Abstract
The accumulation of mutant protein in intracellular aggregates is a common feature of neurodegenerative disease. In Huntington disease, mutant huntingtin leads to inclusion body (IB) formation and neuronal toxicity. Impairment of the ubiquitin-proteasome system (UPS) has been implicated in IB formation and Huntington disease pathogenesis. However, IBs form asynchronously in only a subset of cells with mutant huntingtin, and the relationship between IB formation and UPS function has been difficult to elucidate. Here, we applied single-cell longitudinal acquisition and analysis to monitor mutant huntingtin IB formation, UPS function, and neuronal toxicity. We found that proteasome inhibition is toxic to striatal neurons in a dose-dependent fashion. Before IB formation, the UPS is more impaired in neurons that go on to form IBs than in those that do not. After forming IBs, impairment is lower in neurons with IBs than in those without. These findings suggest IBs are a protective cellular response to mutant protein mediated in part by improving intracellular protein degradation.
Huntington disease (HD)4 is a progressive incurable neurodegenerative disorder caused by the expansion of a polyglutamine (polyQ) stretch in the N-terminal end of the huntingtin (htt) protein above a threshold length of ∼36 (1). The deposition of polyQ-expanded aggregated mutant htt in inclusion bodies (IBs) is a hallmark of HD, and IBs are found in human post-mortem samples, transgenic mouse brain, and cell-culture models (2). The accumulation of ubiquitinated proteins in IBs has implicated the ubiquitin-proteasome system (UPS) in the pathogenesis of HD, amyotrophic lateral sclerosis, Parkinson disease, and polyQ-mediated disorders (3).
The UPS is a major pathway of intracellular protein degradation. After a series of three reactions, each catalyzed by a different set of enzymes, ubiquitin, a 76-amino acid polypeptide, forms an isopeptide bond with the amino group of lysine residues on substrate proteins. Several lysine residues within ubiquitin are sites for more ubiquitin additions. Once a protein accumulates four or more ubiquitins, it is efficiently targeted to the proteasome for degradation. The proteasome binds polyubiquitinated substrates and hydrolyzes ubiquitin isopeptide bonds, releasing ubiquitin moieties before degrading substrate proteins through chymotrypsin-like, trypsin-like, and post-glutamyl peptidase activities (3).
Increased polyubiquitin levels and changes in ubiquitin linkages accompany the accumulation of UPS substrates in the brains of HD patients and transgenic mice and in cellular HD models (4). UPS substrates accumulate throughout the cell in polyQ models, even before IB formation (5, 6). This has added to the confusion over whether polyQ expansion leads to toxicity through direct impairment of proteasomal degradation. Proteasomes have been reported to cleave polyQ stretches efficiently (7), inefficiently (8), or essentially not at all (9). In vivo, polyQ-dependent degeneration occurs with no detectable proteasome inhibition (10, 11) or is tightly linked to it (12, 13). The inability of some studies to detect UPS impairment in HD models may be due to the limited sensitivity of conventional approaches to identify cell-to-cell variations in UPS function.
The relationship between IB formation and UPS function has been difficult to determine. Protein turnover in cells with IBs is evidently reduced and accompanied by the accumulation of cellular proteins (14–16); HEK293 cells containing mutant htt IBs have a greater degree of UPS impairment than those without IBs (5). Proteasome subunits and heat shock proteins colocalize with IBs, but it is unclear if this colocalization facilitates protein delivery or unfolding at the mouth of active proteasomes, or if it harms proteasome function by sequestering essential cellular machinery (18). Some IBs are relatively static (8, 25), but the proteins in others are dynamically exchanged with cytoplasmic and nuclear pools (19, 20).
UPS function is critical to cellular homeostasis. Deletion of one of the two inducible polyubiquitin genes in mice leads to lower intracellular ubiquitin levels in germ cells and hypothalamic neurons. These same populations undergo cell-cycle arrest and hypothalamic neurodegeneration, respectively (22, 23). Cell lines expressing mutant huntingtin accumulate ubiquitinated proteins and undergo cell-cycle arrest in G2/M (5). In neurons, UPS impairment may lead to cell death through an accumulation of signals for apoptosis, a decrease in NF-κB signaling, sensitization to other toxic stimuli, remodeling of synapses, retraction of neurites, or other unidentified mechanisms (24). The effect of UPS impairment depends on cell type and cell cycle, and the relationship between UPS impairment and striatal neuronal survival is largely unknown.
Diffuse species of mutant htt induce IB formation and neuronal death in a protein concentration-dependent manner (2). IB formation delays neuronal death, suggesting that IB formation helps neurons cope with toxic diffuse mutant htt. Whether the effect of IB formation on survival is mediated through UPS function has been difficult to determine. IB formation and neuronal death occur asynchronously in overlapping but distinct subsets of neurons that express mutant htt. The observation that IB formation is not required for UPS impairment also complicates population analysis (6, 26).
To explore this problem, we applied single-cell analysis. We tracked single neurons over their entire lifetimes, gaining spatial and temporal resolution while simultaneously monitoring IB formation, UPS inhibition, and neuronal toxicity.
EXPERIMENTAL PROCEDURES
Plasmids—mRFP (27), pCS2-Venus (28), and pEGFP-CL1(5), pGW1-GFP, pGW1-httQ72-eGFP, pGW1-mRFP (2) have been described. pGW1-httQ72-CFP was generated from pGW1-httQ72-eGFP. pGW1-mRFPu (mRFPu) was generated by subcloning mRFP1 from pcDNA3.1(+) into pEGFP-CL; mRFP1-CL1 was then subcloned into pGW1. pGW1-GFPu was constructed by excising EGFP-CL1 from pEGFP-CL1 and inserting it into pGW1. pGW1-Venus-CL1 (Venusu) was generated by subcloning Venus from pCS2-Venus into pcDNA3.1(+). The stop codon from Venus was removed and replaced by the sequence AGATCTCG. The CL1 sequence (5) was introduced at the 3′-end of Venus. Venus-CL1 was then subcloned into pGW1. pCS2-UbG76V-Venus (UbG76V-Venus) was generated by PCR of UbG76V from UbG76V-GFP (29).
Cell Culture—Striata from rat embryos (E17–18) were dissected, dissociated, and plated on 24-well tissue-culture plates (5.8 × 105/well) coated with poly-d-lysine and laminin (BD Biosciences, San Jose, CA) as described (2, 30). The cells were grown in 1 ml of modified neuronal culture medium (NCM). Cells were fed every 5–7 days by replacement with equal measures of conditioned and fresh neuronal culture medium.
Transfection, Pharmacology, and Colocalization—Primary cultures were transfected 5–7 days in vitro with combinations of pGW1-GFPu and pGW1-mRFP, pGW1-mRFPu, and pGW1-GFP, pGW1-Venusu, or pCS2-UbG76V-Venus, and pGW1-CFP, and pGW1-httQ72-eGFP, pGW1-mRFPu, and pGW1-BFP, or pGW1-httQ72-CFP and pYFP-LMP2 in a 1:1 or 1:1:1 molar ratio with 2–4 μg of total plasmid DNA per well. Our transfection protocol was described (2). MG132 (Sigma-Aldrich), epoxomicin (Boston Biochem, Cambridge, MA), and Bafilomycin A1 (Sigma-Aldrich) were added in 1 ml of conditioned NCM per well 12–60 h after transfection.
Colocalization of fluorescence was calculated using Metamorph. Briefly, images of fluorescence from CFP-htt, ubiquitin staining, or LMP-YFP were analyzed, and pixels were classified as “positive” if their intensity was 3× greater than background pixels. The fraction of positive pixels for CFP-htt IBs that overlapped positive pixels of ubiquitin staining or LMP-YFP fluorescence was calculated with Metamorph for (n = 20 neurons).
Live-Cell Imaging and Analysis—Images of cells were obtained with a robotic microscope system as described (2, 32). Briefly, the imaging was performed with a Nikon TE300 inverted microscope with a long working-distance Nikon 20× (NA 0.45) objective. Stage movements and focusing were performed using a Proscan II stage controller (Prior Scientific, Rockland, MA). Samples were illuminated with a 175 watt Xenon Lambda LS illuminator (Sutter Instruments, Novato, CA). Blue, green, and red fluorescent protein (BFP, GFP, and RFP, respectively) images were captured using an 86014 beamsplitter and 350/50×; 465/30m, 480/40×; 517/30m and 580/20×; 630/60m fluorescence filters respectively. CFP, Venus, and RFP images were captured using a 86006 beamsplitter and 420/35×; 470/30m, 500/20×; 535/30m, and 580/20×; 630/60m fluorescence filters (Chroma Corp, Rockingham, VT). Algorithms for plate registration, stage movements, filter movements, focusing, and acquisition were generated with Metamorph imaging software (Molecular Devices, Sunnyvale, CA). Images were analyzed manually using Metamorph software. Fully automated acquisition and analysis algorithms have been created (Media Cybernetics, Bethesda, MD). Survival analysis was performed with the Statview software package (SAS Institute, Cary, NC); t tests for comparisons of means and two-sample Kolmogorov-Smirnov tests for comparisons of distributions were performed with Prism (Graphpad Software, San Diego, CA).
RESULTS
Longitudinal Live-Cell Monitoring of UPS Function in Primary Neurons—To monitor dynamic changes in protein degradation in live cells, we used a unique high-throughput image acquisition platform (2, 32) and fluorescent protein substrates of UPS degradation. We used fluorescent proteins with the CL1 peptide fused to the C terminus (34) or a non-hydrolyzable ubiquitin moiety (UbG76V) fused to the N terminus (35) to target them to the UPS for degradation. These destabilized fluorescent proteins were transfected into primary neurons and fluorescence in individual cells was monitored for hours or days to detect changes in the degradation of UPS substrates. To control for nonspecific changes in transcription and protein handling while monitoring cell survival (2), we co-transfected and tracked the fluorescence of unmodified fluorescent proteins in the same cells.
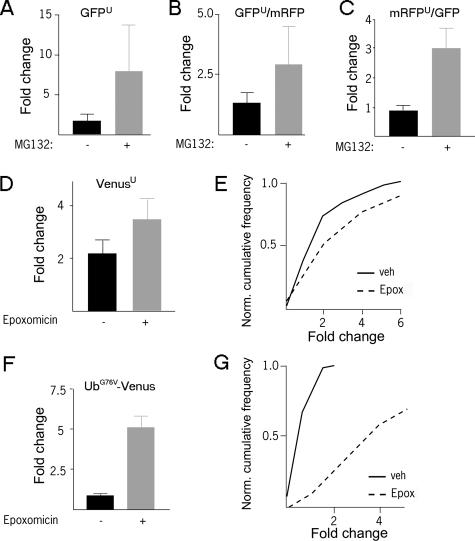
Destabilized Fluorescent Proteins Accumulate after Proteasome Inhibition in Primary Neurons—Fluorescence intensity in live cells is an accurate indicator of the amount of fluorescent protein within the cell (2). Fluorescence levels in primary striatal neurons of a destabilized form of enhanced GFPu (5) (Fig. 1A), monomeric mRFPu (27) (Fig. 1C), or two forms of the enhanced yellow fluorescent protein variant Venus (UbG76V-Venus and Venusu) (28) (Fig. 1, D–G) increased after treatment with proteasome inhibitor, even when changes in fluorescence of unmodified spectrally distinct fluorescent proteins in the same cells was controlled for (Fig. 1, B, C, E, G). The significant and rapid increase in fluorescence of these reporters from low baseline levels after proteasome inhibition in primary neurons is in agreement with previous work in cell lines (5, 6, 26). Addition of the CL1 peptide or UbG76V degron to fluorescent proteins did not cause the proteins to aggregate when they were expressed in neurons, unlike observations from cell lines (36).
FIGURE 1.
Levels of proteasome reporters increase after inhibition of proteasome. A, after transfection with GFPu and mRFP, striatal neurons were treated with 50 μm MG132 for 12 h. GFP fluorescence (A) and the ratio of GFPu/mRFP fluorescence (B) both increase after treatment relative to control. C, after transfection with mRFPu and GFP, striatal neurons were treated with 50 μm MG132 for 12 h. The mRFPu/GFP ratio is significantly greater than the control (p < 0.02). D, after transfection with Venusu and CFP, striatal neurons were treated with 2 μm epoxomycin (solid lines) or vehicle (broken lines) for 10 h. Both mean change in Venusu/CFP fluorescence (D) and single-cell distributions of Venusu/CFP fluorescence (E) are increased relative to control (p < 0.05, p < 0.05). F, after transfection with UbG76V-Venus and CFP, striatal neurons were treated with 2 μm epoxomycin for 10 h. Both mean change in UbG76V-Venus/CFP fluorescence (F) and single-cell distributions of UbG76V-Venus/CFP fluorescence (G) are increased (p < 0.05, p < 0.01). Experiments were repeated twice with over 50 cells analyzed in each condition.
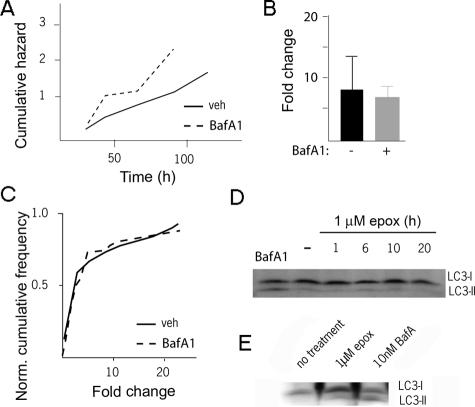
Inhibiting Autophagy Does Not Result in Accumulation of UPS Reporters in Primary Neurons—To ensure that these destabilized proteins were targeted primarily to the UPS for degradation, we used Bafilomycin A1 (BafA1) to inhibit autophagy. BafA1, a vacuolar ATPase inhibitor, prevents autophagosome-lysosome fusion and causes the accumulation of substrates targeted for macroautophagy (37). BafA1 caused a rapid accumulation of the membrane-bound form of microtubule-associated protein 1 light chain 3 (LC3-II) and was toxic to primary neurons (Fig. 2, A and D), but BafA1 did not increase levels of UPS reporters (Fig. 2, B and C).
FIGURE 2.
Limited interaction between the UPS and autophagic pathways in neurons. A, 24 h after cotransfection with UbG76V-Venus and CFP, striatal neurons were treated with vehicle or 50 nm BafA1. BafA1 treatment caused a significant amount of toxicity above control (p < 0.03, top line). Mean UbG76V-Venus/CFP ratio (B) and the distribution of the single-cell changes in UbG76V-Venus/CFP (C) in these cells did not increase above control in 20 h after BafA1 addition. D, neurons or HEK293 cells (E) were treated with BafA1 or epoxomycin, followed by Western blotting with an LC3 antibody. While BafA1 caused accumulation of LC3-II in both neurons and HEK293 cells, epoxomycin increased LC3-II levels only in HEK293 cells. Unlabeled lanes in E are lysates from cells transfected with LC3.
Proteasome Inhibition Does Not Change LC3-II Levels in Primary Neurons—Proteasome inhibition can increase flux through the autophagic pathway in some cells (13). To determine if autophagic activity could be confounding fluorescent reporter measurements of UPS function, we examined the activity of the autophagic pathway after proteasome inhibition. The level of LC3-II is commonly used as a surrogate for the number of autophagosomes and flux through the macroautophagic pathway. After treatment with epoxomicin, primary neurons showed no change in LC3-II levels (Fig. 2D), though as seen in previous reports, LC3-II accumulated in HEK293 cells (Fig. 2E).
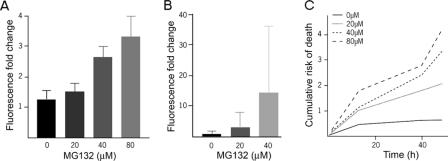
UPS Reporter Fluorescence Demonstrates a Graded Response to Proteasome Inhibition—Having validated the use of destabilized fluorescent proteins as reporters of UPS function in primary neurons, we then examined the nature of their response to varying levels of proteasome impairment. We co-transfected mRFPu and GFP into primary striatal neurons and treated the cells with increasing doses of the proteasome inhibitor MG132. Though fluorescent UPS reporters have been reported to relocalize to IBs, we found that mRFPu fluorescence remained diffuse in striatal neurons after proteasome inhibitor treatment (6). As early as 2.5 h after addition of MG132, reporter fluorescence increased in proportion to the amount of MG132 added (Fig. 3A), and reporter fluorescence continued to increase over time (Fig. 3B). Thus, in primary neurons, the increase in fluorescence of these proteins faithfully reports the extent of proteasome impairment (5, 6).
FIGURE 3.
Inhibition of proteasome activity is toxic in a dose-dependent fashion. A, UPS reporter fluorescence shows a dose-dependent response to MG132 treatment. MG132 at the indicated doses was added to striatal neurons 24 h after transfection with mRFPu and GFP. The change in mRFPu/GFP ratio over the first 2.5 h after MG132 administration is shown. B, UPS reporter fluorescence continues to increase up to 12 h after the addition of MG132. Note the difference in scale with A. Measurements from 80 μm were excluded due to noticeable toxicity. C, MG132 is toxic to neurons in a dose-dependent fashion. The same neurons shown in A and B were observed with the risk of death as shown. Longitudinal analysis was repeated twice on different transfections, with n > 50 for each treatment in each experiment.
By monitoring individual cells treated with MG132 over days, we determined the effect of increasing proteasome inhibition on the survival of primary striatal neurons. When the dose of MG132 increased, neurons died faster (Fig. 3C). These neurons demonstrated a proportional relationship between proteasome impairment and the accumulation of UPS substrates; similarly, there was a proportional relationship between proteasome impairment and neuronal toxicity.
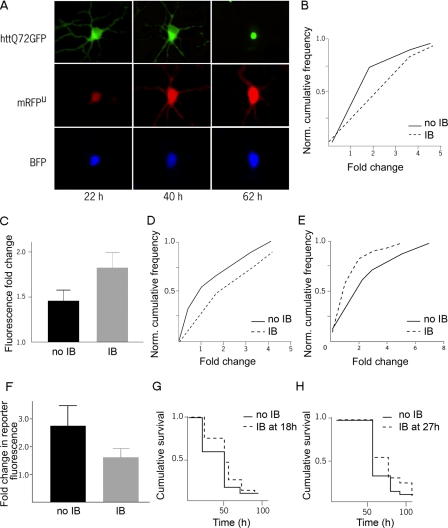
Longitudinal Live Cell Detection of UPS Function in a Primary Neuronal Model of HD—We then examined a primary striatal model of HD (2, 30) and prospectively followed visual markers of UPS function, IB formation, and neuronal viability in single cells. This model reproduces key features of HD, including neuronal subtype specificity (30) and polyQ length-dependent toxicity (2, 30). To induce the HD disease phenotype in this model, we transiently transfected an N-terminal htt fragment with 72 glutamines fused to GFP (httex1-Q72-GFP). We simultaneously introduced mRFPu and BFP into the same neurons to monitor UPS impairment and cell viability, respectively. Virtually all IBs in this model stain with ubiquitin and colocalize with proteasome subunits (supplemental Fig. S1). From series of images of individual neurons, we quantified single-cell changes in UPS reporter fluorescence over the lifetimes of cells expressing the httex1-Q72-GFP protein (Fig. 4A).
FIGURE 4.
IB formation and UPS function in primary neurons. A, GFP-htt, BFP, and mRFPu were imaged over the course of days to follow htt IB formation, UPS impairment, and neuronal survival. Single-cell distributions (B) or population means (C) of the change in mRFPu/GFP fluorescence in the interval preceding IB formation at 54 h. The increase in mRFPu/GFP ratio was higher in neurons that went on to form IBs (p < 0.05, p < 0.05). D, in a parallel experiment, single-cell distributions of the change in mRFPu/GFP fluorescence in the interval preceding IB formation at 76 h also show higher UPS impairment in those neurons that will go on to form IBs (p < 0.05). After 54 h, single-cell distributions (E) or population means (F) show a greater increase in mRFPu/GFP fluorescence in those cells that did not form IBs (p < 0.05, p < 0.05). The survival of those neurons that formed htt IBs at 18 h (G) or 27 h (H) was better than the survival of neurons that survived at least that long but never formed IBs (p < 0.01, p < 0.03). Longitudinal analysis was repeated twice in different experiments with over 300 cells analyzed in each experiment, with n > 30 for each cohort.
Would UPS function differ in neurons that do and do not form IBs? By reviewing images from our longitudinal analysis experiments, we identified neurons that had or had not formed an IB at some point over the course of the experiment. From these two groups, we then chose neurons that were from the same well of the culture dish to form two cohorts based on IB formation. To reduce potential biases introduced by using IB formation as a selection criterion, we included only neurons that had already lived the same length of time in vitro. We then monitored UPS reporter fluorescence in neurons before, during, and after IB formation and compared it to that in the cohort of age-matched neurons that did not form IBs.
UPS Impairment Precedes IB Formation—Those cells that would go on to form IBs had significantly larger increases in UPS reporter fluorescence before IB formation, both in the single-cell distribution of reporter fluorescence (Fig. 4B) and in mean reporter fluorescence (Fig. 4C). This relationship was independent of the time at which IBs formed (Fig. 4D).
UPS Impairment Decreases after IB Formation—To determine if IB formation improves or worsens UPS function, we examined UPS reporter fluorescence in neurons during and after IB formation. We compared these measurements to UPS reporter fluorescence in an otherwise matched cohort of neurons that did not form IBs over the same interval. To again reduce potential biases introduced by using IB formation as a selection criterion, neurons from each cohort were matched for the length of time they lived in vitro. We found that neurons that formed IBs had significantly smaller increases in UPS reporter fluorescence (Fig. 4, E and F), indicating that less UPS impairment occurs in cells after IB formation than in cells that did not form IBs.
IB Formation Improves Neuronal Survival—To determine if this reduced UPS impairment changes neuronal survival, we compared the survival of neurons that we analyzed for UPS function. When we examined matched cohorts of neurons transfected with httex1-Q72-GFP, mRFPu, and BFP that formed or did not form IBs, those cells that formed IBs survived longer (Fig. 4, G and H). This finding agrees with previous results showing that neurons survive longer if they form IBs (2).
DISCUSSION
By applying a high-throughput single-cell longitudinal imaging platform to a neuronal model, we were able to examine the events in the cellular pathogenesis of HD with improved sensitivity and temporal resolution. Through the use of spectrally distinct fluorescent species, we simultaneously monitored neuronal viability, htt IB formation, and intracellular protein degradation. We found that neurons that form IBs have increased UPS impairment preceding IB formation and less UPS impairment after IB formation than cells that do not form IBs. Though tonic UPS inhibition is toxic to primary striatal neurons, neurons that formed IBs survived better than those that did not. These results support a model in which IB formation reflects a beneficial cellular response to mutant protein, mediated in part by restoring UPS function.
Though multiple pathways of intracellular protein degradation may handle aggregation-prone protein, we found that some proteins are likely targeted primarily to the UPS for degradation. In our experience with fluorescent UPS reporters, we found little evidence that they are routinely degraded by autophagy. Though it is clear that autophagy modulates the turnover and toxicity of aggregation prone-proteins, the addition of the CL1 or UbG76V degron does not cause fluorescent proteins to aggregate in neurons. This discrepancy with other reports in cell lines may be due to lower expression levels in neurons after transient transfection.
The finding that proteasome inhibition is not sufficient to change the flux through the autophagic pathway in primary neurons also highlights possible differences between mammalian neurons and other model systems. The difference in behavior of the autophagic pathway in mammalian neurons may be due to a difference in constitutive activity (39). While most non-neuronal cells upregulate autophagy after 24 h of starvation, neurons do not in vivo (40) or in vitro5 even after longer starvation periods. The finding that the deletion of essential autophagic machinery results in a neurodegenerative phenotype points to a critical role in neuronal function and survival (38, 41).
Though it remains unclear how IB formation is functionally linked to an improvement in UPS function, one possibility is that IB formation is a step toward shunting aggregation-prone proteins normally targeted to the UPS to other pathways of intracellular protein degradation. In both yeast and mammalian cells, misfolded and aggregation-prone proteins may be targeted to different intracellular compartments depending on the availability of ubiquitin (31). Differential localization may be one component of targeting proteins to the autophagic pathway of protein degradation, which has been implicated in the clearance of aggregation-prone protein, including mutant htt (17, 21). If expanded polyQ tracts impair the ability of the proteasome to degrade other cellular proteins (9) or if ubiquitination is inadequate due to ubiquitin sequestration by IBs, shifting polyQ degradation from the UPS to the autophagic pathway could effectively increase the flux of other proteins through the UPS.
A second possibility that is not mutually exclusive is that IB formation is part of a cellular program to more efficiently degrade protein through the UPS. The recruitment of chaperones and proteasomal machinery to intracellular inclusions varies based on protein and cell type (19, 25). Though the IBs in our primary neuronal model are long-lived, with fewer than 2% disappearing before the neuron that contains them dies (2), a small proportion of cells can clear IBs, and a detailed longitudinal analysis of these cells will likely be informative.
Previous work suggested that IB formation safely sequesters more toxic forms of mutant htt to improve neuronal survival. This study suggests two additional mechanisms by which IB formation might contribute to improved cell survival after IB formation. First, we found that tonic UPS inhibition is toxic and that IB formation is associated with a relative improvement in UPS function. Thus, IB formation may partially restore longevity by improving UPS throughput and consequently lowering the overall cellular burden of misfolded proteins. A second but related possibility is suggested by reports that transient sublethal proteasome inhibition can induce cells to adapt in ways that protect them against further insults (33). Transient proteasome inhibition might trigger a cell-wide adaptive response in neurons that may involve coordinated changes in molecular chaperones and protein turnover pathways. If so, such an adaptive response may be important in a variety of neurodegenerative diseases that result from misfolded intracellular proteins.
Supplementary Material
Acknowledgments
We thank R. Kopito for pEGFP-CL1, V. Rao for pGW1-GFPu, A. Miyawaki for pCS2-Venus, M. Mancini for pYFP-LMP2, and N. Dantuma for UbG76V-GFP; members of the Finkbeiner laboratory for insightful discussions; S. Ordway and G. Howard for editorial assistance; and K. Nelson for administrative assistance. The animal care facility was supported in part by a National Institutes of Health Extramural Research Facilities Improvement Project (C06 RR018928).
This work was supported, in whole or in part, by National Institutes of Health Grants R01 2NS039074 and R01 NS045191 from the NINDS (to S. F.) and Grant P01 AG022074 from the NIA. This work was also supported by the Taube Family Foundation Program in Huntington Disease, and the Gladstone Institutes (to S. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: HD, Huntington Disease; UPS, ubiquitin-proteasome system; IB, inclusion body.
A. Tsvetkov and S. Finkbeiner, unpublished observations.
References
- 1.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575-621 [DOI] [PubMed] [Google Scholar]
- 2.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R., and Finkbeiner, S. (2004) Nature 431 805-810 [DOI] [PubMed] [Google Scholar]
- 3.Hershko, A., Ciechanover, A., and Varshavsky, A. (2000) Nat. Med. 6 1073-1081 [DOI] [PubMed] [Google Scholar]
- 4.Bennett, E. J., Shaler, T. A., Woodman, B., Ryu, K.-Y., Zaitseva, T. S., Becker, C. H., Bates, G. P., Schulman, H., and Kopito, R. R. (2007) Nature 448 704-708 [DOI] [PubMed] [Google Scholar]
- 5.Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001) Science 292 1552-1555 [DOI] [PubMed] [Google Scholar]
- 6.Bennett, E. J., Bence, N. F., Jayakumar, R., and Kopito, R. R. (2005) Mol Cell 17 351-365 [DOI] [PubMed] [Google Scholar]
- 7.Michalik, A., and Van Broeckhoven, C. (2004) Neurobiol. Dis. 16 202-211 [DOI] [PubMed] [Google Scholar]
- 8.Holmberg, C. I., Staniszewski, K. E., Mensah, K. N., Matouschek, A., and Morimoto, R. I. (2004) EMBO J. 23 4307-4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N., and Goldberg, A. L. (2004) Mol. Cell 14 95-104 [DOI] [PubMed] [Google Scholar]
- 10.Bowman, A. B., Yoo, S.-Y., Dantuma, N. P., and Zoghbi, H. Y. (2005) Hum. Mol. Genet. 14 679-691 [DOI] [PubMed] [Google Scholar]
- 11.Bett, J. S., Goellner, G. M., Woodman, B., Pratt, G., Rechsteiner, M., and Bates, G. P. (2006) Hum. Mol. Genet. 15 33-44 [DOI] [PubMed] [Google Scholar]
- 12.Khan, L. A., Bauer, P. O., Miyazaki, H., Lindenberg, K. S., Landwehrmeyer, B. G., and Nukina, N. (2006) J. Neurochem. 98 576-587 [DOI] [PubMed] [Google Scholar]
- 13.Pandey, U. B., Nie, Z., Batlevi, Y., McCray, B. A., Ritson, G. P., Nedelsky, N. B., Schwartz, S. L., DiProspero, N. A., Knight, M. A., Schuldiner, O., Padmanabhan, R., Hild, M., Berry, D. L., Garza, D., Hubbert, C. C., Yao, T.-P., Baehrecke, E. H., and Taylor, J. P. (2007) Nature 447 859-863 [DOI] [PubMed] [Google Scholar]
- 14.Verhoef, L. G. G. C., Lindsten, K., Masucci, M. G., and Dantuma, N. P. (2002) Hum. Mol. Genet. 11 2689-2700 [DOI] [PubMed] [Google Scholar]
- 15.Chai, Y., Shao, J., Miller, V. M., Williams, A., and Paulson, H. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 9310-9315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, Q., Lewis, J. J., Strum, K. M., Dimayuga, E., Bruce-Keller, A. J., Dunn, J. C., and Keller, J. N. (2002) J. Biol. Chem. 277 13935-13942 [DOI] [PubMed] [Google Scholar]
- 17.Iwata, A., Christianson, J. C., Bucci, M., Ellerby, L. M., Nukina, N., Forno, L. S., and Kopito, R. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13135-13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H., and Wanker, E. E. (2001) Mol. Biol. Cell 12 1393-1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenoien, D. L., Mielke, M., and Mancini, M. A. (2002) Nat Cell Biol. 4 806-810 [DOI] [PubMed] [Google Scholar]
- 20.Taylor, J. P., Tanaka, F., Robitschek, J., Sandoval, C. M., Taye, A., Markovic-Plese, S., and Fischbeck, K. H. (2003) Hum. Mol. Genet. 12 749-757 [DOI] [PubMed] [Google Scholar]
- 21.Rubinsztein, D. C. (2006) Nature 443 780-786 [DOI] [PubMed] [Google Scholar]
- 22.Ryu, K. Y., Sinnar, S. A., Reinholdt, L. G., Vaccari, S., Hall, S., Garcia, M. A., Zaitseva, T. S., Bouley, D. M., Boekelheide, K., Handel, M. A., Conti, M., and Kopito, R. R. (2008) Mol. Cell. Biol. 28 1136-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu, K. Y., Garza, J. C., Lu, X. Y., Barsh, G. S., and Kopito, R. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4016-4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller, J. N., Gee, J., and Ding, Q. (2002) Ageing Res. Rev. 1 279-293 [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, G., Kim, S., and Morimoto, R. I. (2006) J. Biol. Chem. 281 4477-4485 [DOI] [PubMed] [Google Scholar]
- 26.Rusmini, P., Sau, D., Crippa, V., Palazzolo, I., Simonini, F., Onesto, E., Martini, L., and Poletti, A. (2007) Neurobiol. Aging 28 1099-1111 [DOI] [PubMed] [Google Scholar]
- 27.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7877-7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K., and Miyawaki, A. (2002) Nat. Biotechnol. 20 87-90 [DOI] [PubMed] [Google Scholar]
- 29.Lindsten, K., Menendez-Benito, V., Masucci, M. G., and Dantuma, N. P. (2003) Nat. Biotechnol. 21 897-902 [DOI] [PubMed] [Google Scholar]
- 30.Saudou, F., Finkbeiner, S., Devys, D., and Greenberg, M. E. (1998) Cell 95 55-66 [DOI] [PubMed] [Google Scholar]
- 31.Kaganovich, D., Kopito, R., and Frydman, J. (2008) Nature 454 1088-1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arrasate, M., and Finkbeiner, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3840-3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stangl, K., Gunther, C., Frank, T., Lorenz, M., Meiners, S., Ropke, T., Stelter, L., Moobed, M., Baumann, G., Kloetzel, P.-M., and Stangl, V. (2002) Biochem. Biophys. Res. Commun. 291 542-549 [DOI] [PubMed] [Google Scholar]
- 34.Gilon, T., Chomsky, O., and Kulka, R. G. (1998) EMBO J. 17 2759-2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dantuma, N. P., Lindsten, K., Glas, R., Jellne, M., and Masucci, M. G. (2000) Nat. Biotechnol. 18 538-543 [DOI] [PubMed] [Google Scholar]
- 36.Almeida, C. G., Takahashi, R. H., and Gouras, G. K. (2006) J. Neurosci. 26 4277-4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto, A., Tagawa, Y., Yoshimori, T., Moriyama, Y., Masaki, R., and Tashiro, Y. (1998) Cell Struct. Funct. 23 33-42 [DOI] [PubMed] [Google Scholar]
- 38.Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., and Mizushima, N. (2006) Nature 441 885-889 [DOI] [PubMed] [Google Scholar]
- 39.Massey, A. C., Zhang, C., and Cuervo, A. M. (2006) Curr Top Dev. Biol. 73 205-235 [DOI] [PubMed] [Google Scholar]
- 40.Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T., and Ohsumi, Y. (2004) Mol. Biol. Cell 15 1101-1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J.-I., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E., and Tanaka, K. (2006) Nature 441 880-884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.