Abstract
Activation of the Aspergillus nidulans transcription factor PacC, which mediates ambient pH regulation of gene expression and is recruited to ESCRT-III by the Vps32-interacting scaffold PalA, involves its ambient pH-dependent C-terminal proteolysis. This reaction is almost certainly catalyzed by the PalB calpain-like protease. Here we show that PalB associates with membranes and interacts specifically and directly with ESCRT-III Vps24. The PalB N-terminal MIT domain and the Vps24 C-terminal MIM motif are necessary and sufficient for this interaction. PalBΔMIT, a mutant PalB lacking the MIT domain is inefficiently recruited to membranes and impaired in PacC proteolytic processing. Notably, membrane recruitment is promoted and PacC processing largely restored by covalent attachment of Vps24 to mutant PalBΔMIT. This is the first reported evidence that calpain-like recruitment to ESCRT-III lattices plays a physiological role. It unambiguously positions the calpain-like protease PalB within the ESCRT-III-associated pH signaling complex, underlines the positive role of ESCRT-III in ambient pH signal transduction, and suggests a possible mechanism for PalB activation.
The multivesicular body (MVB)3 pathway plays a central role in the delivery of membrane proteins reaching the endosomal system to the lumen of the vacuole/lysosome (1). Four protein complexes, denoted endosomal sorting complex required for transport (ESCRT)-0, -I, -II, and -III, direct the budding of vesicles into the lumen of multivesicular endosomes and mediate the sorting of cargoes in these vesicles (2, 3). In addition, ESCRTs and associated proteins play roles beyond the MVB pathway, including the budding of retroviruses from the plasma membrane and the abscission step during cytokinesis (4, 5).
The ultimate consequence of MVB sorting for an endocytosed membrane cargo is exposure to the late endosome/lysosome lumenal hydrolases, which inevitably ends with proteolytic degradation. Thus, the best characterized role of MVB sorting in signal transduction is the down-regulation of plasma membrane receptors following their endocytic internalization (1, 6). However, work on the fungal ambient pH signaling pathway, which regulates gene expression in response to the pH of the environment, underscored a major positive-acting role of ESCRT proteins in transducing the ambient pH signal to the zinc finger transcription factor PacC/Rim101 (reviewed in Refs. 7, 8), reinforcing the view that ambient pH signaling is one among the increasing number of examples of vesicle-associated signal transduction (VAST) (9).
In A. nidulans, the pH signaling pathway involves six dedicated proteins, denoted PalA, PalB, PalC, PalF, PalH, and PalI. This pal pathway is activated by alkaline ambient pH, which leads to the 2-step proteolytic conversion of the 72-kDa PacC72 translation product into the 53 kDa committed intermediate PacC53 and the 27-kDa final product PacC27 (10–12). PacC27 is a transcriptional activator of alkaline-expressed genes and a repressor of acid-expressed genes (13, 14). The alkaline ambient pH-dependent conversion of PacC72 into PacC53 is almost certainly catalyzed by the calpain-like cysteine protease PalB (10, 15, 16) (see Refs. 7, 8, 17 for reviews). The pH-independent proteolytic processing of PacC53 into PacC27 is in all likelihood mediated by the proteasome (11, 18). The pathway is conserved in Saccharomyces cerevisiae, where the PacC orthologue Rim101p is activated by a single proteolytic step (19), presumably catalyzed by the budding yeast PalB orthologue Rim13p (20, 21).
Work in yeasts (most notably in S. cerevisiae) and in A. nidulans has shown that the fungal ambient pH signaling pathway proteins are organized into two spatially separated complexes, respectively, located at the plasma and endosomal membranes (8). The plasma membrane complex involves the 7-TMD protein and likely ambient pH receptor PalH, the 3-TMD protein PalI and the arrestin-like protein PalF (22–24). The endosomal membrane complex involves the Bro1-domain containing scaffold PalA (Rim20p in S. cerevisiae), a direct interactor of ESCRT-III Vps32 (20, 25, 26) (see also this work) and PacC72/Rim101p, as PalA/Rim20p is able to recruit PacC72/Rim101p to this complex by means of its ability to bind two YPX(L/I) motifs located either side of the respective signaling protease cleavage sites in PacC72 and Rim101p (20, 25). Because PalB/Rim13p would also be a component of this endosomal complex, PalA/Rim20p-mediated recruitment of PacC72/Rim101p to ESCRT-III lattices would facilitate signaling proteolysis (20). As the plasma and endosomal membrane pH signaling complexes are spatially segregated (22, 26), there must exist a mechanism that facilitates their connection to activate signaling proteolysis. The finding that the arrestin-like PalF, which binds strongly to the PalH 7-TMD receptor cytosolic tail, is ubiquitinated in a PalH- and alkaline ambient pH-dependent manner strongly supports the hypothesis that this connection involves endocytic traffic (27). In the current model, the Bro1 domain-like containing protein PalC, which localizes to cortical structures also in a PalH- and alkaline ambient pH-dependent manner and can bind Vps32 directly, is thought to play a bridging role between the two spatially separated complexes (28, 29).
The view that PalA-dependent recruitment of PacC72 would facilitate PalB/Rim13p signaling protease cleavage in association with endosomal membranes (20) is supported by the direct or indirect interaction between the PalB orthologue Rim13p and Vps32p detected in yeast genome-wide two-hybrid screens (30). However, the association of PalB or Rim13p with this endosomal complex has not been studied in any significant detail, despite the key role attributed this calpain-like cysteine protease as the catalyst of the ambient pH-dependent proteolytic cleavage of PacC/Rim101p. PalB and Rim13p contain prototypical calpain domains. However, PalB, unlike Rim13p, contains an N-terminal MIT domain, a hallmark of ESCRT-III-interacting proteins.
The components of ESCRT-III are thought to play key roles in multivesicular body biogenesis through their ability to form polymeric structures on membranes (31, 32). In fungi, ESCRT-III involves six ESCRT-III-related proteins (yeast Vps2p, Vps20p, Vps32p, Vps24p, Did2p and Vps60p). Humans have 10 ESCRT-III-related proteins, denoted CHMP, that can be classified into six families corresponding to the six yeast proteins (2, 3). ESCRT-III-related proteins share a similar structural organization, as determined from structural studies of the human Vps24p orthologue CHMP3 (33, 34). This structural organization involves a basic N-terminal domain formed by four helices and a C-terminal acidic region that, in certain ESCRT-III-related proteins, mediates recruitment of ESCRT-III interactors containing one or more MIT domains (see Ref. 3 for review).
Certain MIT domain-containing ESCRT-III interactors are recruited to ESCRT-III-related proteins somewhat promiscuously. For example, the AAA ATPase Vps4 (35) is recruited, via its N-terminal MIT domain, to CHMP1A, CHMP1B, CHMP2A, and CHMP2B (36–40) in humans and to Vps2p and Did2p in S. cerevisiae (41, 42). In contrast, the AMSH deubiquitinase (43), although interacting with CHMP1 and CHMP2 (44), interacts preferentially with CHMP3 (the Vps24p orthologue) (34, 45–47). Crystallographic studies demonstrated that these interactions involve a MIM (mit interacting motif) located in the C-terminal acidic region of a subclass of ESCRT-III-like proteins including Did2p (CHMP1,2), Vps2p (CHMP2), and Vps24p (CHMP3). These studies provided a structural explanation for the above differences: the MIM amino acid sequences in Did2p, CHMP1A, CHMP1B, CHMP2A, CHMP2B, and Vps2p recognizing the Vps4 MIT domain are nearly identical, conforming to a (D/E)-2XXL-1XXR0L+1XXL+2(K/R)+3 consensus (38, 42). In contrast, the Vps24p (CHMP3) MIM motif contains an aliphatic residue rather than Asp or Glu in position -2 and Met in position -1, suggesting that these changes drive specificity of the Vps24p (CHMP3) MIM toward MIT domains other than that in Vps4.
Here we show that PalB interacts with A. nidulans Vps24 specifically and directly and that the MIT domain plays an important role in this interaction. A mutant PalB deleted for the MIT domain is impaired in the proteolytic processing of PacC but this mutant recovers function by forced recruitment to endosomal membranes. Our data unambiguously position the calpain-like protease PalB in the endosomal membrane signaling complex and underscore a positive role of ESCRT-III in intracellular signaling.
EXPERIMENTAL PROCEDURES
A. nidulans—palB gene replacements were carried out using MAD1764 (pyrG89 pyroA4 inoB2 ΔnkuA::bar pacC900 ΔpalB::pyroAAf), a ΔpalB strain in which the complete coding region of palB had been substituted by pyroAAf. This strain carries pacC900 encoding Myc3-PacC72 (16, 18). palB::(HA)3::pyrGAf, palBΔMIT::HA3::pyrGAf, and vps24:: palBΔMIT::HA3::pyrGAf transgenes, flanked by 5′-UTR and 3′-UTR palB sequences to drive homologous recombination, were obtained as NotI-ClaI linear fragments from plasmids p1725, p1727, and p1786, respectively. The resulting palB gene-replaced alleles were denoted palB800 [palB:: (HA)3::pyrGAf], palB801 [palBΔMIT::HA3::pyrGAf], and vps24::palBΔMIT [vps24::palBΔMIT::HA3::pyrGAf]. Gene-replaced transformants were identified by their pyrimidine-independent, pyridoxine-dependent growth and verified by Southern. The palB801 phenotype (or the wild-type phenotype for palB800 and vps24::palBΔMIT) co-segregated with the recombinant allele, ruling out the possibility of the presence of spurious mutations. pH shift experiments and A. nidulans protein extracts were as described (18). Recombinant strains were confirmed to carry the expected single-copy integration events by Southern analysis. Epitope-tagged transgenes were genotyped by PCR.
Subcellular Fractionation Experiments—These were carried out at 4 °C following a S. cerevisiae protocol (48, 49). Mycelia cultured in minimal medium (50) were protoplasted with Glucanex (Novo CH-4243). Protoplasts were resuspended in lysis buffer (0.2 M sorbitol, 50 mm potassium acetate, 2 mm EDTA, 20 mm HEPES pH 7.2 and protease inhibitor mixture (Roche Applied Science)) and lysed using a Dounce homogenizer. Debris were pelleted after centrifugation at 300 × g for 5 min. The supernatant was then centrifuged at 13,000 × g for 15 min, which resulted in a P13K pellet fraction and a supernatant. This supernatant was centrifuged at 100,000 × g for 60 min, using a Beckman TLA100 micro-ultracentrifuge, which resulted in a P100K pellet fraction and S100 supernatant soluble fraction. The S100, as well as the P13K and P100K pellets, which were resuspended in lysis buffer, were trichloroacetic acid-precipitated and resuspended in SDS-PAGE loading buffer. Equivalent samples of the different fractions were analyzed by Western blotting. Physiological levels of (FLAG)3-Pep12 were driven by the pep12 promoter from a single-copy flag3::pep12::pyrGAf transgene integrated into the pep12 locus. (Strain MAD1404).
Two-hybrid Analyses and GST Pull-down Assays using A. nidulans Extracts—Yeast two-hybrid assays and plasmids encoding GST-GFP and GST-Vps32 fusion protein have been described (28). pGEX-2T-derivatives encoding wild type and mutant (L222D, L223D) GST-Vps24 and GST-DidB were constructed by standard techniques and QuikChange (Stratagene) mutagenesis. GST fusion proteins were expressed in E. coli DH1 for 24 h at 20 °C after induction with isopropyl-1-thio-β-d-galactopyranoside and purified from lysates clarified by centrifugation for 20 min at 13,000 rpm and 4 °C in a Sorvall SS-34 rotor as described (51). GST baits were mixed with 1 mg of total soluble protein extract from palB800 or palB801 strains in BOA buffer (10 mm Tris-HCl pH 8.0, 150 mm NaCl, 1 mm EDTA, 5 mm dithiothreitol, 0.05% (v/v) Triton X-100, and Roche's protease inhibitor mixture), using 0.8-ml Handee Spin columns (Pierce), and rotated at 4 °C in the presence of 25 μl of glutathione-Sepharose (Amersham Biosciences). Beads were washed six times with BOA buffer, and the bound proteins were eluted after boiling in SDS-PAGE loading buffer. Replicate gels were stained with Coomassie Blue and analyzed by Western blot, using anti-HA antibody.
GST Pull-down Assays using Purified Proteins—[35S]PalB or [35S]PalA preys were synthesized in vitro using the Promega TnT system (20 μCi of [35S]Met per reaction). 20 μl of glutathione-Sepharose beads loaded with the corresponding GST bait were incubated at 4 °C with the 35S-labeled prey in a total volume of 0.5 ml of 1% Triton X-100 BOA buffer. Beads were washed, and the bound material eluted as above was run in 10% polyacrylamide gels, which were Coomassie Blue-stained, dried, and autoradiographed, using a Kodak BioMax amplifying screen.
IgG Sepharose Pull-downs with Purified Proteins—ZZ-PalBMIT is a peptide consisting of two protein A IgG binding Z domains fused to the PalB N-terminal 129 residues containing the MIT domain. (His6)-Vps24 and ZZ-PalBMIT were expressed in BLB21 cells. (His6)-Vps24. was purified by Ni2+ affinity chromatography using NTA His-Bind resin (Novagen). ZZ-PalBMIT was purified from cells lysed in the French Press in a buffer containing 20 mm Tris-HCl, pH 7.9, 300 mm NaCl, 5 mm imidazole, 0.1% Triton X-100, 1 mm PFA-block, and Roche Applied Science's EDTA-free protease inhibitor mixture. Clarified bacterial lysates were loaded onto an IgG Sepharose 6 Fast flow column in 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 0.02% Tween 20 (TST). ZZ-PalBMIT was eluted with 0.5 m acetic acid, adjusted to pH 3.5 with ammonium acetate, lyophilized, resuspended in TST buffer, and stored at -20 °C. Purified proteins (or buffer in the case of controls) were mixed in 0.8-ml Handee columns (Pierce) with 30 μl of IgG Sepharose slurry and rotated for 2 h at 4 °C. After five washes with TST, the bound material was eluted with 50 μl of SDS-PAGE loading buffer. Proteins were resolved using 12% polyacrylamide SDS-PAGE and detected with Coomassie Blue.
Western Blot Analyses—The following primary monoclonal antibodies were used: mouse anti-Myc 9E10 (Santa Cruz Biotechnology, 1/1000) for detection of (Myc)3::PacC; rat anti-HA 3F10 (Roche Applied Science, 1/1000) for PalB::(HA)3; mouse anti-FLAG M2 (Sigma, 1/80,000) for (FLAG)3::Pep12; and mouse anti-actin C4 (ICN Biomedicals) for β-actin. Rabbit anti-yeast hexokinase antiserum (Chemicon, 1/80,000) was used for hexokinase. These were combined with peroxidase-coupled sheep anti-mouse IgG (Amersham, 1/4,000), goat anti-rat IgM+IgG (goat, 1/4,000) or donkey anti-rabbit IgG as secondary antibodies, which were reacted with ECL (Amersham Biosciences).
RESULTS
PalB contains a central calpain-like thiol protease catalytic domain, a calpain III domain toward the C terminus and an N-terminal MIT domain (Fig. 1A). MIT domains are hallmarks of ESCRT-III-interacting proteins.
FIGURE 1.
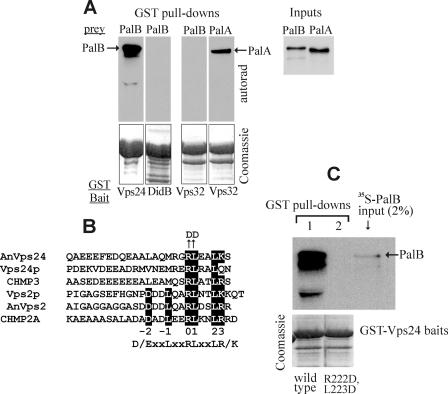
PalB interacts with Vps24 of ESCRT-III. A, schemes of the domain organizations of PalB and of Vps24; B and C, two-hybrid analyses: the indicated GAL4 DNA binding domain (pGBKT7) and activation domain (pACT2AD) constructs were tested in S. cerevisiae Y187, using quantitative β-galactosidase assays. Data represent average values of five clones for each combination. Bars indicate standard errors. Φ indicates empty pGBKT7 or pACT2 vectors. D, GST-Vps24 pull-down assays of A. nidulans extracts expressing physiological levels of PalB or PalBΔMIT, as indicated. Input lanes contained 2% of the pulled-down material. PalB was detected by α-HA Western blotting.
PalB Is an Interactor of ESCRT-III Vps24: the PalB MIT Domain Is Necessary and Sufficient for Binding Vps24—To explore the possible association of PalB with ESCRT-III, we performed two-hybrid assays with full-length PalB. PalB did not interact at all with A. nidulans ESCRT-III components Vps32, Vps2, or Vps20, with the AAA ATPase Vps4 or with ESCRT-II Vps25 (data not shown) (only one of the two possible bait/prey combinations was tested for Vps20 and Vps4, which inhibited growth when fused to the Gal4 DBD). However, PalB showed two-hybrid interaction with Vps24 in either possible orientation (Fig. 1, B and C). In sharp contrast, interaction of PalB with DidBDid2 was barely above detection (Fig. 1B, lane 4) whereas, in agreement with previous reports with S. cerevisiae and human proteins, Vps4 interacted strongly with DidBDid2 (Fig. 1C, lane 15).
As mutant PalB, deleted for the MIT domain, PalB-(77–847) did not interact with Vps24 (Fig. 1B, lane 2) whereas bait PalB-(1–96) and prey PalB-(1–130) constructs encompassing the N-terminal MIT domain showed strong interaction (Fig. 1, B and C), we conclude that the PalB-Vps24 interaction is MIT domain-mediated.
Vps4 contains an intensively investigated ESCRT-III-interacting MIT domain (37, 38, 42, 52). However, in sharp contrast with its strong interaction with DidBDid2, Vps4 interacted very inefficiently with Vps24 in two-hybrid assays, as reflected by the 30-fold lower β-galactosidase activity (Fig. 1C, lanes 14 and 15). These and the above PalB data strongly support the contention that Vps24 specifically recruits the PalB MIT domain whereas DidBDid2 specifically recruits the Vps4 MIT domain. In agreement, the PalB MIT domain prey construct containing the N-terminal 130 PalB residues interacted 10 times more efficiently with Vps24 than with DidB in terms of β-galactosidase activity (Fig. 1C, lanes 13 and 16) whereas the PalB(1–96) bait construct interacted strongly with Vps24 and not at all with DidB (Fig. 1B, lanes 3 and 5).
A gene replacement allele encoding wild-type PalB::(HA)3 was constructed as outlined in supplemental Fig. S1 and denoted palB800. To confirm the Vps24-PalB interaction, we used GST pull-down assays with A. nidulans extracts expressing physiological levels of PalB::(HA)3. Unlike the unrelated GST-GFP fusion protein, GST-Vps24 efficiently pulled-down PalB::(HA)3 (Fig. 1D and supplemental Fig. S2). Of note, several (HA)3 immunoreactive bands showing faster electrophoretic mobility than the full-length protein, almost certainly representing N-terminal PalB degradation products (HA3 is attached to the PalB C terminus), were detectable in the input extract. However, only the full-length protein co-purified with GST-Vps24 beads, in agreement with the Vps24-PalB interaction being mediated by the N-terminal MIT domain (Fig. 1D). Indeed, mutant PalB-(HA)3 lacking the MIT domain was not pulled-down by GST-Vps24 (Fig. 1D).
PalB Interacts Directly with Vps24—The MIT domain dependence displayed by the Vps24-PalB interaction detected by two-hybrid and GST pull-down experiments strongly indicates that this interaction is direct. Definitive evidence that PalB interacts directly with Vps24 was obtained from two types of experiments. Firstly, we used in vitro labeled [35S]PalB and [35S]PalA baits, rather than A. nidulans extracts, in pull-down assays with GST-Vps32, GST-DidB, and GST-Vps24. GST-Vps32 interacted directly with PalA and did not pull-down PalB. In contrast, GST-Vps24 efficiently pulled-down PalB whereas GST-DidB did not (Fig. 2A). Secondly, we confirmed that Vps24 interacts with a ZZ-PalB MIT domain polypeptide using proteins purified from recombinant bacteria. Purified His6-Vps24 was retained on IgG-Sepharose beads only after preincubation with purified ZZ-PalBMIT peptide (supplemental Fig. S3). We conclude that PalB interacts specifically and directly with Vps24 in a MIT domain-dependent manner.
FIGURE 2.
PalB binds directly to Vps24. Key residues in the Vps24 MIM domain are essential for PalB binding. A, autoradiographs of GST pull-down assays with the indicated A. nidulans GST fusion proteins and 35S-labeled PalA and PalB preys synthesized in vitro using a coupled transcription/translation system. Input lanes contained 2% of the material used in the pull-downs. Note that GST-DidB does not pull-down PalB under these conditions. B, amino acid sequence comparison of the MIM motifs of fungal Vps24 and Vps2 proteins and their respective CHMP3 and CHMP2A human orthologues is shown on the left. Shaded residues correspond to the consensus MIM motif recognizing the Vps4 MIT domain (38, 42). AnVps24 Arg-222 and Leu-223 correspond to positions 0 and 1 in the consensus motif. C, GST pull-down assays using wild-type or mutant (R222D and L223D substituted) A. nidulans GST::Vps24 fusion proteins as baits and 35S-labeled wild-type PalB as prey.
The Vps24 MIM Motif Is Necessary and Sufficient for Interaction with PalB—A. nidulans Vps24 is a 228-residue protein containing an N-terminal (residues 12–197) Snf7 domain and a C-terminal potential MIM motif (residues 214–228), conserved in Vps2 and DidB (Figs. 1A and 2B). Two-hybrid assays demonstrated that, in agreement with crystallographic yeast and human protein data, these C-terminal residues mediate the interaction with PalB. A bait construct containing the N-terminal 197 resides of Vps24 did not interact with a PalB prey (data not shown). However, that containing residues 196–228 interacted similarly to full-length Vps24 (Fig. 1C, lanes 11 and 12).
Structural studies demonstrated the key involvement that one Arg and one Leu residue, located at positions 0 and +1, respectively, of yeast and human MIM motifs play in binding to MIT domains (38, 42). These residues correspond to Arg-222 and Leu-223 in A. nidulans Vps24 (Fig. 2B). In agreement with the MIM motif mediating the interaction with PalB, pull-down experiments demonstrated that a double R222D/L223D substitution prevents PalB binding (Fig. 2C).
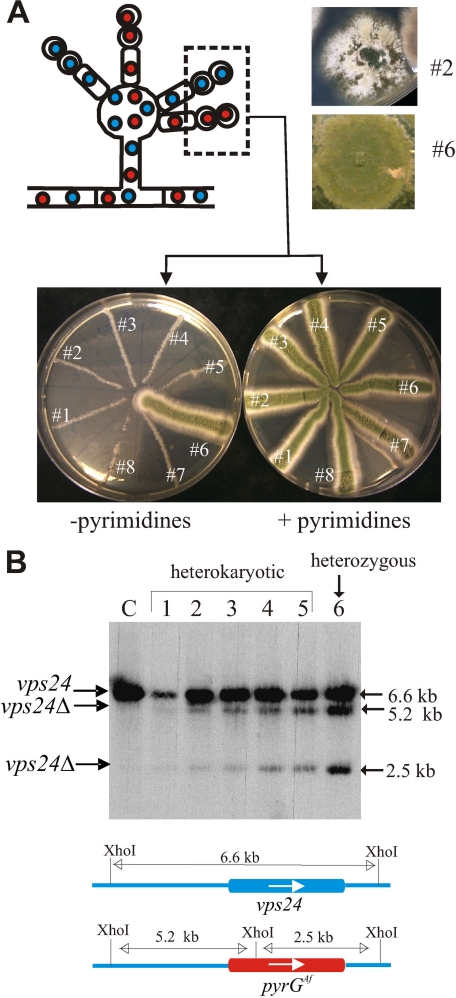
vps24 Is Virtually Essential in A. nidulans—To address the role of Vps24 in pH signaling, we constructed a null allele by reverse genetics, by exploiting that, in multinucleated cells of A. nidulans, lethal or severely debilitating recessive mutations can be maintained in heterokaryosis (53) (Fig. 3). We constructed a vps24 DNA fragment containing the 5′- and 3′-UTR flanking regions, in which the complete coding region had been replaced by the A. fumigatus pyrG gene (54). The construct was transformed into a non-homologous recombination-deficient, pyrimidine-requiring (nkuAΔ pyrG89) double mutant strain. We obtained two types of primary transformants. Type I transformants showed wild-type growth on transfer to pyrimidine-deficient medium and subsequent genotyping showed these to be heterozygous aneuploids or diploids carrying one copy of the vps24+ wild-type allele and one copy of the null vps24Δ allele (Fig. 3, A and B, transformant no. 6). Type II transformants showed typically heterokaryotic growth, indicating that their coenocytic hyphal cells were carrying vps24+, pyrimidine-requiring pyrG89 nuclei and vps24Δ::pyrGAf, pyrimidine prototrophic nuclei. Southern blot analyses demonstrated that these transformants were indeed heterokaryotic and that they contained a relatively higher proportion of wild-type nuclei than of vps24Δ::pyrGAf nuclei (Fig. 3, A and B), as reported previously for other lethal or severely debilitating alleles (53, 55, 56). As individual nuclei segregate into conidiospores (see Fig. 3A, scheme), conidiospores of these heterokaryons were streaked on pyrimidine-sufficient and deficient media. Conidial inoculations led to barely detectable growth in the absence of pyrimidines, which contrasted with the normal growth in the presence of pyrimidines (the latter condition allowing growth of pyrG- nuclei) (Fig. 3A). The fact that growth of pyrimidine-independent vps24Δ::pyrGAf homokaryotic clones is nearly nil led us to conclude that vps24 is a nearly essential gene. A similar situation has been reported for Aspergillus oryzae vps2 and vps24 (57, 58).
FIGURE 3.
A. nidulans vps24 is a virtually essential gene. A, scheme of a conidiophore of an A. nidulans heterokaryotic strain carrying untransformed vps24+, pyrimidine-requiring haploid nuclei (blue) and transformed vps24Δ, pyrimidine-independent nuclei (red) (see also the color scheme in B). During conidiophore development, individual nuclei segregate into conidia, thereby resolving the heterokaryotic situation. Thus, heterokaryotic pyrG- strains carrying nuclei where an essential gene has been replaced by a pyrG+ allele can be propagated from conidiospores in the presence but not in the absence of pyrimidines, as the auxotrophic conidia corresponding to the untransformed nuclei required uracil supplementation for growth. Transformants 1 through 5, 7, and 8 are heterokaryotic, whereas transformant 6 is a spontaneous heterozygous diploid or aneuploid for vps24. B, Southern blot analysis of the transformants with indication of diagnostic bands. Note that, in heterokaryotic transformants 1–5, the intensities of the vps24Δ bands relative to the vps24+ band are lower than in the diploid/aneuploid transformant 6, indicating that the heterokaryons contain fewer vps24Δ than vps24+ nuclei.
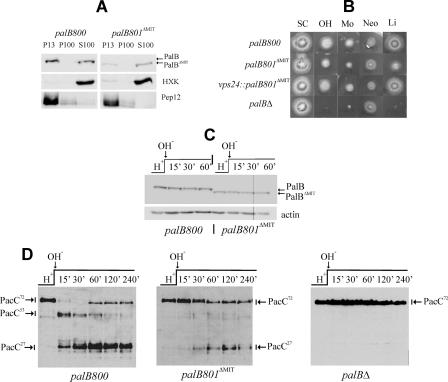
A Proportion of PalB Associates with Membranes—The above data strongly supported the conclusion that PalB is associated with ESCRT-III. To determine whether PalB:(HA)3, expressed at physiological levels, is associated with membranes, we performed subcellular fractionation studies. Protoplast lysates were separated into 13,000 × g pellet (P13) and supernatant fractions, and the supernatant further separated into 100,000 × g soluble (S100) and insoluble (P100) fractions, as described for yeast (35, 48, 49). In agreement with data in S. cerevisiae, FLAG-tagged endosomal syntaxin Pep12 largely localized to P13 membranes, whereas the soluble enzyme hexokinase localized to the S100 fraction. PalB::(HA)3 was distributed between membrane-associated and soluble fractions in approximately equal proportions (Fig. 4A, palB800, left panel). This would be consistent with the view that PalB is associated with endosomal membranes via ESCRT-III.
FIGURE 4.
Deletion of the PalB MIT domain impairs its association with membranes and the processing of PacC. A, MIT domain-mediated association of PalB with membrane fractions. Wild-type palB800 and mutant palB801ΔMIT protoplast lysates were fractionated into 13,000 × g insoluble (P13) and 100,000 × g insoluble (P100) and soluble (S100) fractions, which were analyzed by Western blotting. B, palB801ΔMIT results in weak loss-of-function in diagnostic plate tests of pH signaling (16, 74). palB800 is phenotypically wild-type (data not shown). In contrast, palB801ΔMIT confers some neomycin resistance (Neo), weakly impairs growth on alkaline pH (OH) and molybdate (Mo) plates, and decreases tolerance of LiCl (Li), indicating that deletion of the MIT domain results in weak loss-of-function; SC is synthetic complete medium without any addition. C, similar stability of PalB and PalBΔMIT in pH shift experiments. PalB was detected using α-HA antibody. Similar loading in the different lanes was determined using β-actin. palB800 and palB801ΔMIT cells cultured at acidic ambient pH were shifted to alkaline pH. Cell extracts were analyzed by Western blotting. D, palB801ΔMIT impairs PacC processing in pH shift experiments. Wild-type palB800 and null palBΔ controls are shown. The three forms of PacC are indicated.
Unlike the Pep12p syntaxin or the ESCRT-III-associated protein Bro1p, the S. cerevisiae ESCRT-III components Vps32p or Vps24p form high order oligomers that cannot be extracted from membranes by detergent (1% Triton X-100) treatment (35, 59). The P13-associated fraction of PalB can be solubilized after resuspending the P13 pellet in 1% Triton X-100-containing buffer (supplemental Fig. S4), which suggests that PalB is peripherally associated to membranes. This association appears to be independent of the pal pathway, as the distribution between P13 and S100 fractions was unaffected by the palH17 loss-of-function mutation in the gene encoding the 7-TMD receptor (data not shown).
Deletion of the PalB MIT Domain Impairs but Does Not Prevent the Signaling Proteolysis Step—In view that vps24 is essential, we addressed the physiological role of the MIT domain-dependent PalB-Vps24 interaction by constructing a palB801ΔMIT mutant allele by gene replacement (supplemental Fig. S1). In contrast to palB800, which encodes wild-type PalB::(HA)3, palB801ΔMIT encodes a mutant PalBΔMIT::(HA)3 protein that lacks the N-terminal 76 residues of PalB and thus is deleted for the MIT domain.
In diagnostic plate tests of pH signaling, palB800 is phenotypically indistinguishable from the wild-type (data not shown). In contrast, palB801ΔMIT shows a weak loss-of-function phenotype compared with a null palBΔ mutation (Fig. 4B). As Western blot analysis (Fig. 4C) demonstrated that deletion of the MIT domain has no effect on the levels of PalB, we concluded that deletion of the MIT domain impairs but does not prevent PalB function.
We analyzed the two-step processing of PacC72 in pH shift experiments. In wild-type palB800 cells cultured under acidic conditions, PacC72 is the only detectable form of PacC. Upon shifting cells to alkaline pH, PacC72 is converted to PacC53 within 30 min, and PacC27 becomes the predominant form 30–60 min after the pH shift (Fig. 4D) (10, 16, 18, 22, 28). (After ∼1 h at alkaline pH, newly synthesized PacC72 becomes detectable again, possibly as a result of a negative feedback loop that regulates its processing). palB801ΔMIT reduced, but did not prevent, PacC72 processing to PacC27 (Fig. 4D). The most conspicuous effect of the MIT deletion was the impairment of the signaling proteolysis step, such that PacC72 was the predominating form at all sampled times after the pH shift. PacC53 remained almost undetectable throughout the experiment, whereas PacC27 was only very slowly formed (Fig. 4D). These data indicate that palB801ΔMIT reduces the rate at which PacC72 is converted into PacC53, such that the PacC53 processing activity is in excess and the intermediate does not accumulate at any time point (22). As a palBΔ allele (Fig. 4D) as well as pacC signaling protease box-deficient mutants prevent PacC72 processing (10, 16), these data indicate that deletion of the MIT domain results in partial loss of PalB function, in agreement with diagnostic plate tests. This partial loss-of-function phenotype correlates with the shifting of a large proportion of the membrane-associated PalB pool toward the soluble fraction (Fig. 4A), indicating that PalB function involves its localization to membranes. Most importantly, these data demonstrate that PalB specifically associates with Vps24-containing membranes and that the MIT domain mediates this association. However, a proportion of the mutant protein remained in the P13 fraction, strongly suggesting that MIT domain-dependent association to Vps24 is not the sole mechanism for PalB recruitment to membranes.
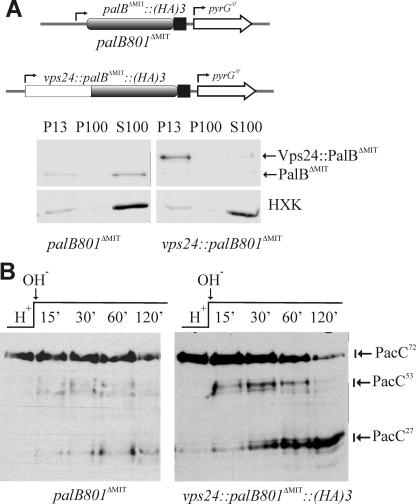
Vps24-mediated Association of PalB with Endosomes Is Important for pH Signaling—To demonstrate that Vps24-mediated recruitment of PalB to endosomal membranes plays a role in pH processing and to circumvent the problems associated with the fact that vps24 is virtually essential, we fused the complete vps24 coding sequence inframe to the sequence encoding residues 77–847 of PalB. The resulting gene, which encodes a Vps24::PalBΔMIT fusion protein, was introduced into the A. nidulans genome using the palB gene replacement procedure. Thus, Vps24::PalBΔMIT, expressed at physiological levels from the palB promoter (scheme in Fig. 5A), was the only source of PalB in strains with this transgene, which contain a wild-type vps24 gene to ensure viability. (C-terminal fusion of PalB to Vps24 almost certainly precludes Vps24 function (60)).
FIGURE 5.
Attachment of Vps24 to the N terminus of PalBΔMIT significantly restores function. A, transgene expressing a Vps24:: PalBΔMIT fusion protein was constructed by gene replacement as described in supplemental Fig. S1. While PalBΔMIT predominates in the soluble fraction, N-terminal attachment of Vps24 redirects the protein to P13 membranes. B, attachment of Vps24 to the N terminus of PalBΔMIT results in a marked increase in the two-step processing of PacC, such that PacC53 becomes clearly detectable in a pH shift experiment.
Western blots demonstrated that, at acidic, neutral and alkaline pH, levels of Vps24::PalBΔMIT were markedly similar to those of PalB and PalBΔMIT, encoded by the palB800 and palB801 alleles described above (data not shown). Thus N-terminal attachment of Vps24 to PalBΔMIT does not lead to protein destabilization/degradation. In fact, diagnostic plate tests demonstrated that Vps24 attachment reversed the partial loss-of-function phenotype associated with PalBΔMIT (Fig. 4B). Subcellular fractionation studies demonstrated that, while a major proportion of PalBΔMIT is cytosolic, Vps24::PalBΔMIT is mostly associated with P13 membranes (Fig. 5A). Finally, sensitive PacC72-processing experiments after an alkaline pH shift demonstrated that Vps24 attachment alleviates the PalBΔMIT-associated impairment in PacC72 processing, markedly increasing levels of PacC27 at late time points after the shift and, most notably, leading to detection of PacC53 (Fig. 5B). However, perhaps due to spatial constrains on PacC signaling protease cleavage resulting from the direct attachment of the PalB catalytic domain to Vps24, the fusion protein did not show fully wild-type PacC72 processing (compare Fig. 5B with Fig. 4D). Thus, the impairment of PalB function resulting from absence of the MIT domain can be alleviated by direct fusion with Vps24. This correlates with the nearly complete association of Vps24:: PalBΔMIT with membranes.
DISCUSSION
We demonstrate that PalB is recruited to ESCRT-III-containing endosomal membranes via specific interaction with Vps24 and that this recruitment is required for normal pH signaling. PalB is the founding member of the family of calpain-like proteases containing a PBH (PalB Homology) domain. The mammalian orthologue of PalB is calpain 7 (61). Although it was originally reported that calpain 7 is nuclearly localized (61), more extensive studies demonstrated that calpain 7 localizes in fact to the cytosol and to endosomes and that this protein, which contains an N-terminal tandem repeat of MIT domains, interacts in vitro with CHMP1B (62). (CHMP1B is the mammalian orthologue of fungal Did2). However, these studies were unable to demonstrate any physiological involvement for the recruitment of calpain 7 to endosomes and therefore this report is first in demonstrating that MIT domain-mediated recruitment of a calpain-like protease to ESCRT-III plays a physiological role.
Unlike calpain 7, PalB has a single MIT domain. We show that PalB binds selectively to Vps24, which suggests that the binding specificity determined by the compound MIT domain differs from that in the single PalB MIT domain. This selectivity for Vps24 is rather suggestive, as work in S. cerevisiae indicated that endosomal, ESCRT-III-containing MVB pathway complexes and pH signaling complexes may be functionally different entities (26) and a recent report strongly implicates Vps24 as a negative regulator of Vps32 oligomerization on endosomal membranes (63). Unfortunately, our attempts to determine the subcellular localization of N- or C-terminally tagged GFP-PalB using different promoters have failed, apparently due to instability of the fusion proteins, and immunolocalization studies of PalB::(HA)3 were inconclusive.
Our work unambiguously places PalB as a member of the fungal endosomal ambient pH signaling complex, associated with the Bro1-domain scaffold PalA (25) and, perhaps transiently, with the Bro1 domain-like containing protein PalC (28). As PalA-dependent recruitment of the zinc-finger transcription factor PacC72 to this complex is essential for pH signaling and PalB is almost certainly the signaling protease, one implication is that the signaling proteolysis step takes place in association with ESCRT-III. Finally, by demonstrating that PalB, like the signaling proteolysis substrate PacC (20, 25), is connected to ESCRT-III, this work constitutes yet further evidence that PalB is the signaling protease.
Analyses of null class E (ESCRT) mutant pH signaling phenotypes in yeasts (64–69) and our unpublished data in A. nidulans4 unambiguously demonstrate the essential involvement of ESCRT-III in pH signaling. Here we show that tailored recruitment to endosomal membranes of a largely cytosolic mutant PalB protein impaired in PacC proteolytic processing significantly restores function, strongly supporting the current view that ambient pH-dependent proteolytic cleavage of PacC and Rim101p takes place in the context of endosomal membranes (8, 9, 20, 25).
The interaction between PalB and ESCRT-III Vps24 reported here is specific, as pull-down assays failed to detect PalB interaction with DidBDid2 or with Vps32 under conditions in which Vps24 very efficiently pulled down PalB. The PalB MIT domain and the Vps24 MIM-containing C-terminal region are necessary and sufficient for the Vps24-PalB interaction. In pull-down assays carried out with purified proteins, the interaction is strictly dependent on the crucial, adjacent Arg and Leu residues of the Vps24 MIM (38, 42), which represents direct evidence that the atypical MIM motif in Vps24 (and, by extension, in CHMP3) is able to bind MIT domains.
Our data strongly indicate that this MIT-mediated interaction cannot be the sole mechanism by which PalB is recruited to endosomal membranes in vivo. This is perhaps not unexpected, as MIT-MIM interactions show relatively weak affinities (Kd in the μm range (34, 38, 42)). The precedent of the de-ubiquitinase AMSH is most relevant. Like PalB, AMSH interacts with CHMP3 (Vps24) and contains a single MIT domain. However, the interaction of AMSH with ESCRT membranes is further reinforced through simultaneous ESCRT-0 STAM binding (45). Moreover, recent work strongly indicated that regions of CHMP3 other than its acidic C-terminal region contribute to the interaction with AMSH, markedly increasing the strength of the interaction (34). We have not yet determined which, if any, additional ESCRT components contribute to PalB recruitment. We cannot formally rule out either a role for undetected Vps24-dependent, PalB MIT-independent interactions in its in vivo recruitment to ESCRT-III, as deletion of A. nidulans vps24 nearly abolishes growth (Fig. 3A), precluding its use.
The possibility that a second interactor reinforces PalB recruitment to ESCRT-III is strongly suggested by data from S. cerevisiae, where the Rim13p calpain-like protease presumably interacts with one or more ESCRT protein(s) other than Vps24p because Rim13p does not contain a MIT domain and vps24Δ does not prevent Rim101p processing (64). Moreover, while S. cerevisiae ESCRT-III Vps32p and Vps20p are essential for pH signaling, the absence of Vps2p or Vps24p results in partially constitutive signaling (67), possibly by increasing the availability of other ESCRT proteins to pH signaling pathway interactors. Vps32 and/or Vps20 are the most obvious candidates to mediate Rim13p recruitment to ESCRT-III, and indeed Rim13p interacts with Vps32 in two-hybrid assays (30), although it remains to be determined whether this interaction is direct and whether it plays a physiological role similar to that demonstrated here for Vps24 and PalB. Thus, PalB and Rim13p would share one anchor in ESCRT-III whereas PalB would have acquired (or Rim13p would have lost) the MIT domain-mediated attachment to Vps24.
Non-compartmentalized intracellular proteases would be harmful for the cell if their activities were not regulated precisely. Calpain-like proteases lack the calmodulin-like Ca2+ binding domain characterizing calpains but contain one calpain III domain, which is structurally related to C2 domains (70). Indeed, the calpain III domain has been shown to bind Ca2+ and, notably, phosphoinositide-containing liposomes (71). This suggests a highly speculative mechanism by which Vps24-mediated recruitment of PalB to endosomal membranes might lead to protease activation in the proximity of endosomal, PalA-recruited PacC72 substrate. Vps24 selectively binds PtIns3,5P2 (72). Vps24-dependent recruitment of PalB might bring the calpain III domain in close proximity to endosomal membrane domains enriched in PtIns3,5P2, facilitating phospholipid binding, which would lead to an activating conformational change in PalB. We note that a similar calpain III domain-mediated, phosphoinositide-dependent mechanism activates m-calpain after its epidermal growth factor-dependent recruitment to the plasma membrane (73).
Supplementary Material
Acknowledgments
We thank A. M. Calcagno for critical reading of the manuscript and E. Reoyo for technical assistance.
This work was supported by DGICYT and Comunidad de Madrid Grants BIO2005-0556 and S2006/SAL-024, respectively, (to M. A. P.) and by Wellcome Trust Grants 067878 and 084660 and BBSRC grant BB/F01189X/1 (to H. N. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: MVB, multivesicular body; 2H, two-hybrid; ESCRT, endosomal sorting complex required for transport; GST, glutathione S-transferase; MIM, MIT domain-interacting motif; MIT, microtubule interacting and trafficking; TMD, transmembrane domain; GFP, green fluorescent protein; HA, hemagglutinin; UTR, untranslated region.
A. M. Calcagno-Pizarelli, A. Hervas-Aguilar, M. A. Penalva, and H. N. Arst, Jr., unpublished data.
References
- 1.Katzmann, D. J., Odorizzi, G., and Emr, S. D. (2002) Nat. Rev. Mol. Cell. Biol. 3 893-905 [DOI] [PubMed] [Google Scholar]
- 2.Hurley, J. H., and Emr, S. D. (2006) Annu. Rev. Biophys. Biomol. Struct. 35 277-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams, R. L., and Urbe, S. (2007) Nat. Rev. Mol. Cell. Biol. 8 355-368 [DOI] [PubMed] [Google Scholar]
- 4.Morita, E., Sandrin, V., Chung, H. Y., Morham, S. G., Gygi, S. P., Rodesch, C. K., and Sundquist, W. I. (2007) EMBO J. 26 4215-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton, J. G., and Martin-Serrano, J. (2007) Science 316 1908-1912 [DOI] [PubMed] [Google Scholar]
- 6.Slagsvold, T., Pattni, K., Malerod, L., and Stenmark, H. (2006) Trends Cell Biol. 16 317-326 [DOI] [PubMed] [Google Scholar]
- 7.Peñalva, M. A., and Arst, H. N., Jr. (2004) Annu. Rev. Microbiol. 58 425-451 [DOI] [PubMed] [Google Scholar]
- 8.Peñalva, M. A., Tilburn, J., Bignell, E., and Arst, H. N., Jr. (2008) Trends Microbiol. 16 291-300 [DOI] [PubMed] [Google Scholar]
- 9.Mitchell, A. P. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 7111-7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díez, E.,Álvaro, J., Espeso, E. A., Rainbow, L., Suárez, T., Tilburn, J., Arst, H. N., Jr., and Peñalva, M. A. (2002) EMBO J. 21 1350-1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mingot, J. M., Tilburn, J., Díez, E., Bignell, E., Orejas, M., Widdick, D. A., Sarkar, S., Brown, C. V., Caddick, M. X., Espeso, E. A., Arst, H. N., Jr., and Peñalva, M. A. (1999) Mol. Cell. Biol. 19 1390-1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orejas, M., Espeso, E. A., Tilburn, J., Sarkar, S., Arst, H. N., Jr., and Peñalva, M. A. (1995) Genes Dev. 9 1622-1632 [DOI] [PubMed] [Google Scholar]
- 13.Espeso, E. A., and Peñalva, M. A. (1996) J. Biol. Chem. 271 28825-28830 [DOI] [PubMed] [Google Scholar]
- 14.Espeso, E. A., and Arst, H. N., Jr. (2000) Mol. Cell. Biol. 20 3355-3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison, S. H., Orejas, M., and Arst, H. N., Jr. (1995) J. Biol. Chem. 270 28519-28522 [DOI] [PubMed] [Google Scholar]
- 16.Peñas, M. M., Hervás-Aguilar, A., Múnera-Huertas, T., Reoyo, E., Peñalva, M. A., Arst, H. N., Jr., and Tilburn, J. (2007) Eukaryot. Cell 6 960-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arst, H. N., Jr., and Peñalva, M. A. (2003) Trends Genet. 19 224-231 [DOI] [PubMed] [Google Scholar]
- 18.Hervás-Aguilar, A., Rodríguez, J. M., Tilburn, J., Arst, H. N., Jr., and Peñalva, M. A. (2007) J. Biol. Chem. 282 34735-34747 [DOI] [PubMed] [Google Scholar]
- 19.Li, W. S., and Mitchell, A. P. (1997) Genetics 145 63-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, W., and Mitchell, A. P. (2001) J. Bacteriol. 183 6917-6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Futai, E., Maeda, T., Sorimachi, H., Kitamoto, K., Ishiura, S., and Suzuki, K. (1999) Mol. Gen. Genet. 260 559-568 [DOI] [PubMed] [Google Scholar]
- 22.Calcagno-Pizarelli, A. M., Negrete-Urtasun, S., Denison, S. H., Rudnicka, J. D., Bussink, H. J., Munera-Huertas, T., Stanton, L., Hervás-Aguilar, A., Espeso, E. A., Tilburn, J., Arst, H. N., Jr., and Peñalva, M. A. (2007) Eukaryot. Cell 6 2365-2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denison, S. H., Negrete-Urtasun, S., Mingot, J. M., Tilburn, J., Mayer, W. A., Goel, A., Espeso, E. A., Peñalva, M. A., and Arst, H. N., Jr. (1998) Mol. Microbiol. 30 259-264 [DOI] [PubMed] [Google Scholar]
- 24.Negrete-Urtasun, S., Reiter, W., Díez, E., Denison, S. H., Tilburn, J., Espeso, E. A., Peñalva, M. A., and Arst, H. N., Jr. (1999) Mol. Microbiol. 33 994-1003 [DOI] [PubMed] [Google Scholar]
- 25.Vincent, O., Rainbow, L., Tilburn, J., Arst, H. N., Jr., and Peñalva, M. A. (2003) Mol. Cell. Biol. 23 1647-1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boysen, J. H., and Mitchell, A. P. (2006) Mol. Biol. Cell 17 1344-1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herranz, S., Rodríguez, J. M., Bussink, H. J., Sánchez-Ferrero, J. C., Arst, H. N., Jr., Peñalva, M. A., and Vincent, O. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12141-12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galindo, A., Hervás-Aguilar, A., Rodríguez-Galán, O., Vincent, O., Arst, H. N., Jr., Tilburn, J., and Peñalva, M. A. (2007) Traffic 8 1346-1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchin-Roland, S., Da Costa, G., and Gaillardin, C. (2008) Microbiology 154 1668-1676 [DOI] [PubMed] [Google Scholar]
- 30.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 4569-4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lata, S., Schoehn, G., Jain, A., Pires, R., Piehler, J., Gottlinger, H. G., and Weissenhorn, W. (2008) Science 321 1354-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson, P. I., Roth, R., Lin, Y., and Heuser, J. E. (2008) J. Cell Biol. 180 389-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muziol, T., Pineda-Molina, E., Ravelli, R. B., Zamborlini, A., Usami, Y., Gottlinger, H., and Weissenhorn, W. (2006) Dev. Cell. 10 821-830 [DOI] [PubMed] [Google Scholar]
- 34.Lata, S., Roessle, M., Solomons, J., Jamin, M., Gottlinger, H. G., Svergun, D. I., and Weissenhorn, W. (2008) J. Mol. Biol. 378 818-829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babst, M., Wendland, B., Estepa, E. J., and Emr, S. D. (1998) EMBO J. 17 2982-2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard, T. L., Stauffer, D. R., Degnin, C. R., and Hollenberg, S. M. (2001) J. Cell Sci. 114 2395-2404 [DOI] [PubMed] [Google Scholar]
- 37.Scott, A., Gaspar, J., Stuchell-Brereton, M. D., Alam, S. L., Skalicky, J. J., and Sundquist, W. I. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13813-13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuchell-Brereton, M. D., Skalicky, J. J., Kieffer, C., Karren, M. A., Ghaffarian, S., and Sundquist, W. I. (2007) Nature 449 740-744 [DOI] [PubMed] [Google Scholar]
- 39.Martín-Serrano, J., Yarovoy, A., Pérez-Caballero, D., and Bieniasz, P. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12414-12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang, H. T., Connell, J. W., Brown, S. E., Thompson, A., Reid, E., and Sanderson, C. M. (2006) Genomics 88 333-346 [DOI] [PubMed] [Google Scholar]
- 41.Nickerson, D. P., West, M., and Odorizzi, G. (2006) J. Cell Biol. 175 715-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obita, T., Saksena, S., Ghazi-Tabatabai, S., Gill, D. J., Perisic, O., Emr, S. D., and Williams, R. L. (2007) Nature 449 735-739 [DOI] [PubMed] [Google Scholar]
- 43.McCullough, J., Clague, M. J., and Urbe, S. (2004) J. Cell Biol. 166 487-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agromayor, M., and Martín-Serrano, J. (2006) J. Biol. Chem. 281 23083-23091 [DOI] [PubMed] [Google Scholar]
- 45.McCullough, J., Row, P. E., Lorenzo, O., Doherty, M., Beynon, R., Clague, M. J., and Urbè, S. (2006) Curr. Biol. 16 160-165 [DOI] [PubMed] [Google Scholar]
- 46.Kyuuma, M., Kikuchi, K., Kojima, K., Sugawara, Y., Sato, M., Mano, N., Goto, J., Takeshita, T., Yamamoto, A., Sugamura, K., and Tanaka, N. (2007) Cell Struct. Funct. 31 159-172 [DOI] [PubMed] [Google Scholar]
- 47.Ma, Y. M., Boucrot, E., Villen, J., Affar, e. B., Gygi, S. P., Gottlinger, H. G., and Kirchhausen, T. (2007) J. Biol. Chem. 282 9805-9812 [DOI] [PubMed] [Google Scholar]
- 48.Rieder, S. E., and Emr, S. D. (1997) Mol. Biol. Cell 8 2307-2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rieder, S. E., and Emr, S. D. (2001) Curr. Protoc. Cell Biol. 3.7.1-3.7.25 [DOI] [PubMed]
- 50.Cove, D. J. (1966) Biochim. Biophys. Acta 113 51-56 [DOI] [PubMed] [Google Scholar]
- 51.Espeso, E. A., Tilburn, J., Sánchez-Pulido, L., Brown, C. V., Valencia, A., Arst, H. N., Jr., and Peñalva, M. A. (1997) J. Mol. Biol. 274 466-480 [DOI] [PubMed] [Google Scholar]
- 52.Scott, A., Chung, H. Y., Gonciarz-Swiatek, M., Hill, G. C., Whitby, F. G., Gaspar, J., Holton, J. M., Viswanathan, R., Ghaffarian, S., Hill, C. P., and Sundquist, W. I. (2005) EMBO J. 24 3658-3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osmani, A. H., Oakley, B. R., and Osmani, S. A. (2006) Nat. Protoc. 1 2517-2526 [DOI] [PubMed] [Google Scholar]
- 54.Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., Osmani, S. A., and Oakley, B. R. (2006) Nat Protoc. 1 3111-3120 [DOI] [PubMed] [Google Scholar]
- 55.Osmani, S. A., Engle, D. B., Doonan, J. H., and Morris, N. R. (1988) Cell 52 241-251 [DOI] [PubMed] [Google Scholar]
- 56.Araujo-Bazán, L., Peñalva, M. A., and Espeso, E. A. (2008) Mol. Microbiol. 67 891-905 [DOI] [PubMed] [Google Scholar]
- 57.Tatsumi, A., Kikuma, T., Arioka, M., and Kitamoto, K. (2006) Biochem. Biophys. Res. Commun. 347 970-978 [DOI] [PubMed] [Google Scholar]
- 58.Tatsumi, A., Shoji, J. Y., Kikuma, T., Arioka, M., and Kitamoto, K. (2007) Biochem. Biophys. Res. Commun. 362 474-479 [DOI] [PubMed] [Google Scholar]
- 59.Odorizzi, G., Katzmann, D. J., Babst, M., Audhya, A., and Emr, S. D. (2003) J. Cell Sci. 116 1893-1903 [DOI] [PubMed] [Google Scholar]
- 60.Strack, B., Calistri, A., Craig, S., Popova, E., and Gottlinger, H. G. (2003) Cell 114 689-699 [DOI] [PubMed] [Google Scholar]
- 61.Futai, E., Kubo, T., Sorimachi, H., Suzuki, K., and Maeda, T. (2001) Biochim. Biophys. Acta 1517 316-319 [DOI] [PubMed] [Google Scholar]
- 62.Yorikawa, C., Takaya, E., Osako, Y., Tanaka, R., Terasawa, Y., Hamakubo, T., Mochizuki, Y., Iwanari, H., Kodama, T., Maeda, T., Hitomi, K., Shibata, H., and Maki, M. (2008) J. Biochem. 143 731-745 [DOI] [PubMed] [Google Scholar]
- 63.Teis, D., Saksena, S., and Emr, S. D. (2008) Dev. Cell 15 578-589 [DOI] [PubMed] [Google Scholar]
- 64.Xu, W., Smith, F. J., Jr., Subaran, R., and Mitchell, A. P. (2004) Mol. Biol. Cell 15 5528-5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kullas, A. L., Li, M., and Davis, D. A. (2004) Eukaryot. Cell 3 1609-1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothfels, K., Tanny, J. C., Molnar, E., Friesen, H., Commisso, C., and Segall, J. (2005) Mol. Cell. Biol. 25 6772-6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayashi, M., Fukuzawa, T., Sorimachi, H., and Maeda, T. (2005) Mol. Cell. Biol. 25 9478-9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanchin-Roland, S., Da Costa, G., and Gaillardin, C. (2005) Microbiology. 151 3627-3637 [DOI] [PubMed] [Google Scholar]
- 69.Cornet, M., Bidard, F., Schwarz, P., Da Costa, G., Blanchin-Roland, S., Dromer, F., and Gaillardin, C. (2005) Infect. Immun. 73 7977-7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sorimachi, H., and Suzuki, K. (2001) J. Biochem. 129 653-664 [DOI] [PubMed] [Google Scholar]
- 71.Tompa, P., Emori, Y., Sorimachi, H., Suzuki, K., and Friedrich, P. (2001) Biochem. Biophys. Res. Commun. 280 1333-1339 [DOI] [PubMed] [Google Scholar]
- 72.Whitley, P., Reaves, B. J., Hashimoto, M., Riley, A. M., Potter, B. V., and Holman, G. D. (2003) J. Biol. Chem. 278 38786-38795 [DOI] [PubMed] [Google Scholar]
- 73.Shao, H., Chou, J., Baty, C. J., Burke, N. A., Watkins, S. C., Stolz, D. B., and Wells, A. (2006) Mol. Cell. Biol. 26 5481-5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tilburn, J., Sarkar, S., Widdick, D. A., Espeso, E. A., Orejas, M., Mungroo, J., Peñalva, M. A., and Arst, H. N., Jr. (1995) EMBO J. 14 779-790 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.