Abstract
Viral matrix (M) proteins bind the nucleoprotein core (nucleocapsid) to host membranes during the process of virus assembly by budding. Previous studies using truncated M proteins had implicated the N-terminal 50 amino acids of the vesicular stomatitis virus M protein in binding both membranes and nucleocapsids and a sequence from amino acids 75-106 as an additional membrane binding region. Structure-based mutations were introduced into these two regions, and their effects on membrane association and incorporation into nucleocapsid-M protein complexes were determined using quantitative assays. The results confirmed that the N terminus of M protein is involved in association with plasma membranes as well as nucleocapsids, although these two activities were differentially affected by individual mutations. Mutations in the 75-106 region affected incorporation into nucleocapsid-M complexes but had only minor effects on association with membranes. The ability of site-specific mutant M proteins to complement growth of temperature-sensitive M mutant virus did not correlate well with the ability to associate with membranes or nucleocapsids, suggesting that complementation involves an additional activity of M protein. Mutants with similar abilities to associate with membranes and nucleocapsids but differing in complementation activity were incorporated into infectious cDNA clones. Infectious virus was repeatedly recovered containing mutant M proteins capable of complementation but was never recovered with mutant M proteins that lacked complementation activity, providing further evidence for a separate activity of M protein that is essential for virus replication.
Most viruses that have a membrane or envelope as part of their structure acquire their envelopes by budding from the plasma membrane of the host cell. For budding to occur, the nucleoprotein core of the virus (nucleocapsid) must interact with the cytoplasmic surface of the host membrane. For many viruses this interaction is mediated by a matrix (M)2 protein that binds to both the nucleocapsid and the host membrane (1, 2). Despite the similarity in the functions of viral M proteins, there is little structural or sequence similarity among the M proteins of different virus families (3). Thus, understanding the relationship of structure to function must be undertaken for individual M proteins before the general principles involved in virus budding can be understood. The goal of the experiments described here was to determine sequences in the M protein of vesicular stomatitis virus (VSV) involved in binding to membranes and nucleocapsids.
VSV is the prototype member of the Rhabdoviridae family and has been widely studied to determine mechanisms involved in virus budding (2). The core of the virus contains an ∼11-kilobase negative-stranded RNA genome covered by 1300 copies of a single nucleocapsid protein (4). The nucleocapsid also contains lesser amounts of two proteins, P and L, which constitute the viral RNA-dependent RNA polymerase. The envelope contains a single species of transmembrane glycoprotein (G protein) that mediates virus attachment and entry into host cells. The virion contains ∼2000 copies of the M protein (4), which binds the nucleocapsid to the envelope and condenses the nucleocapsid into a tightly coiled helical nucleocapsid-M protein (NCM) complex that gives the virion its bullet-like shape (5-8). In cells infected with VSV and in transfected cells that express M protein in the absence of other VSV components, M protein is present both in a soluble form and bound to the cytoplasmic surface of the host plasma membrane (9-18). Mutagenesis studies, affinity labeling, and membrane reconstitution experiments have suggested that a combination of hydrophobic and ionic interactions mediate M protein binding to membranes by binding acidic phospholipids on the inner surface of the host plasma membrane (for review, see Ref. 19). Binding of M protein to nucleocapsids is less well understood than its binding to membranes. Most of the M protein in isolated NCM complexes is bound in a rapidly reversible equilibrium (20). However, M protein does not bind to nucleocapsids from which all of the M protein has been dissociated or to intracellular nucleocapsids that have never been assembled with M protein (11, 20). This suggests that binding of M protein to nucleocapsids in infected cells must be initiated in a separate step, after which most of the M protein is recruited into the NCM complex through the reversible binding step.
M protein does not have separately folded domains that mediate binding to membranes versus nucleocapsids. The 229-amino acid (aa) M protein contains a positively charged N terminus (aa 1-50) that is highly exposed to proteolysis. The remainder of M protein (aa 51-229) is compactly folded to form a protease-resistant core (16, 21-23). The ability to obtain crystals of M protein required proteolytic removal of both the N-terminal sequence (aa 1-47) and a hydrophobic sequence (aa 121-124) to prevent M protein self-association (21, 22); however, the resulting structure showed a single-domain fold for the crystallized portion of M. In the present study we focused on two regions of the M protein structure that had been suggested to be involved in binding to either membranes or nucleocapsids; 1) previous data had implicated the N-terminal sequence in binding to both nucleocapsids and membranes (9, 10, 16, 22-25) and 2) deletion mutagenesis studies had implicated an additional region from aa 75-106 in membrane binding (16).
In the experiments described here, M protein sequence substitutions were made using a scanning approach in the N-terminal sequence, and substitutions were based on the crystal structure in the 75-106-aa region. These mutants were used to determine the specific amino acids involved in these interactions. The results confirm that the N terminus of M protein is involved in association with plasma membranes as well as nucleocapsids, although these two activities are differentially affected by individual mutations. Mutations in the 75-106-aa region affected incorporation into NCM complexes but had only minor effects on association with membranes. Furthermore, the ability of mutant M proteins to function in the context of virus infection suggested that a new activity of M protein that is separate from its ability to associate with membranes or NCM complexes is critical for virus assembly.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Transfection—The M gene of the wild-type VSV was subcloned into pET21d vector as described (10). A glycine at position 2 that was changed during cloning was mutagenized back to wild-type serine using the QuikChange II XL (Stratagene) site-directed mutagenesis kit. The mutations described in Fig. 1 were made using the corrected pET21d-M protein expressing plasmid using the QuikChange II XL site-directed mutagenesis kit. The M protein sequences were verified by automated sequence analysis. BHK cells were infected with vaccinia virus expressing T7 polymerase (VV-T7) at a multiplicity of infection of 5 pfu/cell for 1 h and then transiently transfected using Lipofectin reagent (Invitrogen) with empty vector or with the M protein expression plasmid under the control of the T7 promoter for the times indicated in each of the figures (10).
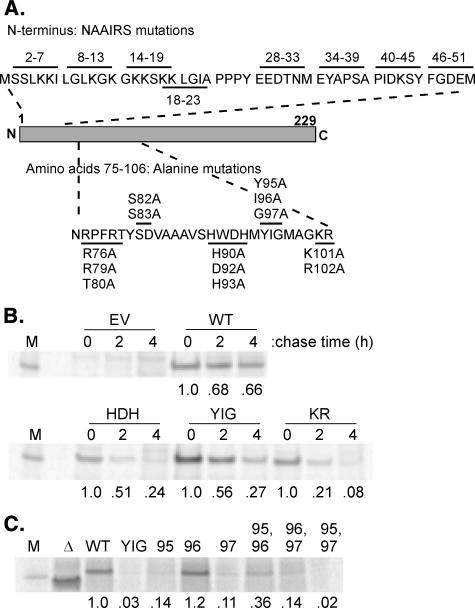
FIGURE 1.
A, strategy for mutagenesis of M protein. Top row, amino acids from positions 2-51 were replaced in blocks of six amino acids with the hexamer sequence Asn, Ala, Ala, Ile, Arg, Ser (NAAIRS). Bottom row, surface-exposed residues in the region from positions 75 to 106 were substituted with Ala in groups of 2 or 3 amino acids. Solid lines indicate sequences whose amino acids were replaced. B, turnover of mutant M proteins. Cells were transfected with plasmids encoding wild-type (WT) or mutant M proteins, HDH, YIG, or KR, or the empty vector (EV) control. Cells were pulse-labeled with [35S]methionine and chased in non-radiolabeled media for the indicated times. Labeled M proteins were immunoprecipitated and analyzed by SDS-PAGE and phosphorimaging, an image of which is shown (M, virion M protein standard). Intensity of M protein bands is shown below, normalized to the intensity at time 0. C, expression of mutant M proteins with single or double amino acid substitutions of Tyr-95, Ile-96, and Gly-97. Cells were transfected with plasmids encoding wild-type or Δ4-21 mutant or mutant M proteins with substitutions at the indicated position. Cells were continuously labeled with [35S]methionine, and labeled M proteins were immunoprecipitated and analyzed by SDS-PAGE and phosphorimaging.
Immunolabeling and Confocal Microscopy—BHK cells were seeded at a density of 7 × 105 cells/well in 2-well chamber slides (Nunc). After 24 h the cells were mock-infected, infected with a multiplicity of infection of 20 pfu/cell of VSV to ensure that cells were synchronously infected, or infected with VV-T7 and transfected with 1 μg of plasmid DNA encoding wild-type or mutant M protein. After 4 h (infection) or 8 h (transfection), cells were fixed with 4% paraformaldehyde (Sigma) for 10 min and processed for immunofluorescence microscopy as described (26). Cellular DNA was labeled with 0.5 μm topro-3-iodide (Invitrogen). Confocal images of labeled cells were used to analyze the plasma membrane and cytoplasmic labeling with laser-scanning microscope 5.0 software (Zeiss). Intersecting lines in the shape of an X were placed through each cell. Rhodamine labeling pixel intensity plots were made for each cross-section. The peak pixel intensity of the plasma membrane was collected at the four points where the X crossed the plasma membrane. Two cytoplasmic pixel intensity readings were collected at the 1/3 and the 2/3 distance between each plasma membrane reading and the nucleus. To avoid bias in selection of which points to quantify, the center of the X was placed in the center of the nucleus without rotating the original image.
Metabolic Labeling and Determination of M Protein Turnover—BHK cells were seeded at a density of 1 × 106 cells/well in 6-well tissue culture plates (Corning Incorporated) and incubated at 37 °C overnight. The cells were infected with VV-T7 at a multiplicity of infection of 5 pfu/cell and then 60 min of post-infection, and the cells were transfected with 1.5 μg of the appropriate pET21d-M plasmid. To determine total levels of M protein expression, cells were radiolabeled with media containing [35S]methionine (25 μCi/ml) for 16 h. M protein was immunoprecipitated from cell lysates as described (27), or cell lysates were analyzed directly by SDS-PAGE and phosphorimaging. No differences among mutant M proteins in immunoprecipitation efficiency were noted in these experiments. For analysis of M protein turnover cells were allowed to express M protein for 5 h. The cells were starved for methionine for 10 min and then pulsed with methionine-deficient medium supplemented with [35S]methionine (100 μCi/ml) for 30 min. The radioactive medium was replaced with nonradioactive medium, and at 0, 2, and 4 h post-labeling, the medium was removed, and M protein was immunoprecipitated from cell lysates as described (27).
Membrane Flotation Assay—Analysis of M protein association with membranes by flotation in sucrose gradients was performed as described (10). Briefly, BHK cells at a density of 1 × 107 cells in 100-mm tissue culture dishes were infected with VV-T7, and then 60 min post-infection, the cells were transfected with 15 μg of the appropriate pET21d-M plasmid. At 5 h post-transfection, cells were labeled with [35S]methionine for 1 h and then lysed in hypotonic buffer. Cell lysates were adjusted to 60% sucrose and were fractionated on a 60-40-10% sucrose gradient by centrifugation at 35,000 × g for 16 h at 4 °C. Fractions were collected from the top, and M protein was immunoprecipitated and analyzed by SDS-PAGE and phosphorimaging.
Virus-like Particle (VLP) Release Assay—BHK cells were infected with VV-T7, transfected with the appropriate M plasmid, and radiolabeled with media containing [35S]methionine (25 μCi/ml) for 16 h. The VLP-containing supernatants were collected, placed over a 15% sucrose cushion, and centrifuged to pellet the VLPs as described (6). VLPs and cell lysates were analyzed by SDS-PAGE and phosphorimaging analysis.
NCM Complex Exchange Assay—Radiolabeled wild-type or mutant M protein was synthesized by in vitro translation, and the ability to exchange with non-radiolabeled M protein in virion NCM complexes was assayed as described (11). Briefly, radiolabeled wild-type or mutant M protein was added to purified virions in buffer containing 120 mm NaCl. Triton X-100 was added to a final concentration of 0.1% to solubilize the viral envelope. This concentration was found to be as effective as the 0.5% Triton X-100 used previously. The mixture was incubated for 5 min at room temperature to allow M protein exchange. Samples were centrifuged at 100,000 × g using a Beckman Airfuge (Beckman Instruments). The supernatant and pellet fractions were analyzed by SDS-PAGE and phosphorimaging analysis.
Complementation of Temperature-sensitive M Protein (tsM) Mutant Virus—The ability of plasmid-derived wild-type and mutant M protein to complement growth of tsO23 virus was assayed as described (6). Briefly, BHK cells were infected with VV-T7 virus and transfected with plasmid DNA encoding wild-type or mutant M protein under control of the T7 promoter (1.5 μg/106 cells). Cells were coinfected with tsO23 virus at 39 °C, and the yields of infectious tsO23 virus progeny were determined by plaque assay at 32 °C.
Incorporation of M Protein Mutations into Recombinant Viruses—M protein mutations were introduced into plasmids containing a full-length infectious cDNA clone of VSV, and recombinant viruses were isolated as described (27).
RESULTS
Mutagenesis Strategy—Previous studies of VSV matrix function have used either protease-treated M protein or M protein deletion mutants to map the sequences involved in binding to nucleocapsids or membranes. In the studies reported here substitution mutagenesis was used to fine-map the putative binding sites more precisely while reducing the probability that the effects of mutation were due to misfolding (Fig. 1A). Our mutagenesis strategies were based on what was known from the crystal structure. Because the crystal structure did not contain the N terminus, amino acids starting after the initiator methionine were replaced in blocks of six aa with the hexamer sequence Asn, Ala, Ala, Ile, Arg, Ser (NAAIRS). The NAAIRS sequence has the ability to form different secondary structures, either an α-helix or a β-sheet, depending on the protein environment in which it resides (28). The NAAIRS flexibility in secondary structure has been used in mutational analysis in place of traditional alanine scanning to reduce the probability of mutation-based misfolding (29). The NAAIRS mutants are referred to by their location in the amino acid sequence; for example, 2-7NAAIRS is 2-7, etc. The sequence PPPY (aa 24-27) was not mutated, as it is known to mediate the late-budding function of M protein that results in virus release (2). Mutations at the second proposed plasma membrane binding site (aa 75-106) were based on the x-ray crystal structure of M protein. The mutations were targeted to change the side chains of surface-exposed amino acids to Ala in the proposed binding region. These mutants are referred to by the aa which were substituted with Ala (RRT, SD, HDH, YIG, and KR).
Mutations in the 75-106 Region of M Protein, HDH, YIG, and KR, Cause Increased Turnover of M Protein—M protein mutants were analyzed for their levels of expression in transfected cells using the vaccinia virus T7 polymerase expression system, as this is the only transfection system in mammalian cells that we have found to express levels of M protein comparable with those in VSV-infected cells (6, 24, 26, 30). All of the mutant M proteins were expressed at the same levels as wild-type M protein, as determined by radiolabeling for 16 h and analysis of cell lysates by immunoprecipitation, SDS-PAGE and phosphorimaging, except for three of the mutants containing substitutions in the 75-106 region, HDH, YIG, and KR, which were expressed at levels 10-30% that of wild-type M protein (not shown). Pulse-chase experiments were used to determine whether the lower level of expression was due to increased turnover of the mutant M proteins. BHK cells were infected with recombinant vaccinia virus that expresses T7 RNA polymerase (VV-T7) and then were transfected with plasmids expressing the mutant M protein or wild-type M protein for 5 h. Cells were pulsed with [35S]methionine, then incubated in non-radiolabeled media for 0, 2, and 4 h. Cell lysates were immunoprecipitated with anti-M protein antibody and analyzed by SDS-PAGE and phosphorescence imaging. The only mutant M proteins that had turnover rates that were more than 2-fold that of wild type were the HDH, YIG, and KR mutants, as shown in Fig. 1B. The high turnover rates of these mutants may be due to misfolding. As shown below, these mutant M proteins do retain some ability to associate with membranes or nucleocapsids despite their high turnover.
We were particularly interested in the YIG mutation because of the high degree of conservation of Tyr-95 and Gly-97 among M proteins of members of the Rhabdoviridae family (supplemental Fig. 1). Individual alanine substitutions were generated at positions 95, 96, and 97 (Y95A, I96A, and G97A mutants) as well as double substitutions at these positions. The levels of expression of these mutant M proteins were determined by labeling transfected cells continuously with [35S]methionine for 16 h followed by immunoprecipitation as above (Fig. 1C). Only the substitution of the nonconserved Ile-96 resulted in expression comparable with wild-type M protein, whereas substitution of Tyr-95 or Gly-97 resulted in expression comparable with the original YIG mutant.
M Protein Mutants Have Altered Association with Host Plasma Membranes Compared with Wild-type M Protein—The association with host plasma membranes of wild-type and mutant M proteins was determined in transfected cells by three different experimental approaches. The first was quantitative analysis of confocal immunofluorescence microscopy images. We have recently shown that this approach is highly effective in detecting quantitative differences in plasma membrane association of M protein provided that a sufficiently large number of cells are analyzed (26). The second approach was flotation analysis of radiolabeled cellular membranes and cytosol. This approach has been used for many years to determine the membrane association of M protein (9-13, 18). The third approach was analysis of release of VLPs (6, 31). When M protein is expressed in transfected cells in the absence of other viral components, association with the plasma membrane results in release of membrane vesicles referred to as VLPs (6, 31). This is because of the association of the PPPY sequence (aa 24-27) with host Nedd4-like proteins, which mediate the late-budding function of M protein that results in virus release (2). Thus, the VLP release assay is a functional assay that requires both association of M protein with plasma membranes and association of the PPPY sequence of M protein with host proteins (2).
For confocal microscopy analysis, BHK cells were infected with VV-T7 followed by transfection with plasmids encoding wild-type or mutant M protein for 4 h. The deletion mutant, Δ4-21 (24), which is missing the positively charged N terminus of M protein, was used as a negative control for membrane association. The cells were fixed, permeabilized with saponin, and immunolabeled with primary anti-M protein antibody and rhodamine-conjugated secondary antibody. Cells were labeled with TO-PRO-3-iodide (TO-PRO) to identify the position of the cell nucleus. In addition, TO-PRO also labeled DNA-containing vaccinia virus factories present in the cytoplasm. Confocal images were obtained with the equator of the cells in focus (Fig. 2A). There was little if any labeling of M protein in the nuclei, likely because of the inefficient permeabilization of the nuclear envelope by saponin (32). Wild-type M protein labeling was apparent in both the plasma membrane and in the cytoplasm, with a higher intensity in the membrane. By contrast, Δ4-21 M protein labeling could not be distinguished between the plasma membrane and the cytoplasm. The labeling intensity of the remaining mutated M proteins was greater in the plasma membrane than the cytoplasm, indicating that they all have the ability to associate with host plasma membranes, including the M protein mutants with higher turnover rates, HGH, YIG, and KR. As an example, Fig. 2A shows results with the HDH mutant in which association with the membrane is apparent.
FIGURE 2.
Analysis of association of wild-type and mutant M protein with host plasma membranes. A, cells were transfected with plasmids encoding wild-type (WT), Δ4-21, or HDH mutant M protein or empty vector (EV) control. M protein was labeled by rhodamine immunofluorescence, and DNA was labeled with topro-3-iodide. Confocal microscopy was used to collect images of differential interference contrast (DIC) and M protein and DNA fluorescence. B, histograms of the rhodamine and topro-3-iodide pixel intensity are shown for the cross-sections of the cells from panel A. C, average ratio of M protein plasma membrane/cytoplasm pixel intensity. For each mutant M protein as well as the positive and negative controls, 10 cells were analyzed in each of 4 separate experiments (n = 40). Statistical differences in the pixel intensity ratios were determined using Student's t test. *, significantly different from wild type (p < 0.05). D, analysis of distribution of wild-type and YIG mutant M proteins by flotation in sucrose gradients. Cells were transfected with plasmids encoding wild-type or mutant (YIG) M protein, radiolabeled with [35S]methionine, and lysed in hypotonic buffer. Cell lysates were mixed with dense sucrose and subjected to centrifugation in sucrose gradients to float cellular membranes. M protein in gradient fractions was quantified by immunoprecipitation, SDS-PAGE, and phosphorimaging.
Plasma membrane and cytoplasmic labeling intensities were quantified along intersecting lines in the shape of an X drawn through each cell. Graphs of pixel intensity versus distance along a line through representative cells are shown in Fig. 2B. The peak pixel intensity of the plasma membrane was collected at the four points where the X crossed the plasma membrane. Two cytoplasmic pixel intensity readings were collected at the 1/3 and the 2/3 distance between each plasma membrane reading and the nucleus. Boundaries of the nuclei were visualized from TO-PRO labeling. The ratios of membrane labeling to cytoplasmic labeling were calculated for each cell. For each mutant M protein as well as the positive and negative controls 10 cells were analyzed in each of 4 separate experiments (n = 40). Fig. 2C shows the average ratio of M protein plasma membrane/cytoplasm pixel intensity for the mutant M proteins. The mutant M proteins 2-7, 8-13, 14-19, 40-45, RRT, SD, HDH, YIG, and KR were statistically different from wild-type M protein (p < 0.05), showing a lower association with membranes. These results are consistent with earlier data indicating that both the N-terminal region and the 75-106 region affect association with membranes.
Membrane association of wild-type and mutant M proteins was also determined by flotation analysis of radiolabeled cellular membranes and cytosol. Cells were infected with VV-T7, transfected with plasmids encoding wild-type or mutant M protein, radiolabeled with [35S]methionine at 5 h post-transfection, and lysed in hypotonic buffer. Cell lysates were mixed with dense sucrose and subjected to centrifugation in sucrose gradients to float cellular membranes. M protein in gradient fractions was quantified by immunoprecipitation, SDS-PAGE, and phosphorimaging.
All of the mutant M proteins associated with host membranes to an extent similar to that of wild-type M protein. By contrast to results of confocal microscopy analysis, there was no difference between wild-type and mutant M proteins in the percent of M protein in the membrane fractions. For example, Fig. 2D shows results comparing wild-type and YIG mutant M proteins in three separate experiments. There was no significant difference between wild-type and YIG mutant in the percentage of M protein in the membrane fractions. The reasons for the quantitative discrepancy between the confocal microscopy and membrane flotation approaches were not explored in detail, but a likely explanation is that the membrane flotation assay measures association with other cellular membranes in addition to the plasma membrane, whereas the image analysis specifically analyzes plasma membrane association.
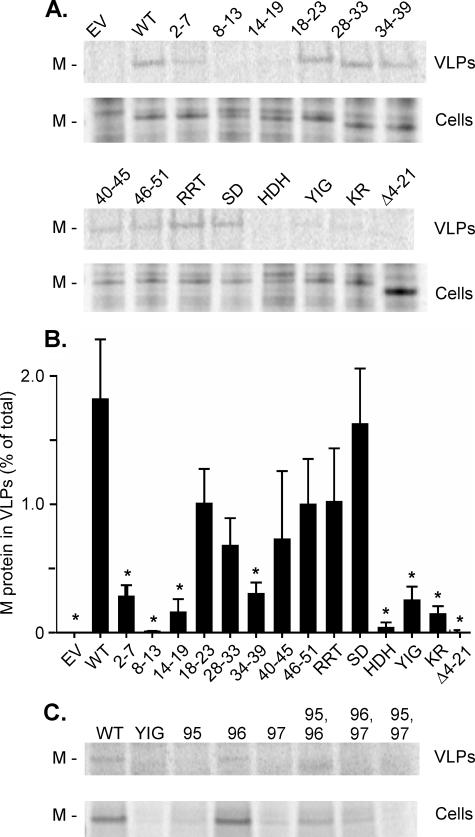
Mutant M Proteins Have a Reduced Activity in Release of VLPs—The ability of mutant M proteins to produce VLPs was assayed by quantifying the release of M protein-containing particles into the media from transfected cells. BHK cells were infected with VV-T7 and then transfected with empty vector or M protein expression plasmid under the control of the T7 promoter. Wild-type M protein was used as a positive control, and the Δ4-21 mutant M protein, which lacks membrane binding activity, was used as a negative control. The cells were radiolabeled with [35S]methionine for 16 h, and VLPs released into the media were concentrated by centrifugation and analyzed by SDS-PAGE and phosphorescence imaging (Fig. 3A). The amounts of wild-type or mutant M protein in VLPs and cell lysates were quantified and expressed as a percent of total M protein in VLPs (Fig. 3B). Mutant M proteins 2-7, 8-13, 14-19, 34-39, HDH, YIG, and KR had statistically different (p < 0.05) VLP release compared with wild-type M protein. The mutants that were significantly different from wild-type M protein in VLP release were those that had significantly different membrane association as determined by confocal microscopy, with the exception of mutant 34-39. This mutation alters a potential late budding sequence in M protein (PSAP, aa 37-40), which plays little role in budding of virions compared with aa 24-27, PPPY (33), but appears to affect VLP release from transfected cells.
FIGURE 3.
Analysis of release of VLPs with wild-type and mutant M proteins. A, cells were transfected with plasmids encoding wild-type (WT) or the indicated mutant M proteins. Cells were continuously labeled with [35S]methionine, labeled VLPs were concentrated by centrifugation through a 15% sucrose cushion, and VLPs and cell lysates (Cells) were analyzed by SDS-PAGE and phosphorescence imaging. EV, empty vector. B, quantification of M protein in VLPs expressed as a percent of total cellular M protein. Data are presented as the averages of three separate experiments ± S.D. Statistical differences were determined using Student's t test. *, significantly different from wild-type (p < 0.05). C, analysis of release of VLPs with single and double mutations in the YIG sequence. VLP release was assayed as in A except that M proteins were immunoprecipitated from cell lysates before SDS-PAGE and phosphorimaging.
In a separate series of experiments, the single and double mutants of the YIG sequence were analyzed for their ability to release VLPs. Of these, the I96A single mutant and Y95A,I96A double mutant released VLPs to a level comparable with that of wild-type M protein, whereas each of the other mutants was defective (Fig. 3C).
M Protein Mutants Have a Reduction in the Ability to Associate with Nucleocapsids—To determine the effect of M protein mutations on the ability to assemble into NCM complexes, we used our M protein exchange assay. This assay measures the reversible exchange of exogenous radiolabeled M protein for nonlabeled endogenous M protein in NCM complexes from virions at physiological ionic strength (10-12, 20). In this assay wild-type or mutant M protein labeled with [35S]methionine by in vitro translation was mixed with purified virions, and the virion envelopes were solubilized with detergent to release the NCM complexes. After a short incubation time to allow the exchange of labeled M protein with non-radiolabeled M protein, NCM complexes were pelleted and analyzed by SDS-PAGE and phosphorimaging. If the labeled M protein is capable of associating with NCM complexes, a portion of the radiolabeled protein will be found in the pellet after centrifugation, whereas the labeled M protein that is not exchanged will remain in the supernatant. With increasing amounts of added NCM complex, more M protein should be found in the pellet.
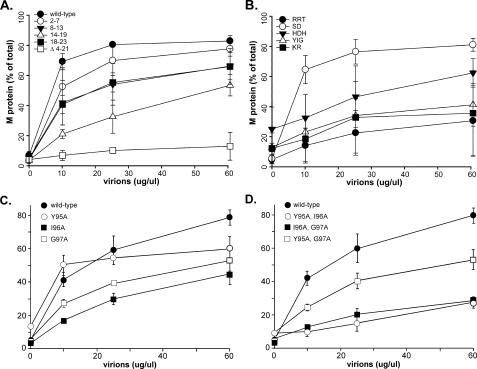
Fig. 4 shows results comparing the assembly into NCM complexes of labeled wild-type and mutant M proteins. For clarity, the results are divided into four separate graphs. Fig. 4A shows results for the most N-terminal NAAIRS mutants (2-7, 8-13, 14-19, and 18-23) as well as the positive and negative controls, wild-type and Δ4-21 M proteins, respectively. All of the mutants had detectable binding activity compared with the negative control Δ4-21 M protein, but the abilities of most of the mutants to be incorporated into NCM complexes were altered compared with wild-type M protein in the order 14-19 < 18-23 = 8-13 < 2-7 < wild type. The remaining N-terminal mutant M proteins (28-33, 34-39, 40-45, and 46-51) assembled into NCM complexes as effectively as wild-type M protein (not shown). These results support the conclusions from previous data that the N terminus of M protein is involved in binding to nucleocapsids (23).
FIGURE 4.
Binding of wild type and mutant M protein to NCM complexes. Radiolabeled wild-type (WT) and mutant M protein translated in vitro were mixed with purified virions as a source of NCM complexes. Virion envelopes were solubilized in buffer containing Triton X-100, radiolabeled M protein was allowed to exchange with the endogenous M protein, and NCM complexes were pelleted. Data shown are the percent of labeled M protein in the pellet from three separate experiments ± S.D. A, WT, 2-7, 8-13, 14-19, 18-23, Δ4-21. B, RRT, SD, HDH, YIG, KR. C, single substitutions of Tyr-95, Ile-96, and Gly-97. D, double substitutions of Tyr-95, Ile-96, and Gly-97.
Fig. 4B shows data for M proteins containing mutations in the 75-106 region. The association of the SD mutant (open circles) with NCM complexes was similar to that of wild-type M protein (shown in Fig. 4A). By contrast, the RRT mutant (closed circles) was dramatically reduced in its ability to associate with NCM complexes. This result indicates that this region (aa 76-80) of M protein is likely involved in interaction with nucleocapsids. The HDH, YIG, and KR mutants were found to have a high background of M protein in the pellet in the absence of added NCM complexes (y intercept > 10% in Fig. 4B), and the association with NCM complexes did not increase markedly above this background. The high background is indicative of aggregation of the mutant M proteins under the conditions of the assay (in which wild-type M protein is monomeric (15)), which may be further evidence of misfolding of these mutant M proteins in addition to their high turnover and inability to support the production of VLPs (Figs. 1 and 3).
Figs. 4, C and D, show the association with NCM complexes of the single and double mutants of the YIG sequence (aa 95-97) analyzed in a separate series of experiments. The single mutant Y95A had a high background of M protein aggregation and an unusual concentration dependence of association with NCM complexes, indicating that this protein was likely to be misfolded. The remaining mutants had low backgrounds of aggregation and were incorporated into NCM complexes to varying extents, although all had reduced activity compared with that of wild-type M protein. These data indicate that the region of M protein around aa 95-97 is likely involved in interaction with nucleocapsids.
Most M Protein Mutants Can Complement Temperature-sensitive M Protein Mutant Virus—tsM mutant viruses produce noninfectious particles containing nucleocapsids but are deficient in M protein at the nonpermissive temperature (6). It has been proposed that the defect in assembly of these mutants is in the initiation step of association of M protein with nucleocapsids (12). The site-specific mutant M proteins in Fig. 1 were tested for their ability to complement a tsM mutant virus to determine whether complementation is correlated with either the ability to bind membranes or the ability to bind nucleocapsids. BHK cells were infected with VV-T7 virus and transfected with plasmid DNA encoding wild-type or mutant M protein under control of the T7 promoter. Cells were coinfected with tsO23 virus at 39 °C, and the yields of infectious tsO23 virus progeny were determined by plaque assay at 32 °C. Data are expressed as a complementation ratio, which is virus yield in the presence of the test M protein divided by virus yield in the presence of the negative control Δ4-21 M protein (Fig. 5).
FIGURE 5.
Complementation of growth of tsM mutant virus by wild-type and mutant M proteins. Cells were transfected with plasmids encoding wild-type (WT) or the indicated mutant M proteins. Cells were coinfected with tsO23 virus at 39 °C. Virus yield was determined by plaque assay at 33 °C. Data are expressed as a complementation ratio calculated as the ratio of the virus yield in the presence of the test M protein divided by the yield in the presence of the negative control Δ4-21 mutant M protein.
All of the N-terminal mutants complemented the growth of tsO23 virus to a level similar to that of wild-type M protein, with the exception of the 14-19 mutant. Of the mutants in the 75-106 region, the SD and RRT mutants complemented tsO23 virus growth, but the I96A and the I96A,G97A mutants did not (the other mutants with high turnover rates were not assayed). These data indicate that the ability to complement tsO23 virus growth did not correlate with the ability to associate with either plasma membranes or NCM complexes. The mutants with the most severe defects in association with membranes or NCM complexes (8-13 and RRT, respectively) were able to complement tsO23 virus growth as effectively as wild-type M protein. The mutants that did not complement tsO23 virus growth (14-19, I96A, and I96A,G97A) were reduced in their ability to associate with NCM complexes but not as severely as the RRT mutant.
Recovery of a Recombinant Virus Containing the RRT M Protein Mutation—The RRT mutant and the two mutants in the YIG sequence that have detectable function (I96A and I96A,G97A) present an interesting contrast. They have similar abilities to release VLP and associate with NCM complexes to similar extents yet differ dramatically in their ability to complement tsO23 virus growth. To further test their effects on virus replication, these mutations were incorporated into plasmids containing full-length infectious VSV cDNAs. Recovery of infectious virus from transfected cells was attempted in multiple experiments. Although RRT M mutant virus was readily recovered in each experiment with an efficiency similar to wild-type M controls, infectious virus was never recovered from plasmid DNA containing the I96A or Y95A,G97A mutations. Similarly, multiple attempts were made without success to recover viruses containing the other single, double, and triple mutations in the YIG sequence.
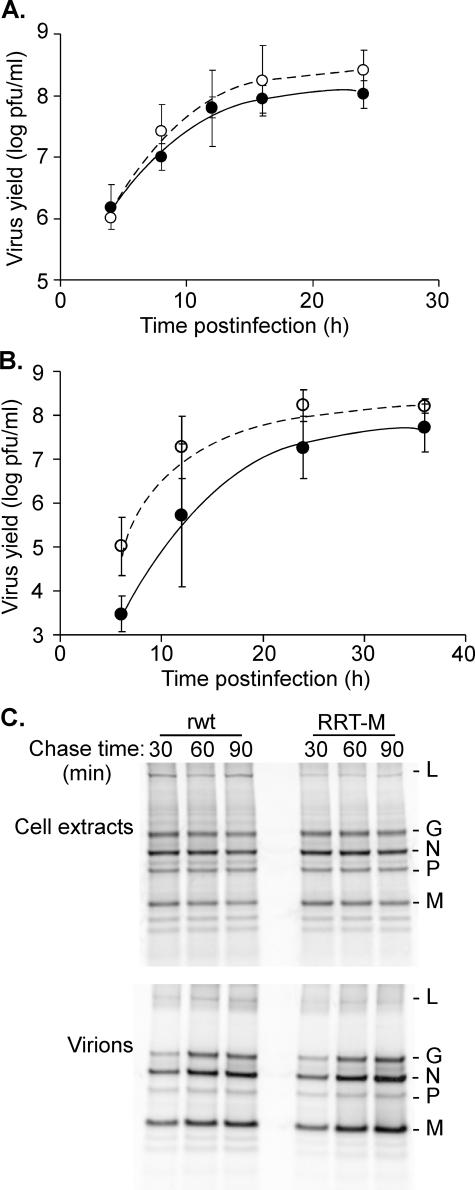
Virus stocks were prepared from plaque-purified virus containing the RRT mutation (RRT-M virus). The presence of the mutation in the recovered virus was confirmed by sequencing reverse transcription-PCR products. BHK cells were infected with RRT-M virus or the isogenic recombinant wild-type (rwt) control virus at a multiplicity of infection of 10 pfu/cell for single-cycle growth curves and 0.01 pfu/cell for multiple-cycle growth curves, and the production of infectious progeny was determined by plaque assay. There was little if any difference between RRT-M and rwt viruses in either single-cycle (Fig. 6A) or multiple-cycle growth curves (Fig. 6B).
FIGURE 6.
Assembly of recombinant virus containing RRT mutant M protein. A, cells were infected with recombinant viruses containing wild-type (rwt) or RRT mutant M proteins (RRT-M) at a multiplicity of 10 pfu/cell to determine single-cycle growth. Virus yield was determined at the indicated times post-infection by plaque assay for RRT-M (open circles) or rwt (closed circles) viruses. B, cells were infected with rwt virus (closed circles) or RRT-M virus (open circles) at a multiplicity of 0.01 pfu/cell to determine multiple-cycle growth. C, infected cells were labeled continuously with [35S]methionine for 6 h. The cells were washed extensively and incubated in non-radiolabeled media for the indicated chase times. The labeled virions and cell lysates were harvested and analyzed by SDS-PAGE and phosphorimaging.
The incorporation of viral proteins into RRT-M virus particles was determined by radiolabeling with [35S]methionine. BHK cells were infected with RRT-M or rwt viruses and labeled continuously for 6 h. The cells were washed extensively, and incubated in non-radiolabeled media for 30, 60, or 90 min. The labeled virions and cell lysates were harvested and analyzed by SDS-PAGE and phosphorimaging (Fig. 6C). There was no obvious difference between rwt and RRT-M viruses in the levels of viral protein expression in cell lysates, the protein composition of the released virions, or the percentage of viral proteins incorporated into virions. Multiple experiments similar to Fig. 6C were quantified, and there was no significant difference in the levels of viral protein expression or assembly into virions despite the low apparent affinity of RRT M protein for incorporation into NCM complexes (Fig. 4B). These results indicate that the affinity of M protein incorporation into NCM complexes is not a rate-limiting step of the efficiency of virus assembly.
DISCUSSION
Table 1 shows a summary of the phenotypes of the mutant M proteins analyzed here. Collectively the data in Figs. 2, 3, 4 confirm earlier studies using deletions and truncations of M protein showing that N-terminal sequences affect association with both host plasma membranes and viral nucleocapsids (9, 16, 22-25). Although there is substantial overlap, the sequences affecting plasma membrane association and VLP release are slightly more N-terminal than those affecting incorporation into NCM complexes. This can be seen by comparing the phenotypes of the 2-7 mutant (more severe effect on membrane association and VLP release) with those of the 18-23 mutant (more severe effect on incorporation into NCM complexes). Together with the RRT mutant, these mutants are the first examples of M protein mutants that clearly separate these two functions of M protein in virus assembly. The key to this achievement was to have quantitative assays for these individual steps.
TABLE 1.
Summary of phenotypes of mutant M proteins
Data from Figs. 3, 4, 5, 6 were categorized as + +, normal (100-80% of wild type); +, reduced (80-40%); −, deficient (40-20%); − −, severely deficient (<20%). NT, not tested.
| Mutant M protein | Membrane association | VLP release | NCM association | Complement tsM virus |
|---|---|---|---|---|
| Wild type | ++ | ++ | ++ | ++ |
| 2-7 | + | − | + | ++ |
| 8-13 | + | −− | − | ++ |
| 14-19 | + | − | − | − |
| 18-23 | ++ | + | − | + |
| 28-33 | ++ | + | ++ | + |
| 34-39 | ++ | − | ++ | ++ |
| 40-45 | + | + | ++ | ++ |
| 46-51 | ++ | + | ++ | ++ |
| RRT | + | + | −− | ++ |
| SDs | + | ++ | ++ | ++ |
| HDH | + | − | − | NT |
| YIG | + | − | − | NT |
| KR | + | − | − | NT |
| 196A | NT | ++ | − | − |
| Y95A, G97A | NT | ++ | − | − |
| Δ4-21 | −− | −− | −− | −− |
It is unclear how similar N-terminal sequences can bind to both the plasma membrane and nucleocapsids. Based on labeling with hydrophobic, photoreactive probes, Lenard and Vanderoef (25) showed that the N terminus of M protein interacts with the hydrophobic region of the envelope lipid bilayer. By analyzing cleavage products, the label was essentially entirely localized within the first 19, and probably within the first 5-10 aa at the N terminus. They suggested that the N terminus of M protein may form an amphipathic helix in which the hydrophobic face associates with lipid fatty acyl chains, and the positively charged face associates with negatively charged lipid head groups. It is possible that positively charged residues in such an amphipathic helix could interact with negative charges on both the membrane and the nucleocapsid protein. Although the stoichiometry is not quite 2:1, there are almost twice as many M proteins as N proteins in virion NCM complexes (4). Thus, another possibility is that the N termini of some M protein molecules interact with the membrane, and the N termini of other M protein molecules interact with the nucleocapsid.
In a previous study M protein amino acids 75-106 were proposed as a second membrane binding site based on deletion mutagenesis (16). However, in the data presented here, mutants in the 75-106-aa region had only slight reductions in membrane localization of M protein. The HDH, YIG, and KR mutants were defective in plasma membrane association, VLP release, and NC binding (Figs. 3, 4, 5). Also, these mutants turned over more rapidly (Fig. 1). It is probable that the previously studied deletion mutations would have had even more severe effects on folding. Unexpectedly, the RRT mutant showed that the 75-106-amino acid region is involved in binding to NCM complexes, but this mutation had little if any effect on membrane association and VLP release. Similarly, the mutant M proteins containing substitutions in the YIG sequence that retain biological activity in VLP release (I96A and Y95A,I96A) were reduced in their apparent affinities for incorporation into NCM complexes.
Neither the association with membranes nor the ability to be incorporated into NCM complexes correlated particularly well with the ability of the site-specific mutant M proteins to complement tsM mutant virus (Table 1). Many different tsM mutant viruses have been isolated, and the amino acid substitutions responsible for their temperature sensitivity have been determined (34, 35). In general, these substitutions are located in the interior of the M protein structure and are thought to affect the conformation of M protein at the nonpermissive temperature (22). However, the tsM protein present in the cytosol at the nonpermissive temperature is fully competent to be incorporated into NCM complexes in our M protein exchange assay, indicating that most of the tsM protein is not misfolded (12). By contrast, the membrane-associated tsM protein appears to be present in nonfunctional aggregates that likely result from misfolding (12). The step in assembly that is blocked in cells infected with tsM mutant viruses appears to be in the initiation of assembly of M protein with nucleocapsids, as cells infected with these viruses at the nonpermissive temperature release VLPs that contain nucleocapsids but are largely devoid of M protein (6).
Most of the site-specific mutant M proteins complemented growth of tsM mutant virus as effectively as wild-type M protein, including those with severe defects in membrane binding and VLP release (e.g. 2-7 and 8-13 mutants) as well as those with defects in binding to NCM complexes (e.g. RRT mutant), indicating that neither of these functions is particularly critical for complementation. This result would be consistent with the idea that the tsM protein is competent both to bind to membranes and to be recruited into NCM complexes but is defective in a function different from either of these two activities, namely the ability to initiate binding to nucleocapsids (12).
The site-specific M proteins that failed to complement growth of tsM mutant virus (14-19, I96A, and the other mutants in the YIG sequence that retained some biological activity) had moderate defects in membrane binding, VLP release, or association with NCM complexes, although not as severe as some of the mutants that did complement tsM virus growth. For example, the I96A virus was incorporated into NCM complexes more effectively than the RRT mutant, and neither of these mutants was defective in VLP release. Further evidence that these two mutants were fundamentally different came from our attempts to incorporate these mutations into recombinant viruses. Although viruses containing the RRT mutation were readily recovered and had no obvious defect in virus growth, we were never able to recover a virus containing the I96A mutation. Although this negative result is not interpretable on its own, when combined with the results of the complementation experiments, these results indicate that the I96A mutant M protein is defective in an essential step in virus assembly.
The recombinant virus containing the RRT mutant M protein had no obvious defect in virus assembly (Fig. 6) despite an ∼10-fold reduction in apparent affinity for association with NCM complexes (Fig. 5). In vitro binding experiments have shown that the binding equilibrium reflected in the M protein exchange assay is responsible for condensing the NCM complex into the tightly coiled helix that gives the virion its bullet-like shape (5, 7, 8). This equilibrium is established rapidly (within seconds) and with a high apparent affinity (apparent Kd ∼ 0.5-1.0 μm) compared with the intracellular concentration of M protein (10-50 μm) (20). This could explain why even a dramatic reduction in the apparent affinity for incorporation into NCM complexes, such as that observed with the RRT mutant, would not have a substantial effect on the efficiency of virus assembly.
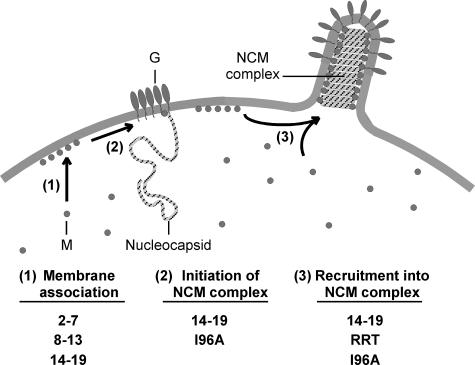
Fig. 7 shows a model of the proposed steps in assembly of M protein into virions and the mutations that have the most pronounced effect on each step. M protein is initially present as a soluble protein, which distributes between the cytosol and the plasma membrane (step 1). It is likely that association of M protein with nucleocapsids is initiated at the plasma membrane and involves nucleocapsids associated with the membrane independently of M protein (step 2). In support of this idea, we have recently demonstrated the presence of membrane-associated nucleocapsids in VSV-infected cells that colocalize with membrane microdomains that contain the viral envelope glycoprotein (G protein), but not M protein, and have proposed that these membrane-associated nucleocapsids are precursors to virus budding sites (26). Thus, initiation of assembly of M protein with these nucleocapsids would be the key step in formation of a virus budding site, and it is this step that is proposed to be defective in cells infected with tsM mutant viruses at the nonpermissive temperature. Once the assembly of M protein with nucleocapsids has been initiated, additional M protein is recruited from the membrane and cytosol to form the NCM complex (step 3). Although this step does not appear to be rate-limiting for virus assembly, it is nonetheless important for forming the bullet-shaped budding site that is characteristic of rhabdoviruses.
FIGURE 7.
Model for three steps in assembly of M protein into virions. M protein mutations that have the most pronounced effect on each step are indicated below.
In the model presented in Fig. 7, steps 1 and 3 are firmly grounded in the previous literature on VSV M protein. The data presented here show that whereas association of M protein with membranes and NCM complexes is no doubt important for virus structure, the critical step in virus assembly that remains to be discovered is the step that initiates assembly of M protein with nucleocapsids, i.e. step 2. We have proposed that this initial step in assembly of M protein with nucleocapsids might involve host factors, analogous to the involvement of the host ESCRT machinery in the late budding step mediated by M protein (11). The requirement for interaction with host proteins might account for the high degree of conservation of the YIG sequence among rhabdovirus M proteins, as this sequence may be involved in the initiation step as well as in the proper folding of M protein. Likewise, the 14-19 M protein mutant might be a useful tool in investigating the proposed initiation step, as this mutant M protein also fails to complement the tsM mutant virus and may be defective in initiation of assembly with nucleocapsids.
Supplementary Material
Acknowledgments
We acknowledge the Microscopy Core Laboratory, especially Ken Grant, for assistance with confocal microscopy and the Biomolecular Resource Laboratory for DNA sequencing. We also thank Maryam Ahmed for critical reading of the manuscript. The Microscopy Core Laboratory and the Biomolecular Resource Laboratory are supported in part by National Institutes of Health Core Grant CA12197 (NCI; to the Comprehensive Cancer Center of Wake Forest University).
This work was supported, in whole or in part, by National Institutes of Health Grant AI15892 (a Public Health Service grant from the NIAID). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: M, matrix; aa, amino acid; NCM, nucleocapsid-M protein complex; pfu, plaque-forming units; rwt, recombinant wild-type; VLP, virus-like particle; VSV, vesicular stomatitis virus; VV-T7, vaccinia virus that expresses T7 RNA polymerase; tsM, temperature-sensitive M protein.
References
- 1.Pornillos, O., Garrus, J. E., and Sundquist, W. I. (2002) Trends Cell Biol. 12 569-579 [DOI] [PubMed] [Google Scholar]
- 2.Jayakar, H. R., Jeetendra, E., and Whitt, M. A. (2004) Virus Res. 106 117-132 [DOI] [PubMed] [Google Scholar]
- 3.Timmins, J., Ruigrok, R. W., and Weissenhorn, W. (2004) FEMS Microbiol. Lett. 233 179-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas, D., Newcomb, W. W., Brown, J. C., Wall, J. S., Hainfeld, J. F., Trus, B. L., and Steven, A. C. (1985) J. Virol. 54 598-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barge, A., Gaudin, Y., Coulon, P., and Ruigrok, R. W. (1993) J. Virol. 67 7246-7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyles, D. S., McKenzie, M. O., Kaptur, P. E., Grant, K. W., and Jerome, W. G. (1996) Virology 217 76-87 [DOI] [PubMed] [Google Scholar]
- 7.Newcomb, W. W., and Brown, J. C. (1981) J. Virol. 39 295-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newcomb, W. W., Tobin, G. J., McGowan, J. J., and Brown, J. C. (1982) J. Virol. 41 1055-1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong, L. D., and Rose, J. K. (1993) J. Virol. 67 407-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, J. H., McKenzie, M. O., and Lyles, D. S. (2006) J. Virol. 80 3701-3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flood, E. A., and Lyles, D. S. (1999) Virology 261 295-308 [DOI] [PubMed] [Google Scholar]
- 12.Flood, E. A., McKenzie, M. O., and Lyles, D. S. (2000) Virology 278 520-533 [DOI] [PubMed] [Google Scholar]
- 13.Knipe, D. M., Baltimore, D., and Lodish, H. F. (1977) J. Virol. 21 1128-1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCreedy, B. J., Jr., and Lyles, D. S. (1989) Virus Res. 14 189-205 [DOI] [PubMed] [Google Scholar]
- 15.McCreedy, B. J., Jr., McKinnon, K. P., and Lyles, D. S. (1990) J. Virol. 64 902-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogden, J. R., Pal, R., and Wagner, R. R. (1986) J. Virol. 58 860-868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono, K., Dubois-Dalcq, M. E., Schubert, M., and Lazzarini, R. A. (1987) J. Virol. 61 1332-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye, Z., Sun, W., Suryanarayana, K., Justice, P., Robinson, D., and Wagner, R. R. (1994) J. Virol. 68 7386-7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenard, J. (1996) Virology 216 289-298 [DOI] [PubMed] [Google Scholar]
- 20.Lyles, D. S., and McKenzie, M. O. (1998) Biochemistry 37 439-450 [DOI] [PubMed] [Google Scholar]
- 21.Gaudier, M., Gaudin, Y., and Knossow, M. (2001) Virology 288 308-314 [DOI] [PubMed] [Google Scholar]
- 22.Gaudier, M., Gaudin, Y., and Knossow, M. (2002) EMBO J. 21 2886-2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaptur, P. E., Rhodes, R. B., and Lyles, D. S. (1991) J. Virol. 65 1057-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black, B. L., Rhodes, R. B., McKenzie, M., and Lyles, D. S. (1993) J. Virol. 67 4814-4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenard, J., and Vanderoef, R. (1990) J. Virol. 64 3486-3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swinteck, B. D., and Lyles, D. S. (2008) J. Virol. 82 5536-5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopecky, S. A., Willingham, M. C., and Lyles, D. S. (2001) J. Virol. 75 12169-12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, I. A., Haft, D. H., Getzoff, E. D., Tainer, J. A., Lerner, R. A., and Brenner, S. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 5255-5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armbruster, B. N., Banik, S. S., Guo, C., Smith, A. C., and Counter, C. M. (2001) Mol. Cell. Biol. 21 7775-7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, E. L., and Lyles, D. S. (2003) J. Virol. 77 3985-3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y., Luo, L., Schubert, M., Wagner, R. R., and Kang, C. Y. (1993) J. Virol. 67 4415-4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyles, D. S., Puddington, L., and McCreedy, B. J., Jr. (1988) J. Virol. 62 4387-4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irie, T., Licata, J. M., Jayakar, H. R., Whitt, M. A., Bell, P., and Harty, R. N. (2004) J. Virol. 78 7823-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopalakrishna, Y., and Lenard, J. (1985) J. Virol. 56 655-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita, K., Vanderoef, R., and Lenard, J. (1987) J. Virol. 61 256-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.