free water. Amplification consisted of 2 min. at 50°C, 2 min. at 95°C, followed by 40 cycles of 95°C for 15 sec. and 60°C for 30 sec.
Total RNA prepared by Qiagen RNEasy column extraction was DNAse treated and reverse transcribed using random hexamer primers and SuperScript III reverse transcription enzyme mix and reagents (Invitrogen) according to the manufacturer’s instructions. Primary transcript expression was quantified using specific forward and reverse primers (see Table 1) at 200 nM concentration in a reaction consisting of SYBRgreen SuperMix plus UDG, 50 nM fluorescein-NIST traceable dye, and nuclease-free water. Cycling parameters were as follows: 2 min. at 50°C, 2 min. at 95°C, followed by 40 cycles of 95°C for 30 sec. and 60°C for 30 sec. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was quantified similarly and was used to normalize the pri-miRNA data; the primer sequences are published elsewhere (West et al., 2004). In the case of the experiments comparing pri-miRNA transcript expression in primary B cells compared to low-passage LCLs, GAPDH was substantially lower in primary B cells, and data was instead normalized to 18S rRNA expression, which was comparably expressed in primary B cells and LCLs.
Table 1.
Primary miRNA transcript quantitative PCR primer sequences
| PRIMER | PRIMER SEQUENCE, 5′-3′ |
|---|---|
| U6 fwd | CTCGCTTCGGCAGCACA |
| U6 rev | AACGCTTCACGAATTTGCGT |
| Pri-miR-21 fwd | GTGACATCTCCATGGCTGTACCA |
| Pri-miR-21 rev | CAAAATGTCAGACAGCCCATCGAC |
| LPPex2 fwd | TCCACCTCTAGATGATTCCAGTG |
| LPPex2-3 rev | GGATTCCCCTGTACTTTGAAAGC |
| Pri-miR-34a fwd | CTTGAACTCCTGGCCTGAAG |
| Pre-miR-34a rev | GCCAAAGAAACACTCACAGCT |
| Pri-miR-146b fwd | GAGCAGCGTCCAGGCTGAA |
| Pre-miR-146b rev | CCGGGCACCAGAACTGAGT |
Melt curve analysis was performed at the end of every qRT-PCR run. Samples were tested in triplicate. Real-time PCR was performed on Bio-Rad iCyclers (MyIQ, IQ4 or IQ5) and data analysis was performed using Bio-Rad IQ5 v2 software. Measures taken to prevent and identify PCR contamination included maintaining a physically separate, nucleic acid-free area with designated equipment for PCR setup, and using both no-template controls and no-reverse transcription controls in every experiment. A standard dilution curve, generated by serially diluting a pooled stock of positive control cDNAs, was included in each run. Experiments were repeated if controls did not behave as predicted.
Relative expression was calculated for each test sample by the standard curve method (for formulas, see ABI Prism 7700 Sequence Detection System User Bulletin #2: Relative Quantitation of Gene Expression, Applied Biosystems, Foster City, CA). Relative quantities of each test sample were extrapolated from the standard curve run concurrently with the sample, and the values were normalized to U6 small nuclear RNA (mature miRNA NCode assays), GAPDH or 18S rRNA (primary transcript assays). The normalized values were then compared to a calibrator sample within the run. Data is presented as expression relative to the calibrator, with the standard error of the mean of the triplicate measures for each test sample.
Cellular miRNA micro-array analysis
Small RNA isolation and hybridization was performed by LC Sciences (Houston, TX). Briefly, small RNA species were isolated from total RNA by column exclusion. The concentrated small RNAs were 3’ poly-adenylated with poly (A) polymerase. A nucleotide tag was then ligated to the poly-A tails. The tagged RNAs were hybridized to a μParaflo superfluidic array chip containing probes for human and viral microRNA sequences, and were subsequently labeled in a second hybridization reaction with Cy3 and Cy5 dendrimer dyes. After overnight hybridization, arrays were stringently washed and scanned on an Axon GenePix 4000B laser scanner (Axon Instruments). Data extraction and imaging were performed using ArrayPro software (Media Cybernetics).
Each array compared an EBV latency type III-expressing cell line (eg, JY, IB4, X50-7, Jijoye) to cells expressing EBV latency type I (Akata, Rael, Mutu) or EBV positive Mutu cells to EBV negative Mutu cells (Mutu E1dn clones). Eight separate arrays were performed. The most up-to-date chip available for analysis was used (containing probes for all human miRNA listed in Sanger Institute miRBase release 9.1, totaling 470 unique miRNAs). Additional probes (108 unique sequences) for viral miRNA expression were included on each chip. Each probe was repeated in sextuplicate. Dye-swap experiments were executed to control for labeling bias. Data within each individual chip was adjusted by subtraction of background fluorescence (calculated as the median of the lower 5–25% of signal intensities) followed by normalization of the data to the statistical median of all detectable signals. Variation among arrays introduced by differences in experimental conditions (ie, sample preparation, dye labeling, chip quality, and scanning variations) was corrected by normalization of the data by the cyclic LOWESS (locally-weighted regression) method (Bolstad et al., 2003). Intensity values were then Log2 transformed and further normalized and centered using the equation: Value = [(Log2 intensity value)-(Mean of values across all samples for the individual gene)]/[(standard deviation of values across all samples for the individual gene)]. The ratio of expression between paired samples was normalized for clustering analysis by the equation: Value = [Log2 (Ratio)]/[(standard deviation (Log2 (Ratio))]. Statistical comparison of paired sample miRNA expression was performed using t-tests, and corresponding p-values were calculated (Pan, 2002). MicroRNA showing differential expression at the p<0.10, p<0.05 and p<0.01 were selected for clustering analysis, which was performed using hierarchical method based on average linkage and Euclidian distance metric (Eisen et al., 1998). All data calculations were performed by LC Sciences. Clustering plots were created using TIGR MeV (Multiple Experimental Viewer) software (The Institute for Genomic Research).
Acknowledgments
We wish to thank Xiaochuan Zhou and LC Sciences for their expertise in miRNA array analysis. Support for this project was provided by funding from the Tulane Cancer Center (J. Cameron) and grants awarded to E. Flemington by the National Institutes of Health (R01-GM48045, R21-DE17008, and R01 CA124311). Support was also provided by a National Institutes of Health COBRE grant, P20 RR020152.
Footnotes
Institution Address: Tulane University Health Sciences Center, 1430 Tulane Ave., New Orleans, LA 70112
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9(3):395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):236–247. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82(4):1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg D, Middeldorp JM, Catalina M, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. Demonstration of the Burkitt’s lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc Natl Acad Sci U S A. 2004;101(1):239–244. doi: 10.1073/pnas.2237267100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg DR, Thorley-Lawson DA. Quantitative detection of viral gene expression in populations of Epstein-Barr virus-infected cells in vivo. Methods Mol Biol. 2005;292:39–56. doi: 10.1385/1-59259-848-x:039. [DOI] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Schmittgen TD. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45(1):103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- Kieff ED, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5. Vol. 2. Lippincott Williams Wilkins; Philadelphia: 2007. pp. 2603–2654. [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45(2):147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, Navarro F, Dykxhoorn D, Lieberman J, Gorospe M. p16(INK4a) translation suppressed by miR-24. PLoS ONE. 2008;3(3):e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Saunders NJ, Soneji S, Palazzo S, Dunlop HM, Cooper CD, Brown PJ, Troussard X, Mossafa H, Enver T, Pezzella CS. MicroRNA expression in lymphocyte development and malignancy. Leukemia. 2008;22(7):1440–1446. doi: 10.1038/sj.leu.2405083. [DOI] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39(2):167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71(7):4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35(10):e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics. 2002;18(4):546–554. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- Parker BD, Bankier A, Satchwell S, Barrell B, Farrell PJ. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179(1):339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA. EBV Persistence and Latent Infection In Vivo. In: Robertson ES, editor. Epstein-Barr Virus. Caister Academic Press; Wymondham, Norfolk, England: 2005. pp. 309–357. [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- West AB, Kapatos G, O’Farrell C, Gonzalez-de-Chavez F, Chiu K, Farrer MJ, Maidment NT. N-myc regulates parkin expression. J Biol Chem. 2004;279(28):28896–28902. doi: 10.1074/jbc.M400126200. [DOI] [PubMed] [Google Scholar]
- Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008a;82(11):5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem. 2008b;283(5):2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]