Abstract
GroEL is an essential Escherichia coli molecular chaperon that uses ATP to facilitate correct folding of a range of proteins in a cell. Central to the GroEL substrate diversity is how GroEL recognizes the substrates. The interaction between GroEL and substrate has been proposed to be largely hydrophobic because GroEL interacts with proteins in non-native conformations but not in native forms. Analysis of GroEL substrate proteins reveals that one of its main substrates are proteins with αβ folding domains, suggesting that GroEL may stabilize the collapsed αβ core by binding to hydrophobic surfaces that are usually buried between the α and β elements. In this study, we characterize the interaction between GroEL and a peptide derived from our previous selection via a phage display method. NMR studies map the peptide-binding site to the region containing Helices H and I, which is consistent with evidence that this region comprises the primary substrate-binding site. The peptide is largely unstructured in solution but adopts a helical conformation when bound to the GroEL apical domain with a moderate affinity (Kd = 17.1 ± 2.5 μm). The helical conformation aligns residues to form an amphipathic structure, and the hydrophobic side of this amphipathic helix interacts with GroEL as suggested by fluorescence quenching studies. Together with previous structural studies on the GroEL-peptide complexes, our work supports the notion that the amphipathic secondary elements in the substrate proteins may be the structural motif recognized by GroEL.
The bacterial chaperonin GroEL and its co-chaperonin GroES are essential for cell viability by assisting folding of a wide range of proteins via an ATP-dependent mechanism (1–3). Structurally, fourteen 57-kDa GroEL subunits assemble into two back-to-back stacking heptameric rings, giving rise to two functionally independent central cavities (4). Each GroEL subunit folds into three distinctive domains: equatorial domain, intermediate domain, and apical domain. The equatorial domains contain the ATP-binding sites and provide most of the intra-ring interactions and all the inter-ring interactions. The apical domains form the rims of the central cavities and contain the binding sites for the substrate proteins and GroES. The intermediate domains link the apical domains and the equatorial domains. For the co-chaperonin GroES, seven GroES subunits, of 10 kDa each, assemble into a heptamer ring (5, 6). In forming the GroEL-GroES complex, GroES caps one end of GroEL, and large structural changes are observed in both GroEL and GroES (7). In GroEL, the apical domain is rotated 90° along its axis and 60° upwards, and the intermediate domain is closed down ∼25° to the equatorial domain. A loop in GroES (residues 17–33) that is unstructured in the isolated GroES adopts a β-turn structure and forms contact with the GroEL apical domain. Compared with the unliganded GroEL, the volume of the enclosed GroEL-GroES cavity is doubled, and the surface lining the wall of the GroEL cavity changes from hydrophobic to hydrophilic.
A wealth of information derived from both intensive biochemical and structural characterizations has revealed a general role of GroEL-GroES in assisting protein folding (see reviews in Refs. 3, 8, and 9). Briefly, GroEL binds the substrate proteins in their aggregation-prone non-native states, preventing them from aggregating. Binding of ATP to the substrate occupied GroEL ring (cis-ring) presumably induces large conformational change in GroEL that promotes binding of GroES to the cis-ring. As a result of ATP and GroES binding, the substrate protein is displaced into the GroEL central cavity, initiating the folding process. Both hydrolysis of ATP in the cis-ring and binding of ATP to the substrate unoccupied ring (trans ring) weaken the GroES-GroEL interaction, and ATP binding to the trans ring results in the dissociation of GroES from GroEL, releasing substrate from the central cavity of GroEL. The released substrate may continue folding into the native state if in a folding competent state or may rebind to GroEL if it is still misfolded.
One of the most intriguing aspects of the GroE-assisted folding is the substrate promiscuity. It has been shown that about 300 Escherichia coli proteins can interact with GroEL, and these proteins are diverse in terms of both structures and functions (10). A range of techniques have been applied to investigate this important yet complex aspect, and salient features regarding GroEL-substrate interactions have emerged. The apical domains, on the rim of the GroEL central cavity, contain the main substrate-binding site (11–13). Structural flexibility, reflected by both high temperature factors of the apical domain in the crystal structure of tetradecameric GroEL (14) and conformational multiplicity around Helix H and I (15), is proposed to account for the diverse spectrum of GroEL substrates. Mutational studies on GroEL suggest that the GroEL-substrate interactions are largely hydrophobic (16). Structural study on GroEL-substrate interaction, however, is hindered mainly because of the multiple conformations of the bound substrate protein. Very recently, NMR techniques have been used to directly investigate the bound conformations of the substrate (17, 18); yet the nature of GroEL-substrate interaction is not revealed. Peptides may mimic segments of substrate proteins, and studies of GroEL-peptide interactions have uncovered detailed intermolecular interactions and provided insights into principles of substrate recognition by GroEL. The bound peptides may adopt α-helix (19–23), β-hairpin (15), or extended conformations (24), and despite different conformations, they all appear to bind to Helix H and I of GroEL. Hydrophobic interaction dominates the interface between GroEL and peptides in either β-hairpin or extended structures and is proposed so between GroEL and α-helical peptides. These detailed structural characterizations on GroEL-peptide interactions have contributed to dissecting the complex nature of the substrate recognition by GroEL (25).
We previously identified a high affinity peptide (strong binding peptide (SBP))2 for GroEL using a phage display method and found that SBP adopts a β-hairpin structure bound to GroEL (15, 26). To investigate the contribution of the β-turn in SBP to the GroEL-SBP interaction, we have created various SBP variants with the intension to disrupt the β-turn structure and have studied their binding to GroEL. One of the peptides (termed SBP-W2DP6V), however, adopts a helical conformation when bound to GroEL by NMR analysis. NMR results also map the peptide-binding site on GroEL to be a region formed by Helix H and I. The helical peptide has an amphipathic feature, and fluorescence studies provide direct evidence that the hydrophobic face is involved in the interaction with GroEL. Our structural analysis, combined with previous studies, suggests that GroEL recognizes the amphipathic property in the secondary structures of the substrate protein and binds preferably to the hydrophobic side of these structural elements to stabilize and preserve their structures.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—Gene encoding sequence from 191 to 345 of GroEL (the apical domain) was cloned, via NdeI/BamHI sites into pET15b (Novagen) to produce fusion proteins with a His6 tag followed by a thrombin site at the N terminus of the apical domain. Tetradecameric GroEL was in a pTrc expression vector. Both the apical domain and tetradecameric GroEL were expressed in E. coli BL21(DE3) (Novagen). Cell growth and protein purification followed previously published procedures (7, 15). To prepare 15N-labeled apical domain, cells expressing His6-apical domain were grown in M9 medium containing 15NH4Cl (Cambridge Isotope). The His6 tag was removed by incubating the purified protein (>95% pure by SDS-PAGE) with biotinylated thrombin (Novagen) at 4 °C for 24 h, followed by further purification using Streptavidinresin (Pierce) and nickel affinity resin (Amersham Biosciences). The MBP-apical domain fusion construct was produced by cloning the gene encoding the GroEL apical domain into pMAL-c2 (New England Biolabs) via BamHI/XbaI sites, resulting in the MBP gene being expressed at the N terminus of the apical domain. The cells were grown in LB medium at 37 °C to an A600 of 1.0 and induced with 0.4 mm isopropyl β-d-thiogalactopyranoside for 5 h. The cells were lysed at 4 °C in 50 mm sodium phosphate, pH 7.4, 200 mm NaCl, and 1 mm EDTA using a continuous flow microfluidizer (Microfluidics). The clear cell lysate was loaded onto an Amylose column (New England Biolabs), fractionated further by Fast Q ion exchange column (Amersham Biosciences) and followed by Superdex 75 gel filtration chromatography (Amersham Biosciences).
Peptide Synthesis and Purification—All of the peptides were synthesized by solid phase synthesis using ABI433A (Applied Biosystems). The fluorescein-labeled peptide was synthesized with the 5,6-fluorescein (Molecular Probe) attached to the N terminus of the sequence via a GGG spacer. The peptides were purified by C18 reversed phase high pressure liquid chromatography (Vydac) with an acetonitrile gradient of 0–80% in 0.1% trifluoroacetic acid and confirmed by mass spectrometry. The purified peptide was lyophilized and stored at -20 °C.
Fluorescent Polarization Measurements—The measurements were carried out at 18 °C using a Beacon 2000 instrument (PanVera). The excitation and emission wavelengths for fluorescein were 345 and 495 nm, respectively. Samples with various concentrations of the apical domain were prepared in 150 mm NaCl, 50 mm Tris-Cl, pH 8.0, whereas the labeled peptide remained 1 nm. The samples were incubated for 30 min prior to measurements. Each data point is an average of 10 readings of the same sample, and the entire experiment was repeated once. fB, the fraction of the apical domain-bound peptide, is related to the observed fluorescence polarization signal FP as follows,
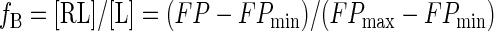 |
(Eq. 1) |
where FPmin and FPmax are the fluorescence polarization signals of the peptide in isolation and in apical domain-bound form, respectively; [L] is the total peptide concentration (1 nm), and [RL] refers to the concentration of the apical domain-peptide complex, which is related to the equilibrium dissociation constant (Kd) as follows.
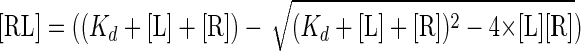 |
(Eq. 2) |
Fluorescence Quenching Experiments—Experiments of fluorescence quenching by acrylamide were carried out at 25 °C using a PerkinElmer Life Sciences 50B luminescence spectrometer with a temperature controller. The samples were excited at 295 nm, and the emission spectra from 310 to 410 nm were recorded. The peptide concentration was 20 μm. A stock solution of freshly prepared 8 m acrylamide in protein buffer was added in 10-μl aliquots to 2 ml of 20 μm peptide-containing solutions (peptide alone, and peptide:proteins in 1:1 and 1:5 molar ratios). The fluorescence intensities were corrected for dilution effect and inner filter effect. Quenching data were fit into the Stern-Volmer equation F0/F = 1 = Ksv[Q], where F0 and F are the fluorescence intensities in the absence and presence of acrylamide, respectively, and [Q] is the acrylamide concentration. The experiments were repeated using proteins (the apical domain and GroEL) from different batches of preparation.
CD Measurements—The measurements were performed on a Jasco J-715 circular dichroism spectropolarimeter (Jasco) using quartz cells with path length of 0.1 cm. The peptide (0.15 mm) was in 10 mm sodium phosphate (pH 6.1), and 10 mm NaCl. The CD signals were taken in steps of 0.2 nm and at 25 °C.
NMR Spectroscopy—All NMR experiments were carried out on a Varian Unity INOVA 500 MHz spectrometer or a Varian NMR System 600 MHz spectrometer with a 5-mm 1H{13C/15N} Triple Resonance Cold Probe (Varian). 2,2-Dimethyl-2-silapentane-5-sulfonic acid was used as chemical shift references. 15N-1H spectra were recorded at 30 °C for 2 h on 15N-labeled apical domain (0.28 mm) in the absence and presence of the peptide (0.56 mm). The data were processed with VNMR software (Varian, CA) and analyzed with Sparky (27). Backbone resonances of the apical domain in heteronuclear single quantum coherence spectrum were assigned according to Kobayashi et al. (23). The average changes in chemical shift were calculated as follows,
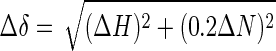 |
(Eq. 3) |
where ΔH and ΔN are the changes of 1H and 15N chemical shift, respectively.
Two-dimensional total correlation spectroscopy and double-quantum filtered correlation spectroscopy experiments were carried out at 25 °C on the peptide only, whereas NOESY spectra were recorded on the peptide alone and in the presence of both MBP-apical domain and tetradecameric GroEL. All of the two-dimensional experiments used a 3-9-19 Watergate sequence (28, 29) for water suppression. All of the homonuclear two-dimensional spectra were acquired with 16 scans and sweep widths of 8000 Hz in both dimensions as matrices of 1024 × 320 complex points. The peptide was dissolved in 50 mm potassium phosphate buffer, pH 6.1, at 2 mm concentration, or into either MBP-apical domain or tetradecameric GroEL containing solution in the same buffer. The two-dimensional NOESY spectra with mixing times of 30, 60, 90, 120, and 150 ms were acquired to analyze the NOE build-up to avoid spin-diffusion artifacts, and to assign the peaks intensity to be strong, medium, or weak. The total correlation spectroscopy spectrum was acquired with a mixing time of 80 ms. For the assignment of trNOE, two-dimensional NOESY data with a 150-ms mixing time were recorded with a spectral width of 8000 Hz and 512 complex points.
Structural Refinement—NOE correlations observed in the NOESY spectra with the 150-ms mixing time were used as distance constraints. Distance constraints derived from sequential NOEs or intraresidue NOEs between NH, α-H and β-H were classified into 2.7, 3.3, and 4.0 Å for strong, medium, and weak NOEs, respectively. Medium range NOEs were assigned as 4.0 Å if they involved exclusively backbone NH and α-H or 5.0 Å if they involved side chain protons. No dihedral angle or hydrogen-bonding constraints were used. A total of 87 (57 sequential and 30 medium range) distance constraints were input for structure determination using the program Discover (30). 50 structures were generated using a distance-geometry/simulated annealing protocol similar to that reported by Nilges et al. (31). Structure quality was analyzed using MOLMOL (32) and PROCHECK-NMR (33). Atomic co-ordinates for the fifteen peptides have been deposited in the Biological Magnetic Resonance Bank (accession number 20063).
RESULTS
SBP-derivatized Peptides—SBP was identified as a peptide with a strong affinity for the apical domain of GroEL from biopanning experiments using a phage display peptide library (15). SBP (SWMTTPWGFLHP) adopts a type I β-hairpin conformation when bound to a groove formed by Helix H and I of GroEL. Interestingly, most of direct contacts of the peptide with GroEL are via the C-terminal half of SBP. A shorter peptide (PWGFLHP), corresponding to the C-terminal half of SBP, was synthesized but was found to have little affinity to the GroEL apical domain (data not shown), suggesting that the internal structure of the peptide (five intrapeptide hydrogen bonds caused by β-hairpin formation) may play an important role in the binding of SBP to the apical domain. To examine the role of β-hairpin structure in binding of SBP to the apical domain, we changed residue at the i + 1 position in SBP from Pro to residues less favorable for a type I turn (Gly, Ala, Val, Met, and Ile) and measured their binding affinities to GroEL with fluorescence polarization assay. The change to Val (termed peptide SBP-P6V) appeared to enhance the peptide affinity to the GroEL apical domain with Kd of 3.8 ± 0.2 μm (supplemental Fig. S1A). (Kd of SBP was reported previously (15) and was re-evaluated to be 11.5 ± 1.7 μm in this study.) We reasoned that peptide SBP-P6V might adopt an extended conformation when bound to the apical domain, thus increasing its interface with the apical domain and resulting in increase in affinity. Our attempts to structurally analyze the peptide-apical domain interaction with SBP-P6V were unsuccessful. We were not able to obtain co-crystals of either the apical domain-peptide or GroEL-peptide complexes, and the limited solubility of SBP-P6V made it unsuitable for NMR experiments. To increase the solubility of the peptide, Trp2 of SBP-P6V was changed to Asp to create SBP-W2DP6V, although the change decreased its affinity to Kd = 17.1 + 2.5 μm; supplemental Fig. S1A). Proton NMR spectra (supplemental Fig. S1B) indicate that binding of the peptide to the apical domain is specific, as the peptide NMR signals in the amide region become significantly broadened in the presence of the apical domain.
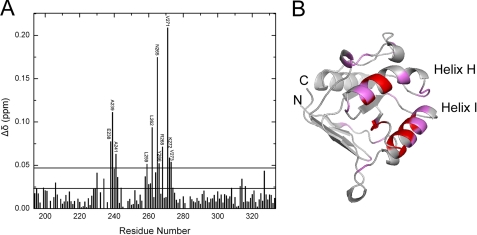
Peptide-binding Site on the Apical Domain—Because the chemical shift is highly sensitive to local or global structural change, 15N-1H correlation NMR spectroscopy (heteronuclear single quantum coherence) is useful in identifying the ligand binding site on the host protein. To map the SBP-W2DP6V binding site on the apical domain, we recorded 15N-1H heteronuclear single quantum coherence spectra of the 15N-labeled apical domain in the absence and presence of the peptide. 127 of 133 published backbone resonances of the apical domain alone (23) were identified, whereas 12 residues (198, 202, 204, 233, 235, 237, 246, 247, 279, 282, 283, and 321) remain unassigned. These unassigned residues distribute throughout the protein sequence. Residues with altered chemical shifts (of either amide protons or 15N nuclei; supplemental Fig. S2) either directly interact with the peptide, or are affected because of the peptide-induced global structural change in the apical domain. The effects of SBP-W2DP6V on the chemical shifts of the apical domain are summarized in Fig. 1A. As shown in Fig. 1B, the affected residues are clustered in helix H and I, suggesting that these two helices comprise the binding site for SBP-W2DP6V. This finding is consistent with previous mutational and structural studies that have identified that helix H and I and the groove between them consist of the primary substrate-binding site (15, 16, 24). The affected residues located outside helix H and I, as also seen in the SBP-apical domain complex (15), may reflect the global structural changes in the apical domain induced by binding of SBP-W2DP6V.
FIGURE 1.
A, summary of changes of the combined chemical shifts (Equation 3) observed for the apical domain in the absence and presence of SBP-W2DP6V. The lower line denotes the mean (0.022), and the upper line represents the mean plus one standard deviation (σ, 0.028) of data. B, mapping of the residues indicated in A on the structure of the apical domain. Residues with Δδ above the mean are colored in magenta, and those with Δδ above the mean plus one standard deviation are in red.
Peptide Conformations in Isolation—The absence of distinct features for secondary structures in the CD spectrum (supplemental Fig. S3) suggest that SBP-W2DP6V in solution is largely unstructured. In NMR, spin-spin couplings (3JHNα) may be correlated with secondary structures. For example, segments with continuous three or more 3JHNα of less than 6.0 Hz are the criteria for formation of helical conformation (34). For SBP-W2DP6V, the 3JHNα coupling constants of nine residues are in the ranges of 6.3 to 7.7 Hz (supplemental Table S1), suggesting that the isolated peptide possesses little helical character. A few crossed peaks are observed in 1H two-dimensional NOESY spectra (Fig. 2A), indicating that the peptide is largely nonstructured in solution. Finally, we used chemical shift index to determine the peptide conformation. In this method, an index (-1, 0, 1) is assigned to each residue if the chemical shift of the α-proton is smaller, within, or larger than the standard values given by Wishart et al. (35). As shown in supplemental Table S2, the lack of at least three consecutive “1” values or four “-1” values, indications for β-strand or helical conformations, respectively, suggests that the peptide in solution is not structured. Taken together, SBP-W2DP6V is mainly unstructured in solution.
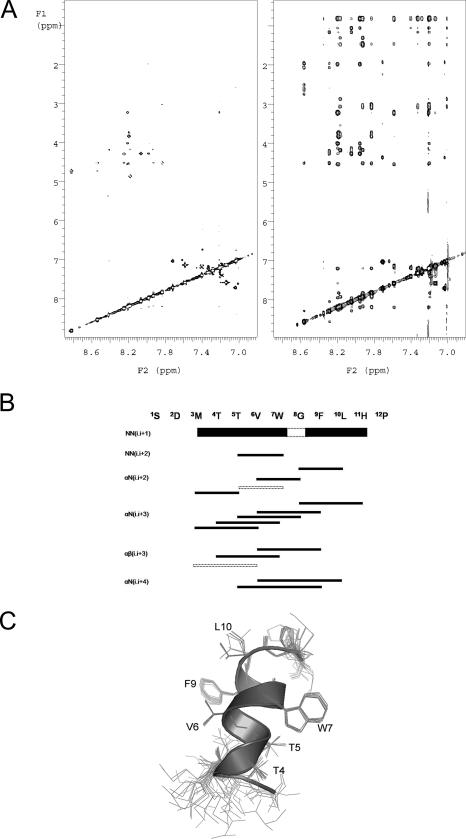
FIGURE 2.
A, two-dimensional NOESY spectra of SBP-W2DP6V in the absence (left panel) and presence (right panel) of MBP-apical domain. B, summary of NOE signals for SBP-W2DP6V in the presence of MBP-apical domain. Line thickness corresponds to the intensity of the NOE cross-peaks. The dashed lines denote NOE signals that overlap with other resonances. C, overlaying 15 NMR structures of the apical domain bound SBP-W2DP6V by fitting the backbone of residues 4–10. Ribbon diagram represents one of the structures.
Peptide Conformations in the Bound States—Transfer NOE experiments are widely used in the study of protein-ligand interactions to characterize the conformation of bound ligand in a fast exchange (weak interaction) system (36, 37). Basically, in the free form, the peptide is characterized by short correlation times and the NOE signals are minimal, whereas in the protein-bound form, the peptide is characterized by long correlation time of the protein and the NOE intensities are large. Two methods were used to maximize the NOE signals. First, the apical domain (18 kDa) was expressed as maltose-binding protein (MBP) fusion protein (with a total molecular weight of 60 kDa), and trNOE spectra were recorded on the sample containing both the peptide and MBP-apical domain. As shown in Fig. 2A, the addition of MBP-apical domain to SBP-W2DP6V led to many additional cross-peaks being observed and the strong intensity of the NOE signals, giving rise to high quality NOESY spectra. NOESY spectra of SBP-W2DP6V in the presence of MBP (data not shown) largely resembled those of the isolated SBP-W2DP6V, suggesting that the peptide does not interact with MBP. This control experiment indicated that the observed trNOE signals result from specific interactions between the peptide and the apical domain.
In a second experiment, we recorded NOESY spectra of a sample of tetradecameric GroEL with SBP-W2DP6V. As expected the NOE signals are sharper and better resolved (supplemental Fig. S4) than those from the MBP-apical domain-peptide sample. No additional cross-peaks were observed in the spectra of GroEL-peptide compared with those of MBP-apical domain-peptide, suggesting that SBP-W2DP6V only binds to the apical domain in the tetradecamic GroEL. The NOE connectivity patterns shown in Fig. 2B indicate that SBP-W2DP6V is helical when bound to the apical domain. Further, the presence of dα,N (i, i + 2) and dα,N (i, i + 4) NOE signals suggests that the helix may sample both 310-helical and α-helical conformations. To compute the NMR structures of the bound peptide, a total of 87 NOE signals were included in the structure calculations, leading to an ensemble of 15 structures that had distance violations smaller than 0.2 Å (the refinement statistics is summarized in supplemental Table S3). As shown in Fig. 2C, a well defined helical conformation is observed for residues Thr4 to Leu10 in all structures and for residues Met3 to His11 in five of the 15 structures. The helix is somewhat deformed: a typical α-helix comprises residues Thr5 to His11 preceded by a partial 310-helix including Thr4 and extending to Met3 in some cases. This mixed helical content is consistent with the observed dα,N NOE patterns above. Side chains of residues in the helical core (Thr5 to Phe9) are converged; in particular, those of the two aromatic residues (Trp7 and Phe9) are well ordered (Fig. 2C).
Binding Surface in Peptide—Acrylamide does not penetrate the protein matrix because it is polar, and it does not bind to the protein in any particular manner. If Trp is accessible by acrylamide, its fluorescence intensity will be reduced or quenched. Thus, the extent of acrylamide accessibility is correlated with how much Trp is exposed to solvent (38). Because GroEL does not contain Trp and Trp7 is the only Trp in the peptide, we reasoned that if Trp7 was secreted within the peptide-GroEL interface, its fluorescence would be less likely quenched by acrylamide than an exposed Trp7 that did not participate in the interaction with GroEL.
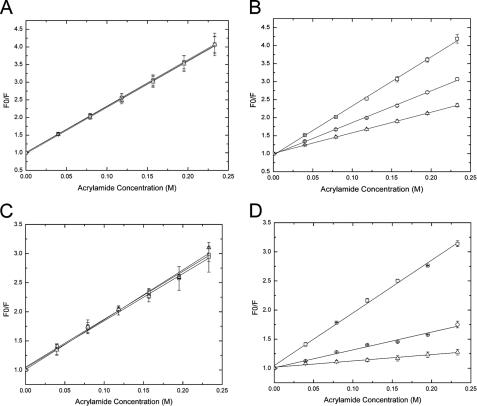
Fig. 3A shows acrylamide titrations of Trp fluorescence from SBP-W2DP6V in the absence and presence of different concentrations of apical domain. The Stern-Volmer quenching constant (KSV) derived from the slope of F0/F versus quencher concentration [Q] is nearly identical for SBP-W2DP6V in the absence and presence of the GroEL apical domain (Table 1). These data suggest that the solvent accessibility of Trp7 is not affected by the presence of GroEL apical domain, i.e. Trp7 is not involved in direct contact with the apical domain. As a control, we also carried out the fluorescence quenching studies using SBP, which contains two Trp residues (Trp2 and Trp7). Crystal structure of SBP-apical domain shows that although Trp2 does not directly interact with the apical domain and remains largely solvent-exposed in the SBP-apical domain complex, Trp7 is mostly buried within the SBP-protein interface (15). As expected, KSV values (Fig. 3B and Table 1) for SBP in the presence of the apical domain are significantly lower, reflecting the decrease of Trp7 solvent accessibility upon complex formation, than that without the apical domain. Therefore, the unaltered KSV of SBP-W2DP6V by the apical domain indicates that Trp7 does not make direct contact with the apical domain in the SBP-W2DP6V-apical domain complex.
FIGURE 3.
Quenching of the peptide tryptophan fluorescence monitored at 360 nm by acrylamide. A, SBP-W2DP6V with and without the apical domain. B, SBP with and without the apical domain. C, SBP-W2DP6V with and without GroEL. D, SBP with and without GroEL. The peptide concentration was 20 μm. Squares, the peptide only; circles, peptide:protein = 1:1; triangles, peptide:protein = 1:5. Multiple independent experiments were carried out. The errors represent discrepancy among different runs and are shown along with the average values. The lines are linear fitting results of data.
TABLE 1.
Acrylamide quenching on Trp fluorescence The quenching constants (KSV) are in M-1.
| SBP-W2DP6V | SBP | |
|---|---|---|
| Peptide only | 13.1 | 13.6 |
| Peptide:apical domain (1:1) | 13.0 | 8.8 |
| Peptide:apical domain (1:5) | 13.1 | 5.7 |
| Peptide only | 8.2 | 9.1 |
| Peptide:GroEL (1:1) | 8.1 | 3.1 |
| Peptide:GroEL (1:5) | 8.5 | 1.1 |
Similar results were observed when tetradecameric GroEL was used in the place of the apical domain (Fig. 3, C and D, and Table 1). KSV of SBP is notably decreased in the presence of GroEL, consistent with the secretion of Trp7 upon complex formation by crystallographic study (26). KSV of SBP-W2DP6V is not affected by GroEL, indicating that Trp7 does not contact with GroEL in the GroEL-peptide complex.
DISCUSSION
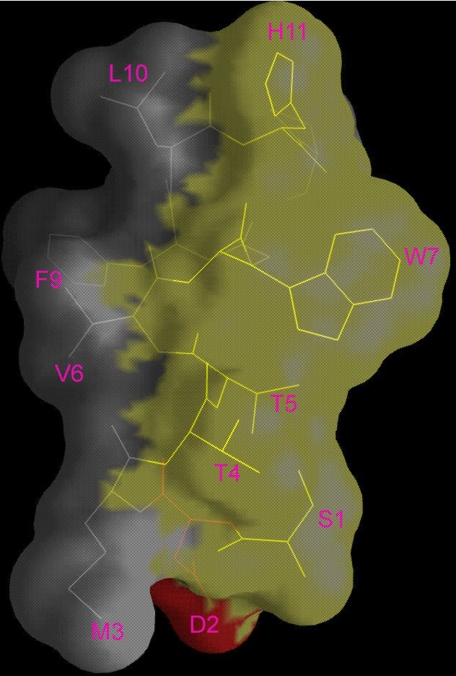
GroEL Interface on the Peptide—As shown in Fig. 4, SBP-W2DP6V is amphipathic. The side chains of the polar residues (Ser1, Thr4, Thr5, Trp7, and His11) are all on one side of the helix, whereas those of the hydrophobic residues (Met3, Val6, Phe9, and Leu10) are on the other side. Because fluorescence studies suggest that Trp7 is not involved in direct contact with the apical domain, we propose that SBP-W2DP6V uses the hydrophobic side of the helix to interact with the GroEL apical domain. The hydrophobic nature of interaction is supported by our observation that binding of SBP-W2DP6V to the GroEL apical domain was enhanced by increasing salt (NaCl) concentration (using fluorescence polarization; data not shown). So far, GroEL has been shown to interact with peptides in α-helical conformations (19–23); however, the interfaces of the peptides with GroEL were not mapped in those studies, and types of interaction between helical peptides and GroEL were not examined directly. In this study, we are able to infer the GroEL interface on the helical peptide and to derive the nature of peptide-GroEL interaction.
FIGURE 4.
Molecular surface of the SBP-W2DP6V helical structure. The polar surface, lined by residues Ser1, Thr4, Thr5, Trp7, and His11, is colored in yellow, and the polar surface formed by residues Met3, Val6, Phe9, and Leu10 is in gray.
In supplemental Fig. S5, SBP-W2DP6V is modeled positioned over the groove formed between Helix H and I, where the C terminus of the peptide situates over the C termini of both GroEL helices. This model is consistent with both the structural dynamics in the N terminus of SBP-W2DP6V (Fig. 2C), and the large chemical shift perturbations concentrated at the C termini of both helices, particularly in that of Helix I (Fig. 1B). The model shows that Val6, Phe9, and Leu10 project their hydrophobic side chains into the groove of the apical domain. In this model, Trp7 is exposed; binding of the peptide to the apical domain does not change the nature of Trp7 environment and its solvent and acrylamide accessibility from its free form in solution, consistent with the observation that acrylamide has identical effect on the fluorescence of the peptide in the complex and in the free form.
Structural Motif Recognition by GroEL—GroEL can interact with a variety of proteins with diverse sequences in their non-native states or molten globular states (39–43). These data suggest that the primary determinant in GroEL substrate recognition does not lie in the substrate protein sequence identity but in the structural features associated with the non-native conformations of proteins. One of the structural characteristics common to the non-native states is the exposure of hydrophobic patches normally buried within the folded proteins. For proteins with an αβ fold, the preferred GroEL substrates (10), misfolding is likely to loosen up the tight packing of the αβ assemblies and expose the hydrophobic interface between the amphipathic α-helices and β-sheets. By binding to these aggregation-prone surfaces, GroEL, in addition to preventing the nonspecific aggregation, serves as a temporary structural scaffold to maintain the conformations in the substrate that are otherwise unstable because of its amphipathicity. The substrate, once released from GroEL, may fold directly with these preserved structural elements to an optimal native arrangement rather than from a polypeptide chain without ordered secondary structures. Because of the rapid time scale (∼μs) of the initial folding event, collapse of a linear polypeptide chain into native-like secondary structures can easily lead to misfolded structures. For example, formation of a hydrophobic/amphipathic β-sheet is challenging, because assembly of β-strands into a β-sheet requires residues that are far away in the sequence to be in close contact, and the β-sheet structure by itself is not stable. By providing a temporary depot for the amphipathic secondary structures and preserving them for the subsequent substrate folding, GroEL reduces the misfolding probability of the substrate and increases the folding yield of the substrate. Thus, it is very likely that the molecular basis of GroEL substrate recognition involves detecting amphipathic structures and binding to the hydrophobic side.
Miller and co-workers (21) found that an amphipathic helical peptide binds 10 times stronger to GroEL than a nonamphipathic version, suggesting that the presence of an amphipathic region in a potential substrate enhances its ability to bind to GroEL. Other peptides that adopt helical conformations when bound to GroEL also appear to be amphipathic, although their interfaces with GroEL were not identified in those studies (19, 20, 22, 23). When bound to GroEL, SBP forms an amphipathic β-hairpin structure, with its hydrophobic strand (7WGFLHP12) in direct contact with the apical domain and the other mainly polar strand (1SWMTT5) exposed to solvent (15, 26). Interestingly, by adopting a β-turn, the GroES mobile loop partitions into an amphipathic structure, where the hydrophobic region (25IVL27) becomes sequestered by direct contact with GroEL and the hydrophilic section (17EVTKSA22) is largely exposed (7). Therefore, these structural analyses along with our work here suggest that amphipathicity of secondary structures plays an important role in substrate recognition by GroEL.
Supplementary Material
Acknowledgments
We thank D. Smiley and R. DiMarchi for peptide synthesis, J. Tomaszewski and Martin Stone for NMR experiments, A. I. Arunkumar for discussions, and J. Richardson for the critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065260-01A1 (to L. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
Footnotes
The abbreviations used are: SBP, strong binding peptide; NOESY, nuclear Overhauser effect spectroscopy; MBP, maltose-binding protein.
References
- 1.Horwich, A. L., Farr, G. W., and Fenton, W. A. (2006) Chem. Rev. 106 1917-1930 [DOI] [PubMed] [Google Scholar]
- 2.Saibil, H. R., and Ranson, N. A. (2002) Trends Biochem. Sci. 27 627-632 [DOI] [PubMed] [Google Scholar]
- 3.Thirumalai, D., and Lorimer, G. H. (2001) Annu. Rev. Biophys. Biomol. Struct. 30 245-269 [DOI] [PubMed] [Google Scholar]
- 4.Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D. C., Joachimiak, A., Horwich, A. L., and Sigler, P. B. (1994) Nature 371 578-586 [DOI] [PubMed] [Google Scholar]
- 5.Hunt, J. F., Weaver, A. J., Landry, S. J., Gierasch, L., and Deisenhofer, J. (1996) Nature 379 37-45 [DOI] [PubMed] [Google Scholar]
- 6.Mande, S. C., Mehra, V., Bloom, B. R., and Hol, W. G. (1996) Science 271 203-207 [DOI] [PubMed] [Google Scholar]
- 7.Xu, Z., Horwich, A. L., and Sigler, P. B. (1997) Nature 388 741-750 [DOI] [PubMed] [Google Scholar]
- 8.Sigler, P. B., Xu, Z., Rye, H. S., Burston, S. G., Fenton, W. A., and Horwich, A. L. (1998) Annu. Rev. Biochem. 67 581-608 [DOI] [PubMed] [Google Scholar]
- 9.Lin, Z., and Rye, H. S. (2006) Crit. Rev. Biochem. Mol. Biol. 41 211-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houry, W. A., Frishman, D., Eckerskorn, C., Lottspeich, F., and Hartl, F. U. (1999) Nature 402 147-154 [DOI] [PubMed] [Google Scholar]
- 11.Chen, S., Roseman, A. M., Hunter, A. S., Wood, S. P., Burston, S. G., Ranson, N. A., Clarke, A. R., and Saibil, H. R. (1994) Nature 371 261-264 [DOI] [PubMed] [Google Scholar]
- 12.Thiyagarajan, P., Henderson, S. J., and Joachimiak, A. (1996) Structure 4 79-88 [DOI] [PubMed] [Google Scholar]
- 13.Elad, N., Farr, G. W., Clare, D. K., Orlova, E. V., Horwich, A. L., and Saibil, H. R. (2007) Mol. Cell 26 415-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braig, K., Adams, P. D., and Brunger, A. T. (1995) Nat. Struct. Biol. 2 1083-1094 [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., and Sigler, P. (1999) Cell 99 757-768 [DOI] [PubMed] [Google Scholar]
- 16.Fenton, W. A., Kashi, Y., Furtak, K., and Horwich, A. L. (1994) Nature 371 614-619 [DOI] [PubMed] [Google Scholar]
- 17.Horst, R., Bertelsen, E. B., Fiaux, J., Wider, G., Horwich, A. L., and Wuthrich, K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12748-12753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horst, R., Fenton, W. A., Englander, S. W., Wuthrich, K., and Horwich, A. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20788-20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry, S. J., and Gierasch, L. M. (1991) Biochemistry 30 7359-7362 [DOI] [PubMed] [Google Scholar]
- 20.Landry, S. J., Jordan, R., McMacken, R., and Gierasch, L. M. (1992) Nature 355 455-457 [DOI] [PubMed] [Google Scholar]
- 21.Preuss, M., Hutchinson, J. P., and Miller, A. D. (1999) Biochemistry 38 10272-10286 [DOI] [PubMed] [Google Scholar]
- 22.Wang, Z., Feng, H., Landry, S. J., Maxwell, J., and Gierasch, L. M. (1999) Biochemistry 38 12537-12546 [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, N., Freund, S. M. V., Chatellier, J., Zahn, R., and Fersht, A. R. (1999) J. Mol. Biol. 292 181-190 [DOI] [PubMed] [Google Scholar]
- 24.Buckle, A. M., Zahn, R., and Fersht, A. R. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3571-3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenton, W. A., and Horwich, A. L. (2003) Q. Rev. Biophys. 36 229-256 [DOI] [PubMed] [Google Scholar]
- 26.Wang, J., and Chen, L. (2003) J. Mol. Biol. 334 489-499 [DOI] [PubMed] [Google Scholar]
- 27.Goddard, T. D., and Kneller, D. G. (2004) SPARKY 3, University of California, San Francisco
- 28.Lippens, G., Dhalluin, C., and Wieruszeski, J. M. (1995) J. Biomol. NMR 5 327-331 [DOI] [PubMed] [Google Scholar]
- 29.Piotto, M., Saudek, V., and Sklenar, V. (1992) J. Biomol. NMR 2 661-665 [DOI] [PubMed] [Google Scholar]
- 30.Accelrys (1996) Discover User Guide, Accelrys, San Diego, CA
- 31.Nilges, M., Clore, G. M., and Gronenborn, A. M. (1988) FEBS Lett. 229 317-324 [DOI] [PubMed] [Google Scholar]
- 32.Koradi, R., Billeter, M., and Wüthrich, K. (1996) J. Mol. Graphics 14 51-55 [DOI] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 34.Pardi, A., Billeter, M., and Wuthrich, K. (1984) J. Mol. Biol. 180 741-751 [DOI] [PubMed] [Google Scholar]
- 35.Wishart, D. S., Sykes, B. D., and Richards, F. M. (1992) Biochemistry 31 1647-1651 [DOI] [PubMed] [Google Scholar]
- 36.Campbell, A. P., and Sykes, B. D. (1993) Annu. Rev. Biophys. Biomol. Struct. 22 99-122 [DOI] [PubMed] [Google Scholar]
- 37.Clore, G. M., and Gronenborn, A. M. (1982) J. Magn. Reson. 48 402-417 [Google Scholar]
- 38.Eftink, M. R., and Ghiron, C. A. (1976) Biochemistry 15 672-680 [DOI] [PubMed] [Google Scholar]
- 39.Martin, J., Langer, T., Boteva, R., Schramel, A., Horwich, A. L., and Hartl, F. U. (1991) Nature 352 36-42 [DOI] [PubMed] [Google Scholar]
- 40.Mendoza, J. A., Butler, M. C., and Horowitz, P. M. (1992) J. Biol. Chem. 267 24648-24654 [PubMed] [Google Scholar]
- 41.Hayer-Hartl, M. K., Ewbank, J. J., Creighton, T. E., and Hartl, F. U. (1994) EMBO J. 13 3192-3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissman, J. S., Kashi, Y., Fenton, W. A., and Horwich, A. L. (1994) Cell 78 693-702 [DOI] [PubMed] [Google Scholar]
- 43.Zahn, R., Spitzfaden, C., Ottiger, M., Wuthrich, K., and Pluckthun, A. (1994) Nature 368 261-265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.