Abstract
Mot1 is an essential TATA-binding protein (TBP)-associated factor and Snf2/Swi2 ATPase that both represses and activates transcription. Biochemical and structural results support a model in which ATP binding and hydrolysis induce a conformational change in Mot1 that drives local translocation along DNA, thus removing TBP. Although this activity explains transcriptional repression, it does not as easily explain Mot1-mediated transcriptional activation, and several different models have been proposed to explain how Mot1 activates transcription. To better understand the function of Mot1 in yeast cells in vivo, particularly with regard to gene activation, TBP mutants were identified that bypass the requirement for Mot1 in vivo. Although TBP has been extensively mutated and analyzed previously, this screen uncovered two novel TBP variants that are unique in their ability to bypass the requirement for Mot1. Surprisingly, in vitro analyses reveal that rather than having acquired an improved biochemical activity, one of the TBPs was defective for interaction with polymerase II preinitiation complex (PIC) components and other regulators of TBP function. The other mutant was defective for DNA binding in vitro yet was still recruited to chromatin in vivo. These results suggest that Mot1-mediated dissociation of TBP (or TBP-containing complexes) from chromatin can explain the Mot1 activation mechanism at some promoters. The results also suggest that PICs can be dynamically unstable and that appropriate PIC instability is critical for the regulation of transcription in vivo.
RNA polymerase II preinitiation complexes (PICs)3 are assembled on promoter DNA from multiple general transcription factors (GTFs) that provide a platform for binding of the RNA polymerase II enzyme itself (1, 2). TATA-binding protein (TBP) is a central component of the PIC, providing stabilizing interactions with TATA box-containing DNA as well as several GTFs (3, 4). In vitro, TBP binds DNA with remarkably high stability but relatively low sequence selectivity (5-7). As TBP binding is rate-limiting for transcription in vivo (8-10), stable binding of TBP to DNA is balanced by the requirement for an adequate pool of unbound TBP to nucleate the assembly of new PICs in response to changes in transcriptional stimuli.
Mot1 is a conserved, essential ATPase in budding yeast that facilitates the redistribution of TBP among DNA sites both in vitro and in vivo (11-15). Mot1-catalyzed TBP·DNA dissociation readily explains the role of Mot1 in transcriptional repression, but how might Mot1 activate transcription? Two classes of models have been invoked to explain how Mot1 activates transcription (14, 16-18). The first idea is that Mot1 activates as well as represses transcription by dissociating TBP from DNA. In such a scenario, the TBP·DNA displacement activity of Mot1 liberates TBP from nonfunctional high affinity sites, ensuring that an adequate pool of TBP is available to activate gene expression (14, 18, 19). TBP bound to an activated promoter would be stabilized against Mot1 action by association with other GTFs (20). Related ideas are that Mot1 facilitates active transcription by clearing TBP bound to spurious sites on promoters that interfere with formation of legitimate PICs or TBP that may bind to promoters in stable but inactive, kinetically trapped complexes. An example of this occurs at the Mot1-activated URA1 promoter, which possesses a TATA box that preferentially directs TBP binding in the wrong orientation for assembly of a functional PIC (21). In this case the hypothesis is that Mot1-mediated removal of inappropriately bound TBP permits additional rounds of TBP binding, with the correct orientation eventually stabilized by other factors. A related idea is that Mot1 collaborates with other factors to restrict TBP to appropriate locations at active promoters (22). Consistent with this idea, the highly dynamic behavior of TBP in vivo is strongly Mot1-dependent (15).
Although the idea that Mot1 utilizes the same enzymatic mechanism to activate and repress transcription is attractive in its simplicity, evidence suggests that TBP·DNA dissociation may not be sufficient to account for the activation mechanism of Mot1. For example, although misoriented TBP appears to contribute to how URA1 transcription is regulated, forcing TBP to interact with the URA1 promoter in the correct orientation does not obviate the requirement for Mot1 in vivo (21). Analyses of other yeast promoters have also led to the suggestion that Mot1 functions as a transcriptional co-activator (17). At the activated GAL1 promoter, chromatin remodeling requires the mutual cooperation of Mot1 and the SAGA histone acetyltransferase complex, which suggests that the Mot1 ATPase is used for an activity other than TBP·DNA dissociation (23). Finally, it has been reported that under some physiological conditions Mot1 is physically associated with active PICs rather than simply dissociating inactive complexes (24).
Taken together, in vivo studies indicate that TBP·DNA dissociation may not be sufficient to explain how Mot1 mediates transcriptional activation. Although much is known about how Mot1 catalyzes TBP·DNA dissociation and how other GTFs influence this activity in vitro, prior work has failed to uncover biochemical evidence for an alternative activity of Mot1 that might be relevant for understanding the above in vivo observations regarding gene activation. To better address the question of how Mot1 activates transcription, we therefore turned to a different approach. Using a library of TBP mutants, we screened for variants that could bypass the requirement for Mot1 in vivo. The rationale was that a variant that had gained the ability to undergo transitions that are normally catalyzed by Mot1 would allow cells to bypass Mot1 function. Here we present the results of the screen and the combined biochemical, molecular, and genetic analyses of the variants that were uncovered. The results show that Mot1 activity is dispensable if certain interactions with TBP are weakened rather than strengthened. These results support a model in which Mot1 ensures dynamic instability in the interactions of GTFs or DNA with TBP and that such instability is important for both gene repression and activation.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions—Saccharomyces cerevisiae strains used in this study were derived from YPH499 (25) and were previously described (26). AY138 (mot1Δ::kanMX, carrying the plasmid pMR13 (MOT1, URA3)), was transformed with plasmid vector (pRS314 (25)) or pRS314-borne alleles of TBP (WT, Y185C, or F207L) that were either untagged or Myc-tagged at the C terminus. After transformation, plasmid-borne MOT1 was shuffled out by plating on 5-FOA (27). Yeast strains were grown at 30 °C to an A600 ∼ 1.0. Cells were harvested for isolation of total RNA or treated for ChIP as described below.
Preparation of Nuclear Extracts and Western Blotting—Nuclear extracts were prepared from 1 liter of yeast cells grown to saturation in YPD. All centrifugation steps were performed at 4000 rpm for 10 min in a Sorvall GSA rotor unless otherwise noted. Cell pellets were resuspended in 50 mm Tris-Cl, pH 7.5, 30 mm DTT, and shaken slowly at 30 °C for 15 min. Cells were then pelleted and resuspended in YPD containing 1 m sorbitol (YPD/S) and incubated with 1.5 mg/ml zymolyase 100T dissolved in 50 mm Tris-Cl, pH 7.5, with protease inhibitors and 1 mm DTT at 30 °C to form spheroplasts. Spheroplast formation was monitored microscopically by estimating the percentage of cell “ghosts” observed in an aliquot of cells placed in 1% SDS. The zymolyase reaction was incubated until 80-90% of cells formed ghosts. Digestion was halted by addition of YPD/S at room temperature, and cells were pelleted and resuspended in YPD/S and allowed to recover for 30 min at 30 °C with gentle shaking. Cells were then washed twice with YPD/S at 4 °C, once with 1 m sorbitol at 4 °C, then resuspended in 18% polysucrose 400, 10 mm Tris-Cl, pH 7.5, 20 mm KCl, 5 mm MgCl2, 1 mm EDTA, 0.15 mm spermine, 0.5 mm spermidine, 3 mm DTT, and protease inhibitors at 4 °C. Spheroplasts were lysed by passing twice through a Yamato LH21 homogenizer at 1000 rpm. The lysate was clarified by centrifugation at 5400 rpm in a Sorvall GSA rotor. Nuclei were then recovered by centrifugation at 13,000 rpm for 30 min in a Sorvall SS-34 rotor. The pelleted nuclei were resuspended in buffer containing 100 mm Tris acetate, pH 7.9, 50 mm potassium acetate, 10 mm MgS04, 20% glycerol, 2 mm EDTA, 3 mm DTT, and protease inhibitors. The nuclei were then lysed by adding 3 m ammonium sulfate, pH 7.6, to a final concentration of 0.5 m followed by centrifugation at 28,000 rpm for 75 min in an SW 28 rotor. Protein yield was quantified by Bradford protein assay using bovine serum albumin as the standard. Epitope-tagged Mot1 was detected by Western blotting using the α-Py monoclonal antibody as previously described (26).
Electrophoretic Mobility Shift Assays (EMSA)—EMSA was performed as previously described (12) using purified yeast TBP, TFIIA, TFIIB, Bur6, Ydr1, and a radiolabeled fragment of the AdMLP unless otherwise noted. Reactions were run on 6% polyacrylamide gels (6% acrylamide from a 20%:0.33% acrylamide:bisacrylamide stock, 2.5% glycerol, 190 mm glycine, 10 mm magnesium acetate, 2.5 mm Tris, pH 8.3, 1 mm EDTA, 0.5 mm DTT) and TG running buffer (2.5 mm Tris-Cl, pH 8.3, 190 mm glycine, 1 mm EDTA) with 5 mm magnesium acetate. TFIIB and NC2 EMSAs were performed using 1× TBE (90 mm Tris, 90 mm boric acid, 2 mm EDTA) in place of the TG running buffer to allow for better detection of the ternary complexes. TBP·DNA complexes were not detectable on TBE gels. Gels were run at 100 V at 4 °C for ∼1 h before loading. 20-μl reactions were incubated in buffer containing 2 μg of bovine serum albumin, 100 ng of poly (dG-dC), 4% glycerol, 0.1% Brij 58, 60 mm KCl, 5 mm Tris-Cl, pH 8, 5 mm MgCl2, 1 mm DTT, 0.025% bromphenol blue, and 1000 cpm of radiolabeled DNA. Complexes were resolved by running the gels at 160 V for 45-60 min, which were then dried and exposed to a PhosphorImager screen (Molecular Dynamics) for 12-36 h.
RNA Isolation and Northern Blotting—Total RNA was isolated using hot acid-phenol extraction (28). Poly(A)+ RNA was prepared from 1 mg of total RNA using the Qiagen Oligotex Midi Kit according to instructions provided by the manufacturer. For Northern blots, 20 μg of total RNA was separated by electrophoresis on 1% formaldehyde gels and transferred to a nylon membrane (Nytran, Schleicher and Schell). DNA probes were generated by random priming of PCR products amplified from a portion of the indicated open reading frames. Blots were hybridized overnight in 30% formamide, 4× SSC (1× SSC = 0.15 m NaCl and 0.015 m sodium citrate), 10 mm EDTA, 0.25 mg/ml RNA, 10% dextran sulfate, 1× Denhardt's, 5% SDS, and washed twice for 15 min each at 24 °C followed by 1 h with 0.1 × SSC, 0.1% SDS at 50 °C. RNA was detected by autoradiography and visualized using a PhosphorImager. Analysis of 35 S RNA levels was performed as previously described (29). Quantitation was performed using ImageQuant software.
Microarray Hybridization and Data Analysis—Gene expression analysis was conducted using Agilent Yeast Oligo arrays (011447) (Agilent Technologies, Palo Alto, CA). Total RNA was amplified using the Agilent Low RNA Input Fluorescent Linear Amplification kit protocol. Starting with 500 ng of total RNA, Cy3- or Cy5-labeled cRNA was produced according to manufacturer's protocol. For each two color comparison, 750 ng of each Cy3- and Cy5-labeled cRNAs were mixed and fragmented using the Agilent in situ hybridization kit protocol. Hybridizations were performed for 17 h in a rotating hybridization oven using the Agilent 60-mer oligo microarray processing protocol. Slides were washed as indicated in this protocol and then scanned with an Agilent Scanner. Each comparison was hybridized to two arrays employing a dye reversal. Data were obtained using the Agilent Feature Extraction software (Version 7.5) using defaults for all parameters. The resulting data were processed using the Rosetta Resolver(r) system (Rosetta Biosoftware, Kirkland, WA). This system combines the replicate hybridizations as described (30) and generates p values that represent the probability that a given probe is differentially expressed. Comparison of microarray data from the strains in this study with results for spt20Δ (31), bur6 (32), and our previous mot1 microarray data (33) was performed using Microsoft Excel. Analysis of overrepresented Gene Ontology terms was conducted using GOstat (34) with a p value cutoff of 0.01. The microarray data have been deposited in NCBI Gene Expression Omnibus (67) and are accessible through GEO Series accession number GSE12371.
Chromatin Immunoprecipitation—ChIP was performed as previously described (14). In brief, cells were grown as described above, then treated with 1% formaldehyde for 15 min followed by the addition of glycine to 125 mm for 5 min. Cells were then washed once with 4 °C TBS containing 125 mm glycine and then and once with 4 °C TBS alone. Cells were resuspended in ChIP lysis buffer with 140 mm NaCl and protease inhibitors (Complete Protease Inhibitor Mixture Tablet, Roche Applied Science) and lysed using acid-washed glass beads (Sigma) in a FastPrep device (MP Biomedicals). Whole cell extracts were sonicated to obtain average DNA fragment sizes of less than 500 bp, and the resulting extract was quantitated using Bradford Reagent. Immunoprecipitations were performed overnight at 4 °C using an anti-Myc (9E10) antibody. Protein A-Sepharose beads (Amersham Biosciences) were added for 2 h, and beads were washed. After reversal of the cross-links, DNA was purified using a QIAquick PCR Purification kit (Qiagen). PCR was performed as described previously (14).
RESULTS
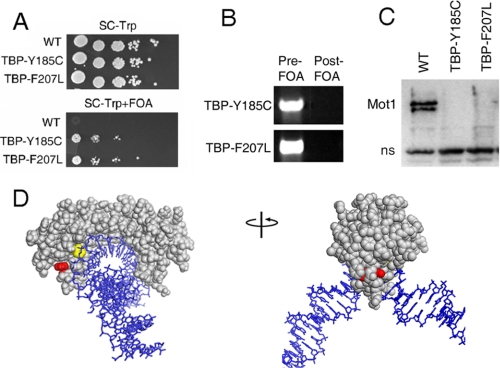
Isolation of TBP Alleles That Bypass the Requirement for Mot1 in Vivo—To identify TBP mutants that support viability in the absence of Mot1, a screen was performed in S. cerevisiae using a TBP mutant library (35) and the plasmid shuffle method (27). Plasmid-borne TBP alleles harboring random base pair changes were introduced into the MOT1 shuffling strain, and colonies were selected on the basis of growth after shuffling out the plasmid-borne copy of MOT1. A screen of about 5000 transformants uncovered two FOA-resistant colonies. After verifying the phenotype of positive colonies by plasmid isolation, retransformation, and restreaking to various selective media, plasmids were isolated and sequenced. The two strains harbored plasmids with two different mutations in TBP encoding Y185C and F207L (Fig. 1A). Note that neither of these alleles restores cell growth to wild-type levels, a point that is addressed below. Comparison of total DNA from pre- and post-FOA cells carrying these alleles confirmed that the MOT1 coding sequence was not detectable after the shuffle (Fig. 1B), and Western analysis showed that Mot1 protein was not detectable in the viable bypass strains (Fig. 1C).
FIGURE 1.
TBP Y185C and TBP F207L bypass the requirement for Mot1 in vivo. A, spot test showing a MOT1 shuffling strain transformed with either WT TBP, TBP Y185C, or TBP F207L. The strains harbored a deletion of the chromosomal copy of MOT1 and a WT MOT1 allele on a URA3-marked plasmid (26). Thus, 5-FOA selected for cells that have lost the URA3-marked copy of MOT1. Spots are 10-fold serial dilutions on synthetic media lacking tryptophan (to select for the TBP plasmids) with (bottom) and without (top) 5-FOA. B, PCR for MOT1 in pre- and post-FOA strains carrying TBP Y185C or TBP F207L as in panel A. C, Western blot of nuclear extracts of post-FOA WT, TBP Y185C, and TBP F207L strains using anti-Mot1 polyclonal antisera (26) to detect Mot1 protein. In this experiment the viability of the WT strain on FOA was maintained by transformation with a plasmid-borne copy of MOT1 on a LEU2-marked plasmid not subject to FOA counter-selection (26). The Mot1 band is indicated, which is notably absent from the bypass strains. A nonspecific band (ns) served as a loading control. D, crystal structure of the TBP·DNA complex (36) illustrating the position of Y185 (red) and F207 (yellow). TBP is shown in gray; DNA is in blue.
Tyr-185 and Phe-207 are solvent-exposed and close to each other, as displayed on the structure of TBP·DNA (Fig. 1D and Ref. 36). However, previous studies suggest that they are involved in different interactions. DNA in the TBP·DNA complex is severely kinked because of intercalation of phenylalanine residues at either end of the TATA box sequence. Phe-207 is one of the kink residues that protrudes from the concave DNA binding surface (36). In contrast, Tyr-185 does not project into the DNA binding surface but instead is located on the C-terminal half of TBP close to the site of TFIIB interaction (37). Interestingly, Tyr-185 is also near the site of TBP interaction with NC2 (38), a global transcriptional regulator whose function overlaps with Mot1 (18, 22, 33, 39). Mapping of the altered residues on the TBP structure, thus, suggested that Mot1 function could be bypassed by perturbing either the TBP·DNA interaction or the interaction of TBP with other proteins, possibly TFIIB or NC2.
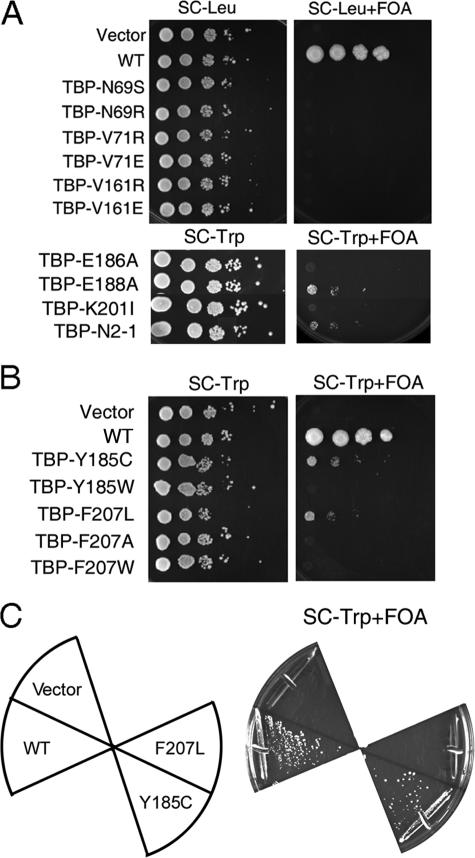
TBP Alleles That Bypass Mot1 Function Are Unusual—To determine whether the ability to bypass Mot1 was a general property of TBPs with related defects, other TBP alleles with previously defined biochemical defects were tested for their ability to bypass Mot1 (Fig. 2A). TBP mutants that impair dimerization and have altered DNA binding activities, either positively as for N69S and N69R (40), or negatively, as for V71R, V71E, V161R, and V161E (41), did not support growth in the absence of Mot1. Therefore, the ability of TBP F207L to bypass Mot1 function was not simply a consequence of a generic effect on DNA binding activity. This conclusion is supported by the observation that the effect of the Phe-207 mutation was specific for leucine, as bypass of Mot1 function was not observed when Phe-207 was converted to either alanine (resulting in loss of the phenyl side chain) or tryptophan (conversion to another bulky hydrophobic side chain; Fig. 2B).
FIGURE 2.
Bypass of Mot1 function by TBP Y185C and TBP F207L is allele-specific. A, spot test performed as in Fig. 1A but with other TBP alleles as indicated. Spots are 10-fold serial dilutions on synthetic media lacking leucine (top) or tryptophan (bottom) with (right) and without (left) 5-FOA. WT refers to cells harboring a wild-type copy of MOT1 not subject to 5-FOA counter-selection. Note that TBP-E188A and TBP-N2-1 bypassed Mot1 function but to a lesser degree than the alleles identified in the screen reported here. B, spot test as in panel A to test the importance of the particular TBP residue at position 185 or 207. The ability to bypass Mot1 function was specific for a cysteine at position 185 or a leucine at position 207. C, a SPT15 (TBP) shuffling strain was transformed with plasmid vector or plasmids expressing WT TBP, TBP Y185C, TBP F207L. Strains were streaked onto synthetic media lacking tryptophan and containing 5-FOA. Growth of the TBP Y185C strain on FOA-containing media shows that this allele supported viability in the absence of WT TBP, whereas TBP F207L did not.
As the TBP Y185C allele suggested that a defect in interaction with TFIIB or NC2 might allow cells to bypass Mot1, TBP alleles defective for association with the general transcription factors TFIIA or TFIIB or activators were tested, including E186A, E188A, K201I, and N2-I (42-44). These mutants were chosen because their side chain alterations are proximal to Tyr-185 and they have previously defined biochemical defects, which facilitated a test of the relationship between the genetics and biochemistry. None of these alleles was able to bypass Mot1 function as effectively as Y185C or F207L, although E188A and N2-1, which have drastically reduced association with TFIIB and TFIIA (42, 44), respectively, were able to bypass the requirement for Mot1 to some extent. This suggests that one way to bypass Mot1 function is to reduce the association of TBP with TFIIB or TFIIA. Mutating Tyr-185 to tryptophan eliminated the bypass phenotype even though both residues are hydrophobic (Fig. 2B). This suggests that the specific loss of the Tyr-185 side chain imparts the ability to bypass Mot1 function.
Next, the mutant alleles were transformed into a TBP shuffling strain to determine whether they could support viability in the absence of WT TBP. As shown in Fig. 2C, TBP Y185C supported viability in the absence of WT TBP, whereas TBP F207L did not. Therefore, although these mutants exhibit similar phenotypes with respect to Mot1, the differences between them suggest that they achieve a similar phenotypic effect through distinct, but perhaps overlapping, mechanisms.
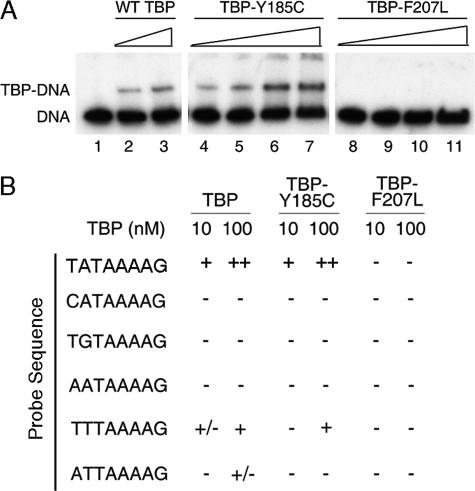
TBP F207L Is Defective for DNA Binding in Vitro—The location of Phe-207 on the TBP DNA binding surface suggested altered DNA binding activity for this protein, which was intriguing in light of the relatively conservative nature of the phenylalanine to leucine amino acid change as well as the allele specificity described above. If the essential function of Mot1 is to catalyze redistribution of TBP in vivo, one potential mechanism by which TBP could bypass the requirement for Mot1 is by altering the DNA binding properties of TBP either by decreasing affinity to facilitate redistribution genome-wide or by decreasing the DNA binding specificity to recognize a wider range of promoter sequences. To test this idea, EMSAs were performed using recombinant protein and a variety of TATA-like sequences as probes. To compare the affinity of the mutant TBP alleles with the WT protein, the binding to a consensus TATA element TATAAAAG was tested (Figs. 3, A and B). TBP Y185C bound to a consensus TATA element with an affinity indistinguishable from WT TBP, as expected, given the location of the mutation and the ability of the allele to support viability. TBP F207L, however, had no detectable DNA binding activity. In the co-crystal structure, Phe-207 intercalates between the T-A base pair step at the 5′ end of the TATA box sequence (36). Thus, F207L alters a residue responsible for DNA kinking at the 5′ end of the TATA box while preserving the overall hydrophobic character of the DNA binding surface. We reasoned that if TBP F207L possessed altered DNA binding specificity, this would most likely manifest as a difference in the ability to bind to sequences with variations at the 5′ end of the TATA box where Phe-207 is positioned. Some sequence variants (e.g. binding sites with consecutive G-C base pairs at the 5′ end of the TATA sequence) were ruled out because structural considerations rendered them unlikely TBP binding sites for any TBP variant (7). As summarized in Fig. 3B, whereas DNA binding activity was detected for WT TBP and TBP Y185C using probes containing TT and AT sequences at the upstream edge of the TATA box, no DNA binding activity was detected for TBP F207L on any of the probes. Some other DNA binding specificity for TBP F207L cannot be ruled out, but the simplest interpretation is that conversion of Phe-207 to leucine reduces TBP binding to DNA sites across the board rather than conferring an altered sequence binding preference.
FIGURE 3.
DNA binding activity of TBP Y185C and TBP F207L in vitro. A, EMSAs showing TBP·DNA complexes using radiolabeled AdMLP DNA (<1 nm) and titrations of WT TBP, TBP Y185C, or TBP F207L. Lane 1 shows free DNA. Concentrations of TBP used were 5 nm (lanes 2, 4, and 8), 10 nm (lanes 3, 5, and 9), 50 nm (lanes 6 and 10), and 100 nm (lanes 7 and 11). Comparison of lanes 2 and 3 with lanes 4 and 5 show no detectable difference in DNA binding between WT TBP and TBP Y185C. B, summary of EMSA results with alternate TATA sequences. Variations from the AdMLP TATA sequence occur in the first two base pairs where residue 207 of TBP makes critical contacts. The top strand of the probe sequence used is written on the left, and the results from reactions containing 10 or 100 nm of the indicated TBP (WT, Y185C, or F207L) are shown. - indicates no detectable binding, +/- indicates binding that was barely detectable, + indicates binding activity seen for 10 nm WT TBP to the consensus probe, and ++ indicates binding seen with 100 nm WT TBP to the consensus probe. Each binding assay was performed at least two independent times.
TBP Y185C Displays Reduced Association with TFIIA, TFIIB, and NC2 in Vitro—The proximity of Tyr-185 to the TFIIB binding site suggested that TBP Y185C has an altered interaction with TFIIB. We also considered the possibility that Y185C might modulate PIC formation by altering or destabilizing the interaction between TBP and TFIIA. Using a radiolabeled consensus TATA element, WT TBP or TBP Y185C was titrated into reactions with either TFIIA or TFIIB. TBP Y185C showed a similar, but perhaps slightly reduced, capacity to form TFIIA·TBP·DNA complexes compared with WT TBP (Fig. 4A). With regard to TBP F207L, we considered the possibility that TFIIA or TFIIB might be able to stabilize interaction with DNA under conditions in which TBP F207L alone was incapable of DNA binding. However, no ternary complexes were detected with TBP F207L (Fig. 4A). Therefore, TFIIA was not able to rescue the defective TBP·DNA interaction for the F207L mutant. In contrast to WT TBP, neither TBP Y185C nor TBP F207L was able to form TFIIB·TBP·DNA ternary complexes (Fig. 4B). Tyr-185 is also near the NC2 binding site. The ability of these TBP mutants to bypass the requirement for Mot1 might be due to, or correlated with, an increased dependence on NC2, especially since there is a strong overlap in the Mot1- and NC2-dependent gene sets (18, 22, 33, 45). As shown in Fig. 4C, TBP Y185C was not as efficiently incorporated into NC2·TBP·DNA complexes as WT TBP, and no ternary complexes were observed using TBP F207L. The reduced interaction of these TBP variants with NC2 indicates that they did not bypass the requirement for Mot1 by augmenting interaction with this functionally related protein. Taken together, the results show that rather than having acquired or strengthened interactions with these other factors to compensate for the absence of Mot1, TBP Y185C and F207L were deficient in any of several different steps in PIC formation in vitro.
FIGURE 4.
Formation of ternary complexes with TBP Y185C and TBP F207L. A, EMSA with TBP alleles and TFIIA. Increasing concentrations of TFIIA (0.5, 1.5, or 5 units indicated by ramps; TBP·DNA binding units as defined previously (65)) were added to 20 nm concentrations of the indicated TBP and DNA (<1 nm). Lane 1 shows free probe; lane 2 shows 5 units of TFIIA alone. The positions of the free DNA, TBP·DNA, and TFIIA·TBP·DNA complexes are shown. B, EMSA with TBP and TFIIB. Increasing concentrations of TFIIB (20, 40, and 100 nm indicated by ramps) were added to 20 nm TBP and DNA (<1 nm). Samples were run on a TBE gel, which allowed for better detection of TFIIB·TBP·DNA ternary complexes than the TG gels used in Figs. 3 and 4A. TBP·DNA complexes are marginally stable in this gel system; the little TBP·DNA complex that was detectable (lane 3) ran indistinguishably from the more stable (and consequently more abundant) TFIIB·TBP·DNA complex (lanes 4-6), as indicated (20). ns refers to a nonspecific complex. The reaction in lane 2 contained 100 nm TFIIB. C, EMSA with TBP and NC2. Equal concentrations of recombinant Bur6 and Ydr1 (100 ng/μl) were combined to permit assembly into the NC2 complex, then 50, 100, 200, or 400 ng of NC2 (indicated by the ramps) was added to 20 nm TBP and radiolabeled DNA as in panels A and B. Binding reactions for TBP F207L are shown only for 200 ng (lane 13) and 400 ng (lane 14) of NC2. TBP·DNA complexes were unstable in this TBE gel system. The position of the NC2·TBP·DNA complex is indicated. ho refers to higher order species as seen previously (66).
Genome-wide Transcriptional Consequences of TBP Y185C and TBP F207L—We first used Northern analysis to compare RNA levels in WT, mot1, and each of the two bypass strains using candidate Mot1-dependent genes analyzed previously (Ref. 33; not shown). No obvious pattern emerged from this small collection of genes to explain how cells survived in the absence of Mot1, so a global approach to gene expression analysis was undertaken. Microarray analysis was performed using RNA isolated from cells containing WT MOT1, a temperature-sensitive allele of MOT1, mot1-14 (46), and either of the two bypass alleles in cells without MOT1. (Henceforth, mot1Δ cells harboring TBP Y185C or TBP F207L are referred to as “bypass” strains.) The genes whose expression was significantly different from the mean value of the WT dataset with 99% confidence were identified from duplicate hybridizations for each. The correlation between the Mot1-dependent gene set derived in this new analysis, and the Mot1-dependent genes identified in our previous study (33) was excellent. Virtually all of the Mot1-dependent genes previously identified were captured in the new dataset. However, with improved arrays and methodology, the data presented here reveal many more Mot1-dependent genes than were identified in the initial study. Of the 185 Mot1-dependent genes we identified previously, 180 were identified in this study, and the majority of the genes were misregulated in the same direction (supplemental Fig. S1). The increased sensitivity of the arrays was demonstrated by the substantial increase in significantly misregulated genes detected in this analysis; a total of 2912 Mot1-regulated genes were identified, which were approximately evenly split between Mot1-repressed and Mot1-activated genes.
Analysis of the bypass mutant data sets delineated the subset of genes that were misregulated in the bypass strains, and comparison of these genes with the Mot1-regulated genes defined by the mot1-14 data set allowed us to determine where the differences in expression occurred and how these TBPs bypass Mot1 function. The results are summarized in Fig. 5. When comparing the misregulated genes from the mot1-14, TBP Y185C, and TBP F207L bypass data sets, the majority of genes were found to be misregulated in all three strains (Fig. 5A). A significant number of genes, however, were uniquely affected in each of the three strains, indicating that the TBP bypass alleles restored expression to WT levels of some Mot1-regulated genes and induced misregulation of new subsets of yeast genes. The overlapping genes between the two bypass data sets highlight the differences and similarities in the functions of the two TBP alleles and support the notion that these alleles bypass Mot1 function by distinct mechanisms. As an estimate of the number of Mot1-regulated genes whose expression was restored to WT levels in the bypass strains, the number of genes significantly misregulated in the mot1-14 data set that were completely absent from one or both of the bypass data sets was determined (Fig. 5B). This calculation is an underestimate of the ability of the TBP alleles to bypass Mot1 function as expression of a number of genes was restored to near WT levels but was still significantly different enough to appear on the misregulated list for that particular allele. Such genes were not taken into consideration for this diagram. A total of 532 genes were restored to WT expression levels in the TBP Y185C bypass strain, whereas the total number for the TBP F207L bypass strain was about twice that, at 936 genes. 398 genes were shared between the two lists of “restored” genes. For both TBP Y185C and TBP F207L strains, the number of Mot1-activated and Mot1-repressed genes whose expression was restored to WT levels was about half (Fig. 5C). Interestingly, the large majority of these genes were misregulated less than 2-fold in mot1 cells, suggesting that the phenotypic effects are the consequence of small, cumulative effects rather than resulting from large changes in a few, highly influential genes.
FIGURE 5.
Summary of the genome-wide transcriptional effects of TBP Y185C and TBP F207L. A, Venn diagram depicting overlap between significantly misregulated genes (>99% confidence) in mot1-14, TBP Y185C, and TBP F207L strain data sets. B, Venn diagram showing the number of Mot1-dependent genes whose expression was restored to WT levels by TBP Y185C or TBP F207L and the overlap between the two. For panels A and B the area of the overlapping region is not proportional to the number of genes contained within that category. C, pie charts breaking down the genes in panel B according to whether they were originally Mot1-activated or Mot1-repressed and by-fold change. The numbers of genes affected less than 2-fold are shown in light blue and light red, and the numbers of genes affected greater than 2-fold are shown in dark blue and dark red, respectively. The slice of the pie is proportional to the number of genes contained within that category.
Gene Ontology (GO) analysis of the genes restored to WT expression levels by TBP Y185C and TBP F207L was conducted using GOstat (34). GOstat calculates the probability that the fraction of genes mapped to a given biological function is higher than expected by chance given the total number of yeast genes that map to that same function. TBP Y185C preferentially restored expression of genes whose products are involved in transport and DNA binding, whereas TBP F207L primarily restored expression of genes implicated in mating and cell growth (data not shown). Analysis of the overlapping genes from Fig. 5B gave no significantly enriched Gene Ontology categories. Thus, the differences in the mechanisms these alleles use to bypass Mot1 function correlate with differences in the biological processes in which the affected genes are involved.
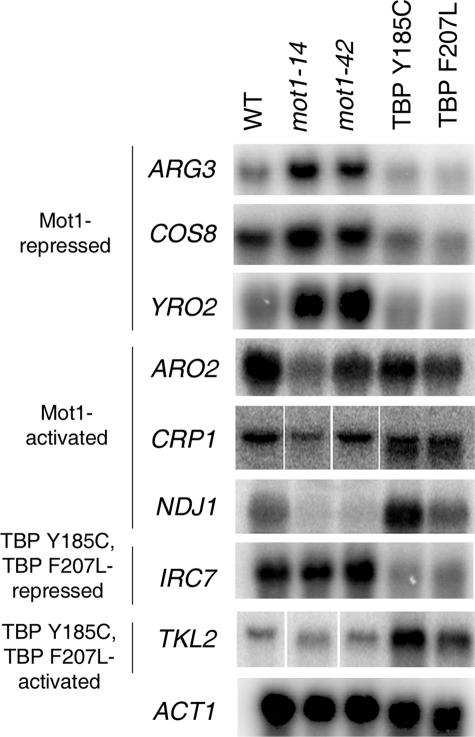
The results of the microarray analysis were validated by Northern blotting of selected genes fitting into several distinct regulatory categories (Fig. 6). These include genes that were originally Mot1-activated or -repressed but restored to WT expression level in the bypass strains and also genes that were newly repressed or activated in the bypass strains. The specific regulatory pattern for each gene is described in the legend. Northern blotting confirmed the expected expression patterns based on the microarray data for all genes tested.
FIGURE 6.
Confirmation of microarray results by Northern blotting. Genes were selected to confirm expression changes based on their regulatory patterns in mot1-14 and/or TBP Y185C and TBP F207L bypass strains. Message levels are shown for WT cells, mot1-14, and mot1-42 cells as well as the bypass strains TBP Y185C and TBP F207L. ARG3, COS8, and YRO2 are Mot1-repressed genes whose expression was restored to near WT levels in TBP Y185C and TBP F207L cells. ARO2, CRP1, and NDJ1 are Mot1-activated genes whose expression was restored to near WT levels in TBP Y185C and TBP F207L cells. IRC7 and TKL2 are genes that were misregulated in the TBP bypass strains only (repressed and activated, respectively). ACT1 is shown as a loading control.
Although the best characterized role for Mot1 is in regulation of RNA polymerase II transcription, Mot1 also regulates polymerase I transcription (29). In mot1 cells, ribosomal RNA synthesis and processing are impaired, and these defects are detectable by quantitation of the level of the unprocessed 35 S RNA species by Northern blotting (29). Given the critical link between cell growth and rRNA synthesis (47), we compared 35 S RNA levels in the bypass strains to the levels in WT and mot1 cells. As shown in Fig. 7, the elevated levels of 35 S RNA detectable in mot1 cells were similarly elevated in each of the bypass strains, indicating that the bypass TBPs do not suppress the ribosomal RNA synthesis and processing defect in mot1 cells. Thus, the effects of the bypass TBPs are RNA polymerase II-specific.
FIGURE 7.
Bypass TBPs do not suppress the defect in polymerase I transcription and RNA processing observed in mot1 cells. A, Northern blot showing the levels of 35 S ribosomal RNA in the indicated strains. TBP Y185C and TBP F207L refer to the bypass strains harboring the mutant TBPs and lacking Mot1. The 35 S label on the left of the upper blot is placed next to the unprocessed ribosomal RNA species detected as previously described (29). The same blot was probed for ACT1 as a loading control. B, quantitation of Northern blots similar the one shown in panel A. Averages of RNA levels in three independent experiments ± standard deviation are shown.
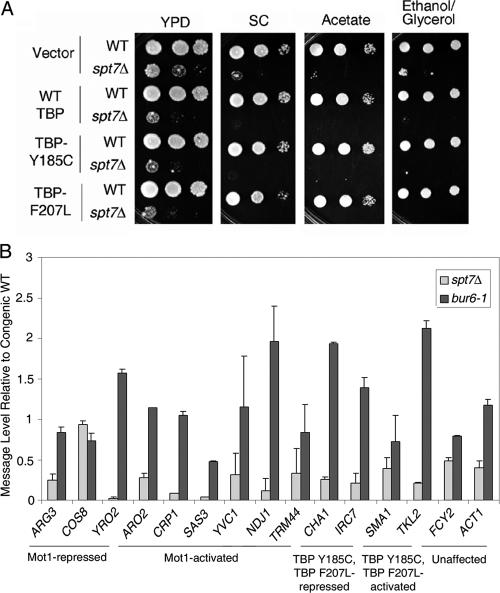
As SAGA, NC2, and Mot1 regulate many of the same genes (14, 18, 22, 33, 45, 48-50), we wondered if the bypass alleles functioned specifically to bypass the function of Mot1 or if they bypassed the requirement for SAGA or NC2 as well. Specificity in their ability to bypass Mot1 function would support the idea that these TBP variants have biochemical properties specifically adapted to obviate Mot1 activity. This idea was tested in several ways. Spt7 is required for structural integrity of the SAGA complex as well as the related SLIK complex (51, 52). Defects in spt7Δ cells can be detected by slow or no growth on various yeast media (53). WT and spt7Δ strains were transformed with the plasmids carrying WT TBP, TBP Y185C, or TBP F207L and cell growth was scored using a spot growth assay. As shown in Fig. 8A, no differences in the spt7Δ strains were detected on any of the media used to assay defects in SAGA function. Notably, expression of TBP Y185C or TBP F207L did not improve the growth of spt7Δ cells as would have been expected if these variants somehow affect SAGA-dependent gene expression globally rather than possessing a property that specifically bypasses Mot1 activity. For technical reasons, an analogous phenotypic experiment was not performed using an NC2 mutant strain. However, selected genes targeted by TBP Y185C and TBP F207L were tested for their dependence on the NC2 subunit Bur6 as well as the SAGA subunit Spt7 (Fig. 8B). Quantitation of message levels in bur6-1 and spt7Δ cells, compared with congenic WT cells, showed that regardless of whether genes were activated or repressed by Mot1 or whether their expression was not dependent on Mot1 but was affected by TBP Y185C or TBP F207L, the selected genes were in general slightly repressed by Bur6 and activated by SAGA. Thus, the genes regulated by TBP Y185C and TBP F207L did not display a diagnostic NC2 or SAGA dependence.
FIGURE 8.
Regulation by SAGA and Bur6 in TBP Y185C and TBP F207L bypass strains. A, spot tests show 10-fold serial dilutions of WT and spt7Δ strains transformed with plasmid vector, WT TBP, TBP Y185C, or TBP F207L plasmids as indicated. Note the slow growth phenotype of spt7Δ cells seen on YPD, synthetic complete media with glucose (SC), or SC containing acetate or ethanol/glycerol rather than glucose; no observable differences were seen in growth rate whether or not the strains harbored TBP Y185C or TBP F207L. B, quantitation of Northern blots to evaluate message levels for selected genes in bur6-1 and spt7Δ strains. Values represent the -fold change in message in the indicated mutant cell compared with its congenic wild type. Results are from two independent experiments; error bars indicate the S.E. between replicates. See Fig. 6 for more information about expression patterns of ARG3, COS8, YRO2, ARO2, CRP1, NDJ1, IRC7, and TKL2. SAS3, YVC1, and TRM44 are Mot1-activated genes whose expression was affected by TBP F207L only. CHA1 expression is repressed by the bypass mutants, and SMA1 is activated by them. FCY2 and ACT1 are included as control genes unaffected by TBP Y185C or F207L.
To gain an understanding of the transcriptional interplay between the TBP bypass alleles and SAGA and NC2 on a more global scale, the microarray data from TBP Y185C and TBP F207L were compared with the previously published data for the SAGA mutant spt20Δ (31) and bur6-1 cells (32). To obtain the most sensitive measure of similarity possible given differences in laboratory techniques and array sensitivities, the values for the overlapping genes from the complete data sets were compared. The correlation coefficients as well as the number and percentage of genes that were commonly up-regulated or down-regulated in the two mutants are shown in supplemental Table 1. For comparison, the gene lists from this study were analyzed in the same manner. This analysis did not provide an absolute measure of the overlap between significantly misregulated genes (because p values were not available for all data sets, this statistical measure of significance could not be considered). However, this analysis showed that ∼55% of the yeast genes were dependent on SAGA function, and ∼70% of the genes were dependent on Bur6 function in all three strains (supplemental Table 1). Thus, the genes whose expression was differentially affected in the bypass strains were not differentially dependent on Spt20 or Bur6. The combined results support the idea that the bypass TBPs specifically overcome loss of Mot1 activity rather than affect the requirement for SAGA or NC2 at some Mot1-regulated genes.
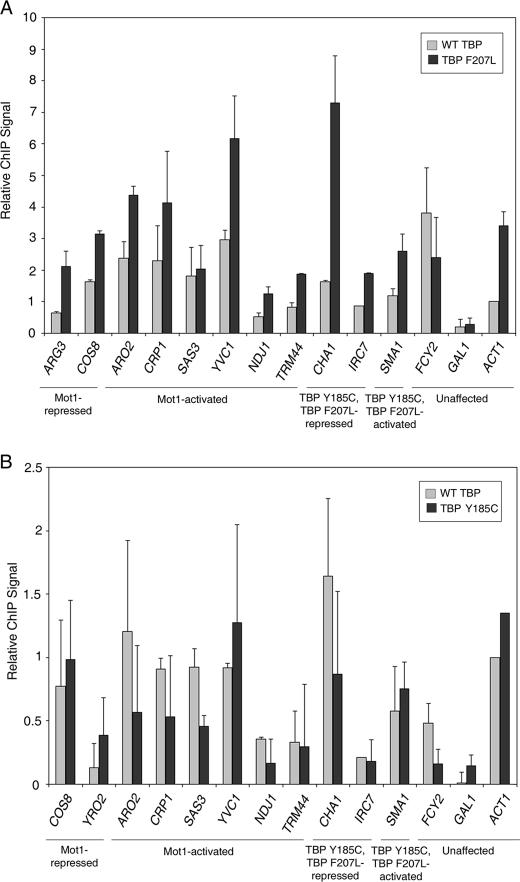
To determine whether the effects of the bypass TBPs on transcription were direct, ChIP was performed to compare WT and mutant TBP localization to promoters in TBP Y185C and TBP F207L strains (Fig. 9). Because TBP Y185C supports viability in the absence of WT TBP (Fig. 2C), its localization to promoters was expected. However, we wondered if the Y185C mutation somehow directed it preferentially to certain genes. Determining whether TBP F207L localized to certain genes in vivo was also critical because its in vitro DNA binding defect (Fig. 3) might reflect a mechanism for gene regulation that did not occur in association with chromatin. ChIP for WT and mutant TBP was performed in the indicated bypass strain in the absence of Mot1 protein by introducing a plasmid carrying Myc-tagged WT or mutant TBP. Myc-tagged WT TBP was assayed in parallel to compare the relative chromatin occupancy of the TBP mutants to WT TBP at the same loci and under the same conditions. Strikingly, with the exception of the repressed GAL1 locus, which served as a negative control, higher levels of chromatin-bound TBP F207L compared with WT TBP were found at most promoters tested (Fig. 9A). Given the DNA binding defect of TBP F207L in vitro (Fig. 3), TBP F207L DNA binding activity was re-examined using a DNA probe for the ARG3 promoter, a Mot1-repressed gene to which TBP F207L localized in vivo. EMSA revealed that WT and Y185C TBP bound similarly but that TBP F207L did not detectably bind to the probe (supplemental Fig. S2). Therefore, consistent with the binding data in Fig. 3, TBP F207L localization to ARG3 in vivo cannot be explained by acquisition of a novel DNA binding specificity for some sequence in this promoter.
FIGURE 9.
Recruitment of TBP Y185C and TBP F207L to chromatin in vivo. A, ChIP of Myc-TBP (WT, gray bars) or Myc-TBP F207L (black bars) to the indicated promoters. ChIP was performed in mot1Δ strains expressing both WT TBP and TBP F207L, one of which was Myc-tagged. B, ChIP of Myc-TBP WT (gray bars) or Myc-TBP Y185C (black bars) to the indicated promoters. ChIP was performed in mot1Δ strains expressing both WT TBP and TBP Y185C, one of which was Myc-tagged. Bars in panels A and B show the average signal obtained from two independent experiments, and error bars denote the S.E. Signal is relative to the average ACT1 signal for WT TBP in the same background.
In comparing the ChIP results (Fig. 9A) to the in vitro data (Fig. 3 and supplemental Fig. S2), the failure to detect in vitro binding of TBP F207L to the variety of DNA probes tested, including a promoter probe to which the protein localizes in vivo, suggests two possibilities. The first is that TBP F207L is directed to chromatin sites in vivo via association with TAFs. Although TAFs might stabilize a binding-defective TBP on chromatin, how could the F207L mutation enhance promoter binding compared with WT TBP? The N-terminal region of TAF1, called TAND1, interacts with the DNA binding surface of TBP (54). If the F207L mutation alleviated the inhibitory interaction of TAND1 with the TBP DNA binding surface, TFIID complexes possessing TBP F207L might bind better to promoters in vivo via stabilizing interactions between TAFs and DNA, without the hindrance of the TAND1-TBP interaction. To test this idea, we reasoned that if TBP F207L compromised the interaction with TAND1, then the genes differentially regulated by TBP F207L would be enriched in TAND1-dependent genes. TAND1-dependent gene expression was previously reported (55). However, as shown in supplemental Table 2, there is no statistically significant enrichment of TAND1-dependent genes in the TBP F207L dataset.
An alternative explanation for the F207L results is that the robust ChIP signals observed for TBP F207L are, paradoxically, not due to more stable promoter binding of TBP F207L to promoters compared with WT TBP. Instead, an increased frequency of transient interactions trapped by formaldehyde cross-linking could also give rise to such a result (for discussion, see Ref. 56). In this case, the TBP F207L ChIP data would be consistent with the in vitro data and would further support the idea that the dynamic behavior of TBP is critical for appropriate gene expression globally.
In agreement with the ability of TBP Y185C to support cell viability when provided as the sole source of TBP (Fig. 2C), no significant differences in the recruitment of WT TBP or TBP Y185C to chromatin were found for any of the genes tested (Fig. 9B). However, the ChIP signals obtained in the TBP Y185C strains were associated with inexplicably large errors, suggesting that TBP binding in this strain was highly variable. These errors were observed in multiple independent ChIP experiments and were in contrast to the much smaller errors observed with WT TBP or TBP F207L. As discussed below, this variability may reflect variable recruitment to promoters or variable resident times once bound to chromatin.
DISCUSSION
The isolation of TBP alleles that bypass the requirement for Mot1 provides insight into the regulation of TBP dynamics and complements other approaches used to study the Mot1 critical function in vivo (15, 22). A role for Mot1 in dissociating TBP from promoters has been well documented, but many questions remain about the nature of the TBP-Mot1 interaction and how this leads to different transcriptional outcomes at different promoters. The relationship between Mot1-catalyzed TBP·DNA dissociation and gene activation is particularly enigmatic. Mot1 might indirectly activate gene expression by displacement of “inactive” TBP complexes from promoters (14) or by ensuring a sufficient pool of free TBP to nucleate the assembly of functional PICs (16). At the Mot1-activated URA1 promoter, TBP has the propensity to bind to the TATA box in the wrong orientation to support functional PIC formation, thus providing a molecular explanation for how TBP can bind to promoters in a transcriptionally inactive form (21). However, changes in the TATA sequence that force TBP binding in the right orientation do not obviate the requirement for Mot1 for URA1 transcription, indicating that Mot1 has another activity as well (21).
Consistent with the above models for how TBP·DNA dissociation can regulate gene expression, the analysis of TBP Y185C and TBP F207L presented here shows that Mot1 function can be bypassed in vivo by proteins that have weakened interactions with DNA or other GTFs. These effects are specific in that Mot1 function cannot be bypassed by simply any TBP with a defect in these interactions. Rather, the allele specificity suggests that the defects must be appropriately tuned to permit transcription but facilitate instability that is somehow required for proper gene expression on a global scale. In the case of TBP F207L, the loss of a critical DNA binding side chain may impart weak but promiscuous DNA binding in vivo both in terms of the number and stability of sites and could allow enough flexibility in PIC formation for transcription to occur well enough to eliminate or reduce the need for Mot1. However, the fine line between generation of more “dynamic” interactions and simply making a defective protein is evident by the large number of genes that are misregulated in the bypass strains. Thus, these alleles restore normal expression at many genes but perturb it at others. Presumably, these differences in expression reflect differences in the requirements for dynamic assembly/disassembly of the PIC at different promoters.
TBP as a Rate-limiting Factor in PIC Formation—The binding of TBP to promoter DNA is the nucleating and often rate-limiting step in PIC formation (11, 57, 58). For this reason TBP is the direct target of both positive and negative regulation (4). With close to 90% of the yeast genome transcribed (59), a readily available pool of TBP is needed to ensure proper regulation of transcription in response to intracellular and extracellular cues.
Although there is a direct relationship genome-wide between TBP occupancy and transcriptional activity (60), simple overexpression of WT TBP has no detectable phenotype in WT cells (61, 62) and results in misregulation of less than 0.5% of the genome (55). Additionally, overexpression of TBP is lethal in cells defective for Mot1 function (62). This suggests that simply making TBP more “available” to bind to additional sites cannot bypass the requirement for Mot1. Overexpression of TBP can suppress defects in growth found by overexpression of Mot1 mutants that are catalytically impaired (46, 61). Therefore, TBP overexpression supports viability when TBP is trapped in catalytically inactive Mot1 complexes. This underscores the unique functions provided by the bypass alleles, which cannot be explained by either simple defects in TBP function or by effects on TBP dosage.
TBP Y185C and TBP F207L Achieve Similar Effects through Different Mechanisms—TBP Y185C and TBP F207L are differentially recruited to chromatin in vivo (Fig. 9) and display different DNA binding activities in vitro (Fig. 3). Furthermore, TBP Y185C can support viability as the only source of TBP (Fig. 2C), and TBP Y185C and TBP F207L have differential effects on genome-wide transcription (Fig. 5). These differences suggest that these two alleles achieve similar effects through distinct mechanisms. Although TBP Y185C is able to bind DNA in vitro similarly to WT TBP, its recruitment to chromatin in vivo is highly variable (Fig. 9B). This observation combined with its decreased ability to form complexes with TFIIA, TFIIB, and NC2 in vitro (Fig. 4) suggests that instability of TBP Y185C PICs is achieved mainly by decreased stability of the interactions with other PIC components. Other TBP alleles that are defective for TFIIB binding also partially bypass the requirement for Mot1 in vivo but to a lesser degree (Fig. 2A), suggesting that TBP Y185C is appropriately tuned for these activities. This is also supported by the elimination of the growth phenotype when another amino acid is substituted for the cysteine (Fig. 2B). Taken together, the results suggest that PICs assembled with TBP Y185C are less stable than PICs assembled with WT TBP.
In contrast to TBP Y185C, TBP F207L had no detectable DNA binding activity in vitro, and neither TFIIA, TFIIB, nor NC2 stabilized the TBP F207L-DNA interaction in vitro (Figs. 3 and 4). This defect in DNA binding is surprising in light of the fact that TBP F207L consistently associated with chromatin at a level higher than WT TBP (Fig. 9A). The ChIP results suggest that TBP F207L bypasses Mot1 function directly by interacting with chromatin at promoters rather than indirectly via interactions that occur in the nucleoplasm. A simple idea is that whereas TBP F207L is grossly defective for DNA binding in vitro, it possesses better promoter binding activity in vivo in association with TAFs. Such a situation is possible because TFIID DNA binding activity is competed by an interaction between the TBP DNA binding surface and TAND1, the N terminus of TAF1 (54). However, computational analyses (supplemental Table S2) show that TBP F207L has no differential effect on TAND1-affected genes (55), as would be expected if the F207L mutation perturbed the interaction between TBP and TAND1. We, therefore, conclude that the mechanistic explanation of the TBP F207L bypass activity is not explained by this TAF1 interaction. An alternative possibility is that like TBP Y185C, TBP F207L also directs the assembly of PICs that are more unstable than those assembled with WT TBP, but that for TBP F207L the instability arises from a defect in DNA binding rather than a defect in interaction with GTFs or regulators that interact with TBP. How could such a model be reconciled with the ChIP data in Fig. 9? Recent results indicate that the robustness of ChIP signals may not correlate with the stability of the interaction being measured. For example, Ace1 occupancy of the yeast CUP1 promoter can be readily demonstrated by ChIP, and yet the interaction is very short-lived as measured by live cell imaging (56). Additionally, TBP binding to promoters can be readily detected, but fluorescence recovery after photobleaching results demonstrate that there is no detectable TBP in yeast nuclei that is bound to chromatin for more than ∼15 s (15). Considered from this standpoint, the elevated ChIP occupancy of TBP F207L compared with WT might reflect more binding events that are nonetheless more transient than occur with WT TBP. If TBP F207L has a general DNA binding defect in vivo as suggested by the in vitro data, then perhaps there is a greater proportion of free TBP F207L than free WT TBP available for such transient promoter interactions. Of course, it is possible the F207L mutation somehow stabilizes TBP binding to chromatin in vivo but not to DNA in vitro. However, there is no biochemical evidence to suggest how such a scenario might occur. Taken together, we feel the likeliest interpretation is that the high ChIP signals for TBP F207L in vivo reflect high but transient occupancy of chromatin rather than stable binding.
Model; Increased PIC Instability Bypasses the Requirement for Mot1—In this study survival of mot1Δ cells is apparently the result of decreased stability of the interaction between TBP and DNA or GTFs. PIC instability could be important for several reasons. Without Mot1 to recycle stable TBP·DNA complexes, PICs may form at inappropriate locations, leading to the synthesis of aberrant RNAs. Inappropriate TBP binding may not lead to the formation of functional PICs but may instead be detrimental by sequestering TBP and/or interfering with the activities of other factors that require access to DNA. A novel, alternative idea is that dynamic instability of legitimate PICs may be important for normal gene expression and regulation. Although biochemical data support a model for PIC activity in which a stable scaffold facilitates reinitiation (63), other factors may make PIC instability at some promoters advantageous in vivo. Dynamic PIC instability may be important for transcription initiation at certain promoters, much like activator degradation or displacement is required for transcriptional activation at some promoters (64). Given the complexity of the interactions involved and the myriad ways in which they are regulated, it seems likely that PICs possess a range of stabilities in vivo. The idea that appropriately tuned PIC instability is required for proper gene expression is supported by the similarities in the behavior of TBP Y185C and F207L in vivo as revealed by the microarray data. Their similar effects on expression across a set of genes are consistent with restoration of appropriate transcription being attributable to their ability to introduce similar decrements in stability of the PIC but by employing different biochemical mechanisms.
Supplementary Material
Acknowledgments
We are grateful to Frank Pugh and Laurie Stargell for providing plasmids, to Sarah Juedes for help with the mutant screen, and to Arindam Dasgupta for the construction of point mutants. Microarray experiments were conducted by the Microarray Group at the National Center for Toxicogenomics/National Institutes of Health (NIEHS). We are grateful to Jennifer Collins for help with the microarray data analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant GM55763 (to D. T. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1 and 2.
Footnotes
The abbreviations used are: PIC, preinitiation complex; GTF, general transcription factor; TBP, TATA-binding protein; EMSA, electrophoretic mobility shift assays; WT, wild type; 5-FOA, 5-fluoro-orotic acid; ChIP, chromatin immunoprecipitation; DTT, dithiothreitol; YPD, yeast extract/peptone/dextrose; YPD/S, YPD with sorbitol; TAF, TBP-associated factor.
References
- 1.Reese, J. C. (2003) Curr. Opin. Genet. Dev. 13 114-118 [DOI] [PubMed] [Google Scholar]
- 2.Hahn, S. (2004) Nat. Struct. Mol. Biol. 11 394-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller, F., and Tora, L. (2004) EMBO J. 23 2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matangkasombut, O., Auty, R., and Buratowski, S. (2004) Adv. Protein Chem. 67 67-92 [DOI] [PubMed] [Google Scholar]
- 5.Hoopes, B. C., LeBlanc, J. F., and Hawley, D. K. (1992) J. Biol. Chem. 267 11539-11547 [PubMed] [Google Scholar]
- 6.Petri, V., Hsieh, M., Jamison, E., and Brenowitz, M. (1998) Biochemistry 37 15842-15849 [DOI] [PubMed] [Google Scholar]
- 7.Patikoglou, G. A., Kim, J. L., Sun, L., Yang, S.-H., Kodadek, T., and Burley, S. K. (1999) Genes Dev. 13 3217-3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson, G. H., Schroeder, S. C., Bai, Y., Weil, P. A., and Piston, D. W. (1997) Yeast 14 813-825 [DOI] [PubMed] [Google Scholar]
- 9.Colgan, J., and Manley, J. L. (1992) Genes Dev. 6 304-315 [DOI] [PubMed] [Google Scholar]
- 10.Davidson, I. (2003) Trends Biochem. Sci. 28 391-398 [DOI] [PubMed] [Google Scholar]
- 11.Li, X.-Y., Virbasius, A., Zhu, X., and Green, M. R. (1999) Nature 399 605-609 [DOI] [PubMed] [Google Scholar]
- 12.Darst, R. P., Wang, D., and Auble, D. T. (2001) EMBO J. 20 2028-2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira, L. A., Klejman, M. P., and Timmers, H. T. M. (2003) Gene (Amst.) 315 1-13 [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta, A., Juedes, S. A., Sprouse, R. O., and Auble, D. T. (2005) EMBO J. 24 1717-1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprouse, R. O., Karpova, T. S., Mueller, F., Dasgupta, A., McNally, J. G., and Auble, D. T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 13304-13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muldrow, T. A., Campbell, A. M., Weil, P. A., and Auble, D. T. (1999) Mol. Cell. Biol. 19 2835-2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrau, J.-C., Van Oevelen, C. J. C., Van Teeffelen, H. A. A. M., Weil, P. A., Holstege, F. C. P., and Timmers, H. T. M. (2002) EMBO J. 21 5173-5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geisberg, J. V., Moqtaderi, Z., Kuras, L., and Struhl, K. (2002) Mol. Cell. Biol. 22 8122-8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huisinga, K. L., and Pugh, B. F. (2007) Genome Biology http://genomebiology.com/2007/8/4/R46 [DOI] [PMC free article] [PubMed]
- 20.Auble, D. T., and Hahn, S. (1993) Genes Dev. 7 844-856 [DOI] [PubMed] [Google Scholar]
- 21.Sprouse, R. O., Shcherbakova, I., Cheng, H., Jamison, E., Brenowitz, M., and Auble, D. T. (2008) J. Biol. Chem. 283 24935-24948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Werven, F. J., van Bakel, H., van Teeffelen, H. A. A. M., Altelaar, A. F. M., Koerkamp, M. G., Heck, A. J. R., Holstege, F. C. P., and Timmers, H. T. M. (2008) Genes Dev. 22 2359-2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topalidou, I., Papamichos-Chronakis, M., Thireos, G., and Tzamarias, D. (2004) EMBO J. 23 1943-1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisberg, J. V., and Struhl, K. (2004) Mol. Cell 14 479-489 [DOI] [PubMed] [Google Scholar]
- 25.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auble, D. T., Wang, D., Post, K. W., and Hahn, S. (1997) Mol. Cell. Biol. 17 4842-4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987) Methods Enzymol. 154 164-175 [DOI] [PubMed] [Google Scholar]
- 28.Schmitt, M. E., Brown, T. A., and Trumpower, B. L. (1990) Nucleic Acids Res. 18 3091-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta, A., Sprouse, R. O., French, S., Aprikian, P., Hontz, R., Juedes, S. A., Smith, J. S., Beyer, A. L., and Auble, D. T. (2007) Mol. Cell. Biol. 27 2886-2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng, L. (2006) Bioinformatics 22 1111-1121 [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. I., Causton, H. C., Holstege, F. C. P., Shen, W.-C., Hannett, N., Jennings, E. G., Winston, F., Green, M. R., and Young, R. A. (2000) Nature 405 701-704 [DOI] [PubMed] [Google Scholar]
- 32.Geisberg, J. V., Holstege, F. C., Young, R. A., and Struhl, K. (2001) Mol. Cell. Biol. 21 2736-2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta, A., Darst, R. P., Martin, K. J., Afshari, C. A., and Auble, D. T. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2666-2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beissbarth, T., and Speed, T. P. (2004) Bioinformatics 20 464-465 [DOI] [PubMed] [Google Scholar]
- 35.Arndt, K. M., Ricupero-Hovasse, S., and Winston, F. (1995) EMBO J. 14 1490-1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, Y., Geiger, J. H., Hahn, S., and Sigler, P. B. (1993) Nature 365 512-520 [DOI] [PubMed] [Google Scholar]
- 37.Kim, T. K., Zhao, Y., Ge, H., Bernstein, R., and Roeder, R. G. (1995) J. Biol. Chem. 270 10976-10981 [DOI] [PubMed] [Google Scholar]
- 38.Kamada, K., Shu, F., Chen, H., Malik, S., Stelzer, G., Roeder, R. G., Meisterernst, M., and Burley, S. K. (2001) Cell 106 71-81 [DOI] [PubMed] [Google Scholar]
- 39.Klejman, M. P., Pereira, L. A., van Zeeburg, H. J. T., Gilfillan, S., Meisterernst, M., and Timmers, H. T. M. (2004) Mol. Cell. Biol. 24 10072-10082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blair, W. S., and Cullen, B. R. (1997) Mol. Cell. Biol. 17 2888-2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson-Fisher, A. J., Chitikila, C., Mitra, M., and Pugh, B. F. (1999) Mol. Cell 3 717-727 [DOI] [PubMed] [Google Scholar]
- 42.Stargell, L. A., and Struhl, K. (1995) Science 269 75-78 [DOI] [PubMed] [Google Scholar]
- 43.Stargell, L. A., and Struhl, K. (1996) Mol. Cell. Biol. 16 4456-4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, M., and Struhl, K. (1997) Mol. Cell. Biol. 17 1336-1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masson, P., Leimgruber, E., Creton, S., and Collart, M. A. (2008) Nucleic Acids Res. 36 539-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darst, R. P., Dasgupta, A., Zhu, C., Hsu, J.-Y., Vroom, A., Muldrow, T. A., and Auble, D. T. (2003) J. Biol. Chem. 278 13216-13226 [DOI] [PubMed] [Google Scholar]
- 47.Moss, T. (2004) Curr. Opin. Genet. Dev. 14 210-217 [DOI] [PubMed] [Google Scholar]
- 48.Huisinga, K. L., and Pugh, B. F. (2004) Mol. Cell 13 573-585 [DOI] [PubMed] [Google Scholar]
- 49.Zanton, S. J., and Pugh, B. F. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16843-16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Oevelen, C. J. C., van Teeffelen, H. A. A. M., and Timmers, H. T. M. (2005) Mol. Cell. Biol. 25 4863-4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pray-Grant, M. G., Schieltz, D., McMahon, S. J., Wood, J. M., Kennedy, E. L., Cook, R. G., Workman, J. L., Yates, J. R. I., and Grant, P. A. (2002) Mol. Cell. Biol. 22 8774-8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterner, D. E., Belotserkovskaya, R., and Berger, S. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11622-11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gansheroff, L. J., Dollard, C., Tan, P., and Winston, F. (1995) Genetics 139 523-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, D., Ishima, R., Tong, K. I., Bagby, S., Kokubo, T., Muhandiram, D. R., Kay, L. E., Nakatani, Y., and Ikura, M. (1998) Cell 94 573-583 [DOI] [PubMed] [Google Scholar]
- 55.Chitikila, C., Huisinga, K. L., Irvin, J. D., Basehoar, A. D., and Pugh, B. F. (2002) Mol. Cell 10 871-882 [DOI] [PubMed] [Google Scholar]
- 56.Karpova, T. S., Kim, M. J., Spriet, C., Nalley, K., Stasevich, T. J., Kherrouche, Z., Heliot, L., and McNally, J. G. (2008) Science 319 466-469 [DOI] [PubMed] [Google Scholar]
- 57.Klein, C., and Struhl, K. (1994) Science 266 280-282 [DOI] [PubMed] [Google Scholar]
- 58.Kuras, L., and Struhl, K. (1999) Nature 399 609-613 [DOI] [PubMed] [Google Scholar]
- 59.David, L., Huber, W., Granovskaia, M., Toedling, J., Palm, C. J., Bofkin, L., Jones, T., Davis, R. W., and Steinmetz, L. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5320-5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim, J., and Iyer, V. R. (2004) Mol. Cell. Biol. 24 8104-8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adamkewicz, J. I., Hansen, K. E., Prud'homme, W. A., Davis, J. L., and Thorner, J. (2001) J. Biol. Chem. 276 11883-11984 [DOI] [PubMed] [Google Scholar]
- 62.Auble, D. T., Hansen, K. E., Mueller, C. G. F., Lane, W. S., Thorner, J., and Hahn, S. (1994) Genes Dev. 8 1920-1934 [DOI] [PubMed] [Google Scholar]
- 63.Yudkovsky, N., Ranish, J. A., and Hahn, S. (2000) Nature 408 225-229 [DOI] [PubMed] [Google Scholar]
- 64.Collins, G. A., and Tansey, W. P. (2006) Curr. Opin. Genet. Dev. 16 197-202 [DOI] [PubMed] [Google Scholar]
- 65.Ranish, J. A., and Hahn, S. (1991) J. Biol. Chem. 266 19320-19327 [PubMed] [Google Scholar]
- 66.Cang, Y., Auble, D. T., and Prelich, G. (1999) EMBO J. 18 6662-6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar, R., Domrachev, M., and Lash, A.E. (2002) Nucleic Acids Res. 30 207-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.