Abstract
Micro-RNAs (miRNAs) are important in regulating cell fate determination because many of their target mRNA transcripts are engaged in cell proliferation, differentiation, and apoptosis. DGCR8, Dicer, and Ago2 are essential factors for miRNA homeostasis. Here we show that these three factors have critical roles in osteoclast differentiation and function. Gene silencing of DGCR8, Dicer, or Ago2 by small interfering RNA revealed global inhibition of osteoclast transcription factor expression and function, decreased osteoclastogenesis, and decreased bone resorption in vitro. In vivo, CD11b+-cre/Dicer-null mice had mild osteopetrosis caused by decreased osteoclast number and bone resorption. These results suggest that miRNAs play important roles in differentiation and function of osteoclasts in vitro and in vivo. We found a novel mechanism mediating these results in which PU.1, miRNA-223, NFI-A, and the macrophage colony-stimulating factor receptor (M-CSFR) are closely linked through a positive feedback loop. PU.1 stimulates miRNA-223 expression, and this up-regulation is implicated in stimulating differentiation and function of osteoclasts through negative regulation of NFI-A levels. Down-regulation of NFI-A levels is important for expression of the M-CSFR, which is critical for osteoclast differentiation and function. NFI-A overexpression decreased osteoclast formation and function with down-regulation of M-CSFR levels. Forced expression of the M-CSFR in M-CSF-dependent bone marrow macrophages from Dicer-deficient mice rescued osteoclast differentiation with up-regulation of PU.1 levels. Our studies provide new molecular mechanisms controlling osteoclast differentiation and function by the miRNA system and specifically by miRNA-223, which regulates NFI-A and the M-CSFR levels.
The skeleton in vertebrates is comprised of bone and cartilage that include three specific cell types: osteoblasts and osteoclasts in bone and chondrocytes in cartilage (1). Bone metabolism is maintained by a well organized balance of old bone resorption by osteoclasts and new bone formation by osteoblasts (1, 2). Osteoblasts are bone-forming cells derived from multipotent mesenchymal stem cells (1). In contrast, multipotent hematopoietic stem cells give rise to myeloid stem cells, which further differentiate to megakaryocytes, granulocytes, and monocyte-macrophages (2). Osteoclasts are derived from monocyte-macrophage precursors (2). The osteoclast is the primary bone-resorbing cell, and it is formed from fusion of precursors to form multinucleated osteoclasts (2). Osteoclastogenesis is regulated by exogenous hormones and cytokines that positively or negatively modulate osteoclast proliferation, survival, differentiation, and function (3). Two essential cytokines, macrophage colony-stimulating factor-1 (M-CSF)2 and receptor activator of NFκB ligand (RANKL), are produced by osteoblasts or activated T cells and are required for osteoclast differentiation, function, and survival (1-3). Both M-CSF, as in the op/op mouse, and M-CSF receptor deficiency are characterized by osteopetrosis and the lack of macrophages and osteoclasts (1, 2).
Many transcription factors involved in osteoclastogenesis have been identified through studies in genetically engineered mice (4-10). Mouse mutants lacking factors that function early in the lineage including PU.1, c-Fos, and NFκB (p50 and p52) are osteopetrotic and lack either macrophages and osteoclasts or only osteoclasts (4-7). In particular, c-Fos and NFκB (p50 and p52) are required for the differentiation of monocyte precursors into osteoclasts (5-7), and c-Fos induces a second transcription factor, NFATc1, that is essential for osteoclastogenesis (8, 9). Other transcription factors, such as MITF, are also involved in osteoclastogenesis (10). Although much is known about the transcriptional regulation that controls osteoclast differentiation, potential roles for post-transcriptional gene regulation are not as well defined.
Micro-RNAs (miRNAs) are single-stranded RNAs 19-25 nucleotides in length that regulate several pathways including the development timing, hematopoiesis, organogenesis, apoptosis, cell proliferation, and tumorigenesis (11). The first known miRNA, the lin-4 small temporal RNA, was discovered in the nematode Caenorhabditis elegans (12). Since then, several groups identified hundreds of miRNAs in different organisms (13-15). miRNA genes are transcribed by RNA polymerase II to generate the primary transcripts (pri-miRNAs) (16). pri-miRNAs are processed by the double-stranded RNA-binding protein DiGeorge syndrome critical region gene 8 (DGCR8) and the nuclear RNase III enzyme Drosha into stem-loop-structured miRNA precursors (pre-miRNAs) (16). pre-miRNAs are exported by the nuclear export factor exportin-5 into the cytoplasm (16) and are processed by the cytoplasmic RNase III enzyme Dicer into mature miRNAs (17). Mature miRNAs are incorporated into the multiprotein RNA-induced silencing complex (RISC) with Dicer and Argonaute proteins (16). Argonaute2 (Ago2) binds miRNA and mediates gene silencing (18). Binding of miRNA-RISC to a partially complementary mRNA results in silencing by the inhibition of translation or mRNA degradation (18). On the other hand, direct cleavage of mRNA needs perfect or nearly perfect complementarity between the miRNA and the its target mRNA (16). Some miRNAs have been implicated in mouse hematopoietic differentiation (19-22). For example, miRNA-223, a mouse bone marrow-specific miRNA, is specifically expressed in myeloid lineage, and its expression is regulated by PU.1 and CAAT enhancer-binding proteins (19, 20). miRNA-181a expression is up-regulated only in differentiated B cells (19). miRNA-150 expression is elevated during developmental stages of B and T cell maturation (21). In transgenic mice expressing mouse pre-miRNA-155 in a B cell-specific manner, pre-B cell proliferation was markedly stimulated in the spleen and bone marrow, indicating that miRNA-155 regulates mouse pre-B cell proliferation (22). miRNA-146 is up-regulated by proinflammatory mediators in human monocytes (23).
Recent studies have shown that miRNAs play critical roles in osteoblast differentiation and bone development (24-27). We reported the first evidence that miRNA-223 is important for osteoclastogenesis (24). Expression of miRNA-26a is up-regulated in late osteoblast differentiation of human adipose tissue-derived stem cells, and miRNA-26a modulates this process by targeting the SMAD1 transcription factor (25). miRNA-125b controls osteoblast differentiation in mouse mesenchymal stem cells, ST2, by regulating cell proliferation (26). Dicer regulates chondrocyte proliferation and differentiation during skeletal development (27). Despite accumulating evidence that post-transcriptional gene regulation by miRNAs is essential for osteoclast, osteoblast, and chondrocyte differentiation in vitro and in vivo study, their roles in osteoclast differentiation and function are largely unknown compared with osteoblasts and chondrocytes. To study the global functions of miRNAs in osteoclast differentiation and bone resorbing activity, we employed a retroviral system for delivery of DGCR8, Dicer1, or Ago2 small interfering RNA into osteoclast precursors and CD11b+-cre/Dicer-null mice and found that the miRNA pathway for generation of miRNAs including miRNA-223 is critical for osteoclast differentiation and function, and miRNAs may be implicated in regulation of the bone remodeling cycle by osteoclasts. Thus, miRNAs including miRNA-223 would be expected to be potential targets of therapeutic strategies applied to drug discovery in bone metabolic disorders with excessive osteoclast activity such as osteoporosis.
EXPERIMENTAL PROCEDURES
Construction of Retroviral Vectors—The mouse DGCR8, Dicer1, and Ago2 siRNA target sequences were designed with search of off target effect by Enhanced siDirect (RNAi Co., Ltd.) (Table 1). The 63-mer sense and antisense strands of DNA oligonucleotides were synthesized by Integrated DNA Technologies. Construction of a retroviral vector for delivery of small interfering (siRNA) and generation of virus were performed using the pSilencer 5.1-H retroviral system (Ambion) according to the manufacturer's instructions. Scrambled siRNA retroviral vectors were obtained from Ambion. PU.1, M-CSFR, and NFI-A cDNA (Open Biosystems) were inserted into pMX retroviral vector (Cell Biolabs, Inc.) digested with BamHI and NotI restriction enzymes. Clones containing the cDNA insertion were verified by DNA sequencing.
TABLE 1.
Mouse siRNA targeted sequences
Circular, negative control pSilencer 5.1-H1 Retro scrambled was purchased from Ambion.
| siRNA | Targeted region | Sequences |
|---|---|---|
| DGCR8 | 526-548 | 5′-CTC CTG CCG ACG ACC CAT T-3′ |
| Dicer | 4221-4243 | 5′-GCA TGC TAT CAC CAC ATA T-3′ |
| Ago2 | 1179-1201 | 5′-GCG AAG TGC AAG TTT CAA T-3′ |
Cell Culture and Infection—Subconfluent Plat-E packaging cells (Dr. Kitamura, University of Tokyo, Tokyo, Japan) were transfected with retroviral vectors using FuGENE (Roche Applied Science). After 48 h, the supernatant was collected, filtered through a 0.45-μm syringe filter, and used to transduce target cells. Mouse primary M-CSF-dependent bone marrow macrophages (osteoclast precursors) were prepared from the femur and tibia of 4-6-week-old C57BL/6J mice (Jackson Lab) and Dicer-null osteoclast precursors and incubated in Petri dishes (100-mm dish) (Fisher) in αMEM (Sigma) containing 10% FBS (Atlas) in the presence of conditioned medium (1:10) from a M-CSF-producing CMG14-12 cell line that was a gift from Dr. Teitelbaum (Washington University School of Medicine, St. Louis, MO) (28). After 3 days in culture, the cells were plated in tissue culture dishes (100-mm dish) (Fisher) in αMEM containing 10% FBS in the presence of M-CSF conditioned medium (1:10). After 24 h in culture, the cells were infected with retrovirus for 6 h in αMEM containing 10% FBS in the presence of M-CSF conditioned medium (1:10) and 4 μg/ml polybren (Sigma). Infected cells were cultured for an additional 3 days for puromycin (Sigma) selection, and the cells were harvested for assay.
Real Time RT-PCR—Total RNA was isolated from infected cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Two micrograms of total RNA were reverse transcribed to cDNA using SuperScript II Reverse Transcriptase (Invitrogen). Real time PCR analysis was performed using the SYBR green PCR method according to the manufacturer's suggestions (Applied Biosystems). The primer sequences are as follows: mouse DGCR8, forward, 5′-AGT TCA ACC GTG AGA TGA AGC GGA-3′, and reverse, 5′-AGG ATG CAA ACC TCA GAC TTC CCA-3′; mouse Dicer1, forward, 5′-GCA GGC TTT TAC ACA CGC CT-3′, and reverse, 5′-GGG TCT TCA TAA AGG TGC TT-3′; mouse Ago2, forward, 5′-AGT TCT TGC TTC TCC TGC TCC G-3′, and reverse, 5′-GAT GCG ATC TTT GCC TTC TCC C-3′; mouse PU.1, forward, 5′-AGA AGC TGA TGG CTT GGA GC-3′, and reverse, 5′-GCG AAT CTT TTT CTT GCT GCC-3′; mouse MITF, forward, 5′-GGA CTT TCC CTT ATC CCA TTC A-3′, and reverse, 5′-GTT CTT GCT TGA TGA TCC GAT TC-3′; mouse c-Fos, forward, 5′-CCA AGC GGA GAC AGA TCA ACT T-3′, and reverse, 5′-TCC AGT TTT TCC TTC TCT TTC AGC AGA-3′; mouse NFATc1, forward, 5′-TCA GAA TGA GGC TGG ATA A-3′, and reverse, CGC TAC ATC ACT GAA CCT C-3′; mouse tartrate-resistant acid phosphatase (TRAP), forward, 5′-TCC TCG GAG AAA ATG CAT CAT-3′, and reverse, 5′-GCA GTT AAG CTC CTG GAC CAA-3′; mouse matrix metalloproteinase 9 (MMP9), forward, 5′-CAC CTT CAC CCG CGT GTA C-3′, and reverse, 5′-GCT CCG CGA CAC CAA ACT-3′; mouse cathepsin K (Ctsk), forward, 5′-GCT GTG GAG GCG GCT ATA TG-3′, and reverse, AGA GTC AAT GCC TCC GTT CTG-3′; mouse calcitonin receptor (CTR), forward, 5′-AGT TGC CCT CTT ATG AAG GAG AAG-3′, and reverse, 5′-GGA GTG TCG TCC CAG CAC AT-3′; mouse integrin β3, forward, GGA AGC AGC GCC CAG ATC AC-3′, and reverse, 5′-TTG TCC ACG AAG GCC CCA AA-3′; mouse receptor activator of NFκB (RANK), forward, 5′-TGC CTA CAG CAT GGG CTT T-3′, and reverse, 5′-AGA GAT GAA CGT GGA GTT ACT GTT T-3′; and mouse GAPDH, forward, 5′-ACC ACA GTC CAT GCC ATC AC-3′, and reverse, 5′-TCC ACC ACC CTG TTG CTG TA-3′. PCR products were normalized to GAPDH level for each reaction.
Immunoblotting—To evaluate the effect of knock down by siRNA on expression of transcription factors in infected cells treated with recombinant human RANKL (50 ng/ml) (Pepro Tech), whole cell or nuclear lysates of cells were assayed by immunoblotting using specific antibodies for DGCR8, Dicer1, Ago2, PU.1, MITF, c-Fos, NFATc1, Dicer, GAPDH (Santa Cruz Biotechnology, Inc.), histone H3, M-CSF receptor (M-CSFR) (Cell Signaling), and NFI-A (Abcam). Quantitative image analysis of DGCR8, Dicer1, Ago2, PU.1, MITF, c-Fos, NFATc1, NFI-A, or M-CSFR expression was normalized to GAPDH or histone H3.
miRNA RT-PCR and Real Time RT-PCR—Total RNA was isolated from osteoclast precursors or infected cells using the mirVana miRNA Isolation Kit (Ambion) that was used according to the manufacturer's protocol. RT-PCR and real time RT-PCR were performed using the mirVana qRT-PCR miRNA Detection Kit (Ambion) that was used according to the manufacturer's suggestions. miRNA-223, miRNA-155, and U6 primer were purchased from Ambion. In RT-PCR analysis, the PCR products were confirmed using a 20% polyacrylamide gel, and U6 small nuclear RNA was used as a loading control. In real time RT-PCR analysis, the PCR products were normalized to U6 levels for each reaction.
Chromatin Immunoprecipitation (ChIP)—ChIP was performed with the ChIP assay kit (Upstate Biotechnology, Inc.) according to the manufacturer's suggestions, using antibodies against NFATc1, c-Fos, PU.1, MITF, and normal IgG (Santa Cruz Biotechnology, Inc.). The purified DNA was analyzed by PCR using primers that detect sequences containing the mouse NFATc1 promoter: forward, 5′-CCG GGA CGC CCA TGC AAT CTG TTA GTA ATT-3′, and reverse, 5′-GCG GGT GCC CTG AGA AAG CTA CTC TCC CTT-3′; and mouse TRAP promoter: forward, 5′-CGG CTC AGT CTG GCC TGG GT-3′, reverse, 5′-GCT TAA CTG CCT CTT GCA GC-3′. The data were analyzed by Typhoon 9410 variable mode imager (Amersham Bioscience) according to the manufacturer's suggestions.
Osteoclast Formation Assay—Infected cells and Dicer-null cells were plated in 24-well plates (Corning) in αMEM containing 10% FBS in the presence of M-CSF-conditioned medium (1:20) and RANKL (50 ng/ml). After 5 days of culture, the cells were stained for TRAP activity. TRAP-positive multinucleated cells containing more than three nuclei were counted as osteoclasts under microscopic examination. Osteoclasts bone resorbing activity was measured using the OsteoLyse™ assay kit (Lonza) according to the manufacturer's protocol.
Generation of the Hematopoietic Myeloid-Osteoclast Lineage-specific Dicer KO Mice—The Dicer conditional mice (29) were crossed with CD11b+-cre mice (30) to generate CD11b+-cre/Dicerwt/f mice. CD11b+-cre/Dicerwt/wt (Dicerwt/wt) and CD11b+-cre/Dicerf/f (Dicerf/f) mice were obtained by crossing CD11b+-cre/Dicerwt/f with Dicer conditional mouse lines. In this study, only male Dicerf/f mice were used because transgene integration is in the Y chromosome (30).
Bone Histomorphometry—Bone histomorphometry analysis of femurs from Dicerwt/wt and Dicerf/f littermates (3 months old) was conducted at the University of Alabama at Birmingham Center for Metabolic Bone Disease Core Laboratory. The femurs were decalcified with 20% EDTA in PBS, embedded in paraffin, sectioned, and stained with hematoxylin and eosin and TRAP.
Transient Transfection—Antisense inhibitors of miRNA-223 and negative control oligonucleotides were obtained from Ambion. For antisense inhibition of miRNAs, RAW264.7 cells, a murine osteoclast progenitor cell line, were plated in 24-well plates (40% confluence) (Corning) and transfected with each antisense miRNA inhibitors or negative control using the siPORT NeoX transfection reagent (Ambion) according to the manufacturer's instructions. Subsequently, the cells were incubated in αMEM containing 10% FBS in the presence of RANKL (100 ng/ml) for 5 days to induce TRAP-positive multinucleated osteoclasts, and then the cells were stained for TRAP. TRAP-positive multinucleated cells containing more than three nuclei were counted as osteoclasts under microscopic examination.
Micro-RNA Northern Blotting—Total RNA was isolated from infected cells using the mirVana miRNA Isolation Kit (Ambion) that was used according to the manufacturer's protocol. Micro-RNA Northern blot was performed using Micro-RNA Northern blot assay kit (Signosis) according to the manufacturers instructions. Biotin prelabeled micro-RNA-223 and sno234 probe were purchased from Signosis.
RESULTS
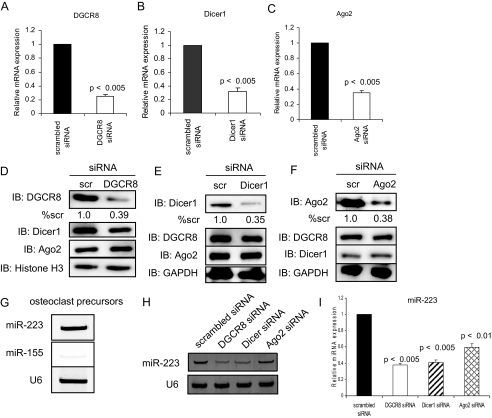
Micro-RNA Regulator Gene Silencing in Osteoclast Precursors—Plasmids containing siRNA sequences to DGCR8, Dicer1, or Ago2 were transduced by retroviruses in osteoclast precursors. We measured mRNA and protein expression levels by real time RT-PCR and Western blotting. Real time RT-PCR demonstrated that siRNA to DGCR8, Dicer1, or Ago2 decreased mRNA expression levels by 66-76% compared with scrambled siRNA at 48 h after infection (Fig. 1, A-C). The levels of the respective proteins were inhibited without effects on levels of the other factors (Fig. 1, D-F). Next, we confirmed that the retroviral vectors to DGCR8, Dicer1, and Ago2 were functional and inhibited miRNAs present during osteoclast differentiation. We have previously reported that miRNA-223 is expressed in RAW264.7 cells, mouse osteoclast precursor cell lines (24). In addition, miRNA microarray (data not shown) showed that miRNA-223 is expressed in osteoclast precursors, and miRNA-155 was barely detectable. Using miRNA RT-PCR and real time RT-PCR with primers specific to miRNA-223 and miRNA-155, we showed that miRNA-223 was expressed in osteoclast precursors in the experiments reported here and that miRNA-155 was barely detectable (Fig. 1G). miRNA RT-PCR and real time RT-PCR analysis showed that DGCR8, Dicer1, and Ago2 gene silencing by siRNA significantly suppressed expression of miRNA-223 (Fig. 1, H and I), suggesting that these siRNA vectors work in osteoclast precursors. The effect of Ago2 was surprising because it is generally believed that Ago2 proteins bind miRNAs and mediate downstream gene silencing (11). However, consistent with our data, miRNA biogenesis in hematopoietic cells from precursormi-RNAs was impaired in Ago2-deficient mouse (31). This and our data indicate that the Slicer endonuclease activity of Ago2 may be dispensable for hematopoiesis including osteoclast precursors. Our data show that DGCR8, Dicer1, and Ago2 are important in miRNA biogenesis in osteoclast precursors.
FIGURE 1.
Gene silencing and function of DGCR8, Dicer1 or Ago2 in osteoclast precursors. Quantitative real time PCR was performed with primer for mouse DGCR8 (A), Dicer1 (B), or Ago2 (C). The data represent the means ± S.D. of three experiments in triplicate. The loss of DGCR8 (D), Dicer1 (E), or Ago2 (F) protein expression levels by siRNA gene silencing was analyzed by immunoblotting (IB) using nuclear or whole cell extracts. Quantitative image analysis of these protein expression levels was normalized to GAPDH (% scr). scr, scrambled. miRNA RT-PCR (G and H) and quantitative real time RT-PCR (I) were performed to measure miRNA-223 and miRNA-155 expression levels. The PCR products were confirmed using a 20% polyacrylamide gel, with U6 small nuclear RNA as a loading control (G and H). PCR products were normalized to U6 level for each reaction (I). The data represent the means ± S.D. of three experiments in triplicate.
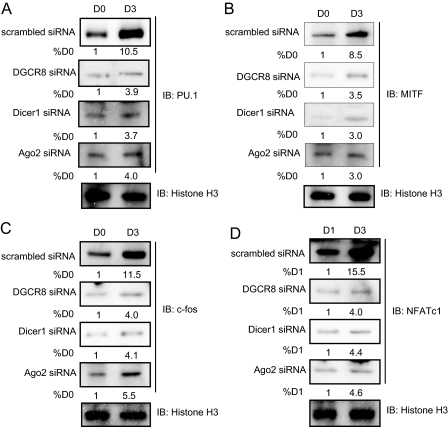
miRNAs Control Expression and Function of Transcription Factors Required in Osteoclastogenesis—It is not known whether miRNAs play a critical role in osteoclast differentiation and function. To elucidate the role of miRNAs in osteoclastogenesis, we utilized RANKL-induced differentiation of osteoclast precursors harboring reduced levels of DGCR8, Dicer1 or Ago2 protein. Analysis of expression of the transcription factors important for osteoclastogenesis such as PU.1, MITF, c-Fos, and NFATc1 (1, 2) revealed increased levels by day 3 during RANKL-stimulated osteoclastogenesis in cells expressing scrambled siRNA (Fig. 2). In DGCR8, Dicer1, or Ago2 siRNA-expressing cells, in contrast, the expression levels of these proteins by Western blotting did not reach normal levels during differentiation with RANKL stimulation, although there was slight up-regulation by day 3 (Fig. 2).
FIGURE 2.
Expression of transcription factors in osteoclast differentiation. Infected cells were treated with RANKL (50 ng/ml) for 3 days, and nuclear extracts were analyzed by immunoblotting with antibodies against PU.1 (A), MITF (B), c-Fos (C), and NFATc1 (D). Quantitative image analysis of the protein expression levels (values below bands) was normalized to histone H3. IB, immunoblotting.
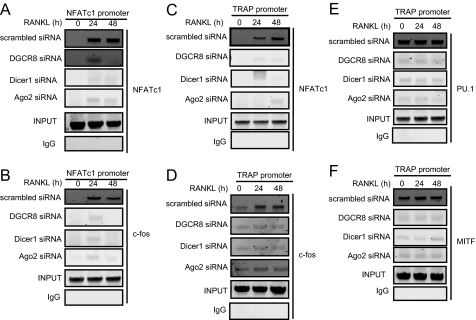
The transcription factors required in osteoclastogenesis shown in Fig. 2 are recruited to a variety of osteoclast-specific marker gene promoters such as TRAP, MMP-9, Ctsk, CTR, and integrin β3 and NFATc1 itself for the formation and activity of multinucleated osteoclasts (33-43). We examined whether PU.1, MITF, c-Fos, or NFATc1 are recruited to the NFATc1 or TRAP promoter. ChIP analysis showed that NFATc1 and c-Fos were recruited to the NFATc1 promoter by 24 and 48 h after RANKL stimulation in scrambled siRNA-expressing cells (Fig. 3, A and B). NFATc1 was also recruited to the TRAP promoter by 24 and 48 h after RANKL stimulation in scrambled siRNA-expressing cells (Fig. 3C). Low levels of c-Fos were already bound to the TRAP promoter in osteoclast precursors without RANKL stimulation (0 h), and increased binding was observed at 24 and 48 h in scrambled siRNA-expressing cells (Fig. 3D). PU.1 and MITF were more strongly recruited to the TRAP promoter in the same binding pattern as c-Fos in scrambled siRNA-expressing cells (Fig. 3, E and F). The binding of these factors to the NFATc1 or TRAP promoter was strikingly attenuated by a loss of DGCR8, Dicer1, or Ago2 protein (Fig. 3).
FIGURE 3.
ChIP analysis. Shown are ChIP assays to study the association of NFATc1, c-Fos, PU.1, and MITF with NFATc1 or TRAP promoters during osteoclast differentiation with RANKL (50 ng/ml) stimulation for indicated times.
Consistent with the results of the ChIP analysis in Fig. 3, the mRNA expression levels of the osteoclast-specific markers were markedly inhibited during osteoclast differentiation with DGCR8, Dicer1, or Ago2 siRNA gene silencing (Fig. 4). In particular, Ctsk, CTR, and integrin β3 mRNA expression levels had been suppressed by day 1 after RANKL stimulation in DGCR8, Dicer1, or Ago2 siRNA-expressing cells compared with scrambled siRNA-expressing cells (Fig. 4, C-E). These data of Chip assay and real time RT-PCR analysis can be attributed to inhibition of expression of the transcription factor proteins important for osteoclastogenesis by loss of DGCR8, Dicer1, or Ago2 protein during osteoclast differentiation as shown by Fig. 2.
FIGURE 4.
Expression of osteoclast-specific markers during osteoclastogenesis. Infected cells were treated with RANKL (50 ng/ml) for 3 days. Total RNA was extracted at the indicated time points, and expression of TRAP (A), MMP9 (B), Ctsk (C), CTR (D), and integrin β3 (E) mRNA levels were measured by real time RT-PCR. The PCR products were normalized to GAPDH for each reaction. The data represent the means ± S.D. of three experiments in triplicate.
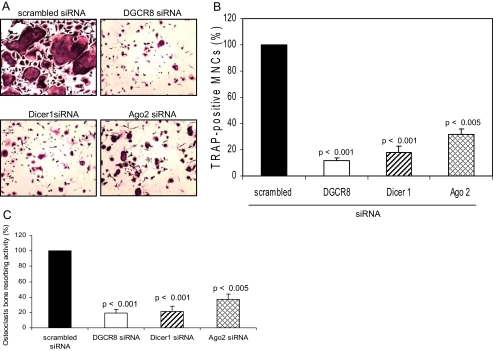
Inhibition of Osteoclast Differentiation and Function—Consistent with data of Figs. 2, 3, 4, DGCR8, Dicer1, or Ago2 siRNA gene silencing inhibited TRAP-positive multinucleated osteoclast number and bone resorbing activity compared with scrambled siRNA-expressing cells (Fig. 5). These data provide direct evidence that the miRNA pathway for the generation of miRNAs play critical roles in osteoclast differentiation and function in vitro.
FIGURE 5.
TRAP-positive osteoclast formation and bone resorbing activity. A and B, infected osteoclast precursors were replated in 24-well plates, and the cells were treated with RANKL (50 ng/ml) and M-CSF conditioned media (1:20). After 5 days, the cells were then fixed and stained for TRAP, and the number of TRAP-positive multinucleated cells was scored. Similar findings were obtained in four independent sets of experiments. C, infected osteoclast precursors were cultured on europium-labeled human collagen-coated bone for 5 days with RANKL stimulation. Osteoclasts bone resorbing activity was measured using each cell culture supernatant by the OsteoLyse™ assay kit. The data represent the means ± S.D. of three experiments in duplicate.
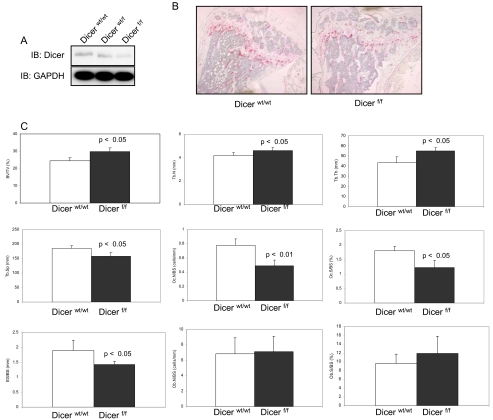
Dicer-null Mice Show Mild Osteopetrosis—We generated Dicerwt/wt and Dicerf/f mice using CD11b+-cre mice to examine whether micro-RNAs are involved in differentiation and function of osteoclasts in vivo as well as in vitro. CD11b is expressed during osteoclast differentiation from mononucleated early progenitor cells to mature multinucleated osteoclasts (30). First of all, Dicer protein levels were reduced as shown by Western blot analysis of Dicer-null osteoclast precursors (Fig. 6A). Next, to confirm whether a functional Dicer KO is generated, Dicer-null osteoclast precursors were analyzed for their ability to produce mature miRNA-223 by miRNA real time RT-PCR assay. miRNA-223 was detected in Dicerwt/w osteoclast precursors, but its expression levels were decreased by 58% in Dicer-null osteoclast precursors (data not shown). We next studied the skeletal tissues of Dicerwt/wt and Dicerf/f mice. Histological examination using TRAP stain revealed reduced numbers of TRAP-positive osteoclasts in growth plate and cancellous bone tissues of Dicerf/f mice and expanded trabecular thickness compared with Dicerwt/wt mice (Fig. 6B). Histomorphometry demonstrated that Dicerf/f mice have mild osteoporosis with increased bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and reduced trabecular separation (Tb.Sp), osteoclast number (Oc.N/BS), osteoclast surface (Oc.S/BS) and eroded surface (ES/BS) (Fig. 6C). Osteoblast number (Ob.N/BS) and osteoblast surface (Ob.S/BS) were not different between Dicerwt/wt and Dicerf/f mice (Fig. 6C). These data provide evidence that the impaired miRNA pathway causes aberrant osteoclast differentiation and function in vivo as well as in vitro.
FIGURE 6.
Dicerf/f mice cause mild osteoporosis because of impaired osteoclast formation and activation. A, dicer protein expression levels were analyzed by immunoblotting using whole cell extracts. IB, immunoblotting. B, histological analysis using TRAP stain of femur from three-month-old Dicerwt/wt and Dicerf/f mice. C, histomorphometric analysis. The parameters are measured in the distal femur of 3-month-old Dicerwt/wt and Dicerf/f mice. The data are expressed as the means ± S.D. of five or six mice of each genotype. BV/TV, bone volume; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation; Oc.N/BS, osteoclast number; Oc.S/BS, osteoclast surface; ES/BS, eroded surface; Ob.N/BS, Osteoblast number; Ob.S/BS, osteoblast surface.
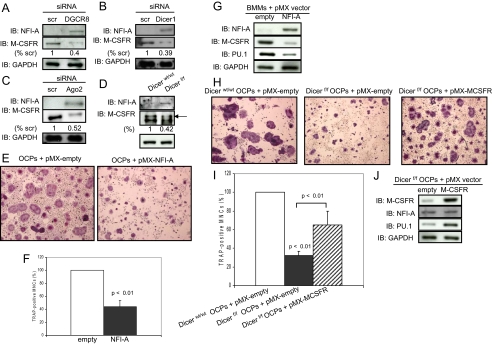
NFI-A Down-regulation by miRNA-223 Is Implicated in Osteoclastogenesis—Thus far we have shown that the miRNA pathway for generation of miRNAs is important for osteoclast differentiation and function in vitro and in vivo. However, the mechanism by which miRNAs control osteoclastogenesis and bone resorbing activity remains unknown. Our miRNA microarray showed that miRNA-223 expression is sustained during osteoclast differentiation (data not shown). Therefore, we pursued the function of miRNA-223 that is needed in osteoclastogenesis. A bioinformatic search indicated a putative target site for miRNA-223 in the NFI-A 3′-untranslated region that is highly conserved among mammals. Recently, it was reported that NFI-A negatively controls expression of the M-CSFR by unknown mechanisms in the NB4 cell line, derived from the bone marrow of a patient with acute promyelocytic leukemia (44). M-CSFR is expressed in osteoclast precursors through osteoclasts and is critical for differentiation, function, and survival of osteoclasts (1, 2). Consistent with these reports, NFI-A was up-regulated, and M-CSFR was down-regulated in DGCR8, Dicer1, or Ago siRNA-expressing cells and Dicer-null osteoclast precursors (Fig. 7, A-D). On the other hand, NFI-A expression was not detectable in scrambled siRNA-expressing cells and Dicerwt/wt osteoclast precursors (Fig. 7, A-D) that expressed miRNA-223 (Fig. 7, G-I). We next studied whether down-regulation of NFI-A expression is needed in osteoclastogenesis and bone resorbing activity. Overexpression of NFI-A in osteoclast precursors significantly suppressed TRAP-positive osteoclast formation compared with control cells (58% down) (Fig. 7, E and F) and bone resorption (data not shown). Expression of M-CSFR and PU.1 were down-regulated in NFI-A-overexpressing cells (Fig. 7G). In addition, pri-miRNA-223 and mature miRNA-223 expression levels were also decreased in this study (data not shown). However, ChIP analysis failed to detect the mechanism of a negative effect of NFI-A on M-CSFR expression, even though two putative NFI-A-binding sites are presented on the M-CSFR promoter (data not shown).
FIGURE 7.
A negative effect of NFI-A on M-CSFR expression. A-D, G, and J, NFI-A, M-CSFR, and PU.1 levels in infected-osteoclast precursors and Dicer-null osteoclast precursors were analyzed by immunoblotting using whole cell extracts. Quantitative image analysis of these protein expression levels were normalized to GAPDH. scr, scrambled. IB, immunoblotting. E, F, H, and I, infected osteoclast precursors were replated in 24-well plates, and the cells were treated with RANKL (50 ng/ml) and M-CSF-conditioned medium (1:20). After 5 days, the cells were then fixed and stained for TRAP, and the number of TRAP-positive multinucleated cells was scored. Similar findings were obtained in four independent sets of experiments. OCP, osteoclast precursors.
We next examined whether overexpression of M-CSFR rescues osteoclast formation and function in Dicer-null osteoclast precursors in response to M-CSF and RANKL. Dicer-null osteoclast precursors prepared from the Dicerf/f mice had impaired osteoclastogenesis compared with control cells (Fig. 7, H and I). Forced expression of M-CSFR in Dicer-null osteoclast precursors significantly rescued TRAP-positive osteoclast formation compared with that of Dicer-null osteoclast precursors (Fig. 7, H and I). Moreover, we found that PU.1 expression is up-regulated by M-CSFR overexpression in rescued cells (Fig. 7J), even though its expression is impaired the miRNA pathway is defective as shown Fig. 2. However, mature miRNA-223 expression levels were down-regulated, although pri-miRNA-223 expression levels were up-regulated in this study (data not shown). In addition, NFI-A expression levels were not altered (Fig. 7J). These results demonstrate that the action of miRNA-223 to suppress NFI-A levels in osteoclast precursors is a prerequisite for induction of M-CSFR expression and osteoclastogenesis.
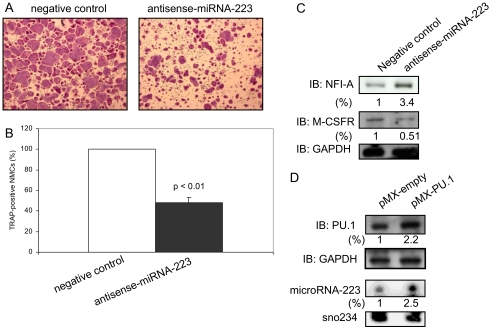
To further investigate whether miRNA-223 is essential for osteoclastogenesis, we performed osteoclast formation assays using antisense miRNA-223 oligonucleotides. Inhibition of miRNA-223 induced down-regulation of TRAP-positive osteoclast formation compared with control in RAW264.7 cells (Fig. 8, A and B). We previously reported that TRAP-positive osteoclast formation was not affected in pre-miRNA-223 siRNA-expressing RAW264.7 cells in which the miRNA-223 was decreased by 66% (24). There were no alterations of NFI-A expression level by this level of miRNA-223 decrease (data not shown). However, in the present studies, the miRNA-223 decrease was 92.0% (data not shown), and NFI-A levels were up-regulated (Fig. 8C). In addition, unlike osteoclast precursors wherein NFI-A is not expressed, NFI-A is moderately expressed in RAW264.7 cells (Fig. 8C). Thereby, we reasoned that the direct suppression of mature miRNAs by antisense oligonucleotides may be able to inhibit more effectively than that of pre-miRNA-223 by siRNA including the target sequence of stem-loop structure. We next analyzed NFI-A and M-CSFR expression levels in RAW264.7 cells with antisense miRNA-223 oligonucleotides. Western blot analysis showed up-regulation of NFI-A levels and a negative effect of NFI-A on M-CSFR levels (Fig. 8C), suggesting that miRNA-223 expression is critically important for osteoclastogenesis through M-CSFR expression. Moreover, we found that overexpression of PU.1 in osteoclast precursors induced expression of miRNA-223 compared with control cells (Fig. 8D). These data are in agreement with the report of two putative PU.1-binding sites present in the mouse miRNA-223 promoter (20).
FIGURE 8.
Inhibition of miRNA-223 expression causes aberrant osteoclastogenesis. A and B, RAW 264.7 cells were transiently transfected with antisense miRNA-223 or negative control oligonucleotides, and the cells were treated with RANKL (100 ng/ml). After 5 days, the cells were then fixed and stained for TRAP, and the number of TRAP-positive multinucleated cells was scored. Similar findings were obtained in four independent sets of experiments. C and D, NFI-A, M-CSFR, and PU.1 levels in RAW264.7 cells with negative control or antisense miRNA-223 and infected osteoclast precursors were analyzed by immunoblotting (IB) using whole cell extracts. Quantitative image analyses of these protein expression levels were normalized to GAPDH. D, The expression levels of miRNA-223 was measured by miRNA Northern blotting using biotin prelabeled probes specific for miRNA-223. Sno234 was used as a loading control. The sno234 normalized fold increase in the miRNA-223 in osteoclast precursors with pMX-PU.1 retroviral vector is shown.
DISCUSSION
In this study, we used DGCR8, Dicer1, and Ago2 siRNA gene silencing and Dicer-null mice to clarify the importance of the miRNA pathway for generation of miRNAs in osteoclast differentiation and function in vitro and in vivo. The first discovery was that loss of Ago2 protein as well as loss of DGCR8 or Dicer1 proteins attenuated expression levels of miRNAs in osteoclast precursors (Fig. 1, H and I). Ago2 is known to be important in regulation of gene silencing (11). Our data are in agreement with a recent report using Ago2-/- hematopoietic bone marrow cells concluding that the Slicer endonuclease activity is dispensable for hematopoiesis and that Ago2 is an important regulator of miRNA homeostasis (31). Ago2 could be very specialized member of the Argonaute family with an important function within RISC in the regulation of miRNA homeostasis.
PU.1, MITF, c-Fos, and NFATc1 are crucial transcription factors in osteoclastogenesis (1, 2), and these transcription factors bind to the TRAP, MMP9, Ctsk, CTR, or integrin β3 promoters for expression of osteoclast-specific markers in activated osteoclasts (33-41). Expression of these molecular markers is essential for osteoclast function such as bone resorption to maintain bone metabolism (1, 2). However, this sequence of events was extremely reduced by impaired miRNA homeostasis. The question became why the levels of these transcription factor proteins were not increased by loss of DGCR8, Dicer1 or Ago2 protein (Fig. 2). We demonstrated that one mechanism by which this occurs is PU.1 down-regulation by the negative control of NFI-A on M-CSFR levels (Fig. 7) because M-CSF induces PU.1 expression through M-CSFR in the hematopoietic myeloid-osteoclast lineage (1-3). As has been mentioned, we were not able to detect NFI-A binding on the M-CSFR promoter by ChIP assay, even though two putative binding sites are present (data not shown), suggesting that this effect may be indirect.
In studies in vitro, we demonstrated that the impaired miRNA pathway causes aberrant osteoclast differentiation and function caused by inhibition of expression and activity of transcription factors required in osteoclastogenesis and consequently reduced osteoclast-specific gene expression (Figs. 2, 3, 4, 5). Moreover, RANK expression was also down-regulated in DGCR8, Dicer1, or Ago siRNA-expressing cells and Dicer-null osteoclast precursors (data not shown). The RANKL-RANK pathway is pivotal in osteoclast differentiation, activity, and survival (1-3). RANK is the receptor for RANKL (1, 2), and RANK is expressed in osteoclast precursors and mature osteoclasts, where it can be up-regulated by PU.1 and M-CSF stimulation (45, 46). Mice lacking RANK are osteopetrotic (47), demonstrating that RANK is essential for osteoclast differentiation. This could be one of the major causes of aberrant osteoclast differentiation by impaired miRNA homeostasis. These data strongly suggest that miRNAs play a critical role in osteoclast differentiation and function in vitro. Moreover, using the cre-loxP system, we showed that CD11b+-cre/Dicerf/f mice have mild osteopetrosis caused by reduced osteoclast numbers and bone resorbing activity without affecting osteoblast number or activity (Fig. 6). This suggests that the miRNAs are involved in bone metabolism, at least a part, by controlling differentiation and function of osteoclasts in vivo.
Unexpectedly, miRNA real time RT-PCR analysis revealed the moderate expression levels, not low expression levels, of miRNA-223 in Dicer-null osteoclast precursors (data not shown) as well as Dicer1 siRNA gene silencing (Fig. 1I). Recent studies have demonstrated that overexpression of each Ago protein (Ago1-Ago4) increases the abundance of mature miRNAs compared with Dicer overexpression (48). In addition, Ago2 is a key regulator of miRNAs biogenesis rather that the Slicer endonuclease activity (31), suggesting that Ago2 would be a more important factor than Dicer for miRNA homeostasis in vivo. Moreover, it has been reported that RISC including Dicer and Ago2 more efficiently cleaves target mRNAs by using pre-miRNAs than the duplex miRNAs that do not have the stem-loop structure (49). For these reasons, we will need to generate Ago2 or DGCR8 osteoclast precursor-specific null mice to further investigate miRNA function and bone metabolism in vivo in a setting with lower levels of the miRNAs including miRNA-223 and perhaps more severe osteopetrosis.
We have shown previously that miRNA-223 expression is important for osteoclastogenesis. However, we were not able to provide detailed molecular mechanisms. First of all, we showed that osteoclast differentiation of RAW264.7 cells was significantly suppressed by antisense to miRNA-223 (Fig. 8, A and B). Moreover, this antisense miRNA markedly suppressed osteoclast bone resorbing activity (data not shown), indicating that miRNA-223 may be important for regulation of differentiation and function of osteoclasts in vitro. However, overexpression of pre-miRNA-223 also suppressed TRAP-positive osteoclast formation (24), suggesting that appropriate expression levels of miRNA-223 would be needed in osteoclastogenesis. Although all the conserved miRNA-223 target mRNAs for osteoclastogenesis or its function have not been identified, we found that NFI-A suppression by miRNA-223 is important (Fig. 8, A-C). This suggests that expression of miRNA-223 is essential for osteoclast differentiation and function by overcoming a constitutive inhibition induced by NFI-A. In addition, we found that PU.1 induces miRNA-223 expression in osteoclast precursors (Fig. 8D). This appears to be important for down-regulation of NFI-A levels by miRNA-223. In fact, PU.1 levels are up-regulated during osteoclastogenesis by M-CSF and RANKL (1, 2).
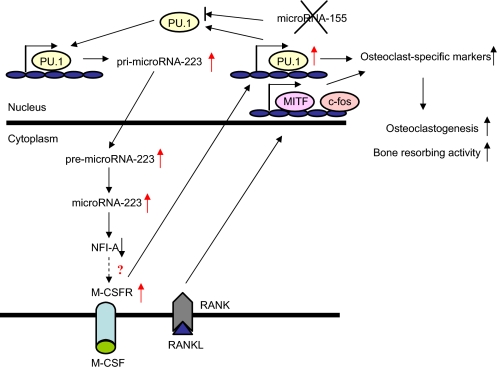
In summary, we propose a mechanism of miRNA-223 action associated with osteoclast differentiation and function (Fig. 9). In osteoclast precursors, PU.1 induced by M-CSF stimulates production of pri-miRNA-223 and RANK. Importantly, expression of miRNA-155, a target of PU.1 (50), is not induced by M-CSF and RANKL stimulation during osteoclastogenesis.3 Next, pri-miRNA223 is processed into mature miRNA-223 from pre-miRNA-223 by RNase III enzymes including Dicer, and miRNA-223 down-regulates the NFI-A levels required for up-regulating of M-CSFR levels in cells, which in turn feed forward to increase expression of transcription factors such as PU.1, MITF, and c-Fos. These transcription factors are induced by M-CSF and RANKL through the up-regulated M-CSFR and RANK, and consequently cells differentiate into activated osteoclasts by up-regulated osteoclast-specific markers.
FIGURE 9.
A model of a novel mechanism for controlling osteoclast differentiation and function by miRNA-223 in osteoclast precursors. PU.1 induced by M-CSF activates the transcription of miRNA-223, which causes a negative control on M-CSFR expression levels. Also, induced PU.1 stimulates RANK expression, and consequently osteoclast differentiation and function are induced by M-CSF and RANKL stimulation.
Our results and those of others in aggregate indicate that the functions of DGCR8, Dicer, and Ago2 are necessary to maintain bone metabolism, including the differentiation and function of osteoclasts. Understanding the role of post-transcriptional gene regulation by miRNAs and identification of target mRNAs negatively regulated by miRNAs in normal bone biology is critical to understanding bone metabolism disorders. Identification of osteoclast-specific miRNAs may provide potential targets for drug development in bone metabolic disorders with abnormal osteoclast activity.
Acknowledgments
We greatly appreciate the gift of the M-CSF-producing CMG14-12 cell line from Dr. Teitelbaum (Washington University School of Medicine, St. Louis, MO), the Plat-E cell line from Dr. Kitamura (University of Tokyo, Tokyo, Japan), Dicer flox mice from Dr. Harfe (University of Florida College of Medicine, Gainesville, FL), and CD11b-cre mice from Dr. Vacher (Institute de Recherches Cliniques de Montréal, Montréal, Canada). We are also grateful to the University of Alabama at Birmingham, Center for the Metabolic Bone Disease-Histomorphometry Core Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grants DK070790 and AR41677 (to K. A. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: M-CSF, macrophage colony-stimulating factor; miRNA, micro-RNA; pri-miRNA, primary micro-RNA; pre-miRNA, precursor micro-RNA; DGCR8, DiGeorge syndrome critical region gene 8; RISC, RNA-induced silencing complex; Ago2, Argonaute2; siRNA, small interfering RNA; RANKL, receptor activator of NFκB ligand; MITF, microphthalmia-associated transcription factor; NFATc1, nuclear factor of activated T cells, cytoplasmic 1; TRAP, tartrate-resistant acid phosphatase; MMP9, matrix metalloproteinase 9; Ctsk, cathepsin K; CTR, calcitonin receptor; NFI-A, nuclear factor I-A; M-CSFR, M-CSF receptor; αMEM, α-minimum essential medium; FBS, fetal bovine serum; RT, reverse transcription; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
T. Sugatani and K. A. Hruska, unpublished data.
References
- 1.Karsenty, G., and Wagner, E. F. (2002) Dev. Cell 2 389-406 [DOI] [PubMed] [Google Scholar]
- 2.Tanaka, S., Nakamura, K., Takahasi, N., and Suda, T. (2005) Immunol. Rev. 208 30-49 [DOI] [PubMed] [Google Scholar]
- 3.Del, F. A., Teti, A., and Rucci, N. (2008) Arch. Biochem. Biophys. 473 147-160 [DOI] [PubMed] [Google Scholar]
- 4.Tondravi, M. M., McKercher, S. R., Anderson, K., Erdmann, J. M., Quiroz, M., Maki, R., and Teitelbaum, S. L. (1997) Nature 386 81-84 [DOI] [PubMed] [Google Scholar]
- 5.Grigoriadis, A. E., Wang, Z. Q., Cecchini, M. G., Hofstetter, W., Felix, R., Fleisch, H. A., and Wagner, E. F. (1994) Science 266 443-448 [DOI] [PubMed] [Google Scholar]
- 6.Iotsova, V., Caamano, J., Loy, J., Yang, Y., Lewin, A., and Bravo, R. (1997) Nat. Med. 3 1285-1289 [DOI] [PubMed] [Google Scholar]
- 7.Franzoso, G., Carison, L., Poljak, L., Shores, E. W., Brown, K. D., Leonardi, A., Tran, T., Boyce, B. F., and Siebenlist, U. (1997) Genes Dev. 11 3482-3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida, N., Hayashi, K., Hoshijima, M., Ogawa, T., Koga, S., Miyatake, Y., Kumegawa, M., Kimura, T., and Takeya, T. (2002) J. Biol. Chem. 277 41147-41156 [DOI] [PubMed] [Google Scholar]
- 9.Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., Saiura, A., Isobe, M., Yokochi, T., Inoue, J., Wagner, E. F., Mak, T. W., Kodama, T., and Taniguchi, T. (2002) Dev. Cell 3 889-901 [DOI] [PubMed] [Google Scholar]
- 10.Weilbaecher, K. N., Motyckova, G., Huber, W. E., Takemoto, C. M., Hemesath, T. J., Xu, Y., Hershey, C. L., Downland, N. R., Wells, A. G., and Fisher, D. E. (2001) Mol. Cell 8 749-758 [DOI] [PubMed] [Google Scholar]
- 11.Kim, V. N. (2005) Nat. Rev. Mol. Cell. Biol. 6 376-385 [DOI] [PubMed] [Google Scholar]
- 12.Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993) Cell 75 843-854 [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001) Science 294 853-858 [DOI] [PubMed] [Google Scholar]
- 14.Lau, N. C., Lim, L. P., Weinstein, E. G., and Bartel, D. P. (2001) Science 294 858-862 [DOI] [PubMed] [Google Scholar]
- 15.Lee, R. C., and Ambros, V. (2001) Science 294 862-864 [DOI] [PubMed] [Google Scholar]
- 16.Bushati, N., and Cohen, S. M. (2007) Annu. Rev. Cell Dev. Biol. 23 175-205 [DOI] [PubMed] [Google Scholar]
- 17.Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001) Nature 409 363-366 [DOI] [PubMed] [Google Scholar]
- 18.Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R., and Hannon, G. J. (2001) Science 293 1146-1150 [DOI] [PubMed] [Google Scholar]
- 19.Chen, C. Z., Li, L., Lodish, H. F., and Bartel, D. P. (2004) Science 303 83-86 [DOI] [PubMed] [Google Scholar]
- 20.Fukao, T., Fukuda, Y., Kiga, K., Sharif, J., Hino, K., Enomoto, Y., Kawamura, A., Nakamura, K., Takeuchi, T., and Tanabe, M. (2007) Cell 129 617-631 [DOI] [PubMed] [Google Scholar]
- 21.Zhou, B., Wang, S., Mayr, C., Bartel, D. P., and Lodish, H. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7080-7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costinean, S., Zanesi, N., Pekarsky, Y., Tili, E., Volinia, S., Heerema, N., and Croce, C. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7024-7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taganov, K. D., Boldin, M. P., Chang, K.-J., and Baltimore, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 12481-12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugatani, T., and Hruska, K. A. (2007) J. Cell. Biochem. 101 996-999 [DOI] [PubMed] [Google Scholar]
- 25.Luzi, E., Marini, F., Sala, S. C., Tognarini, I., Galli, G., and Brandi, M. L. (2008) J. Bone Miner. Res. 23 287-295 [DOI] [PubMed] [Google Scholar]
- 26.Mizuno, Y., Yagi, K., Tokuzawa, Y., Kanesaki-Yatsuka, Y., Suda, T., Katagiri, T., Fukuda, T., Maruyama, M., Okuda, A., Amemiya, T., Kondoh, Y., Tashiro, H., and Okazaki, Y. (2008) Biochem. Biophys. Res. Commun. 368 267-272 [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, T., Lu, J., Cobb, B. S., Rodda, S. J., McMahon, A. P., Schipani, E., Merkenschlager, M., and Kronenberg H. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1949-1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshita, S., Kaji, K., and Kudo, A. (2000) J. Bone Miner. Res. 15 1477-1488 [DOI] [PubMed] [Google Scholar]
- 29.Harfe, B. D., McManus, M. T., Mansfield, J. H., Hornstein, E., and Tabin, C. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10898-10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferron, M., and Vacher, J. (2005) Genesis 41 138-145 [DOI] [PubMed] [Google Scholar]
- 31.O'Carroll, D., Mecklenbrauker, I., Das, P. P., Santana, A., Koenig, U., Enright, A. J., Miska, E. A., and Tarakhovsky, A. (2007) Genes Dev. 21 1999-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleted in proof
- 33.Matsuo, K., Galson, D. L., Zhao, C., Peng, L., Laplace, C., Wang, K. Z. Q., Bachler, M. A., Amano, H., Aburatani, H., Ishikawa, H., and Wagner, E. F. (2004) J. Biol. Chem. 279 26475-26480 [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto, M., Kogawa, M., Wada, S., Takayanagi, H., Tsujimoto, M., Katayama, S., Hisatake, K., and Nogi, Y. (2004) J. Biol. Chem. 279 45969-45979 [DOI] [PubMed] [Google Scholar]
- 35.Asagiri, M., Sato, K., Usami, T., Ochi, S., Nishina, H., Yoshida, H., Morita, I., Wagner, E. F., Mak, T. W., Serfiling, E., and Takayanagi, H. (2005) J. Exp. Med. 202 1261-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundaram, K., Nishimura, R., Senn, J., Yousef, R. F., London, S. D., and Reddy, S. V. (2007) Exp. Cell Res. 313 168-178 [DOI] [PubMed] [Google Scholar]
- 37.Crotti, T. N., Flannery, M., Walsh, N. C., Fleming, J. D., Goldring, S. R., and McHugh, K. P. (2006) Gene (Amst.) 372 92-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma, S. M., Bronisz, A., Hu, R., Patel, K., Mansky, K. C., Sif, S., and Ostrowski, M. C. (2007) J. Biol. Chem. 282 15921-15929 [DOI] [PubMed] [Google Scholar]
- 39.Hu, R., Sharma, S. M., Bronisz, A., Srinivasan, R., Sankar, U., and Ostrowski, M. C. (2007) Mol. Cell. Biol. 27 4018-4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang, M., Martinez, A. F., Fernandez, I., Balkan, W., and Troen, B. R. (2007) Gene (Amst.) 403 151-158 [DOI] [PubMed] [Google Scholar]
- 41.Shen, Z., Crotti, T. N., Flannery, M. R., Matsuzaki, K., Goldring, S. R., and McHugh, K. P. (2007) Biochim. Biophys. Acta 1769 659-667 [DOI] [PubMed] [Google Scholar]
- 42.Asagiri, M., and Takayanagi, H. (2007) Bone 40 251-264 [DOI] [PubMed] [Google Scholar]
- 43.Walsh, N. C., Cahill, M., Carninci, P., Kawai, J., Okazaki, Y., Hayashizaki, Y., Hume, D. A., and Cassady, A. I. (2003) Gene (Amst.) 307 111-123 [DOI] [PubMed] [Google Scholar]
- 44.Rosa, A., Ballarino, M., Sorrentino, A., Sthandier, O., De Angelis, F. G., Marchioni, M., Masella, B., Guarini, A., Fatica, A., Peschle, C., and Bozzoni, I. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19849-19854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon, O. H., Lee, C. K., Lee, Y. I., Paik, S. G., and Lee, H. J. (2005) Biochem. Biophys. Res. Commun. 335 437-446 [DOI] [PubMed] [Google Scholar]
- 46.Arai, F., Miyamoto, T., Ohneda, O., Inada, T., Sudo, T., Brasel, K., Miyata, T., Anderson, D. M., and Suda, T. (1999) J. Exp. Med. 190 1741-1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dougall, W. C., Glaccum, M., Charrier, K., Rohrbach, K., Brasel, K., De Smedt, T., Daro, E., Smith, J., Tometsko, M. E., Maliszewski, C. R., Armstrong, A., Shen, V., Bain, S., Cosman, D., Anderson, D., Morrissey, P. J., Peschon, J. J., and Schuh, J. (1999) Genes Dev. 13 2412-2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diederichs, S., and Haber, D. A. (2007) Cell 131 1097-1108 [DOI] [PubMed] [Google Scholar]
- 49.Gregory, R. I., Chendrimada, T. P., Cooch, N., and Shiekhattar, R. (2005) Cell 123 631-640 [DOI] [PubMed] [Google Scholar]
- 50.Vigorito, E., Perks, K. L., Abreu-Goodger, C., Bunting, S., Xiang, Z., Kohlhaas, S., Das, P. P., Miska, E. A., Rodriguez, A., Bradley, A., Smith, K. G., Rada, C., Enright, A. J., Toellner, K. M., Maclennan, I. C., and Turner, M. (2007) Immunity 27 847-859 [DOI] [PMC free article] [PubMed] [Google Scholar]