Abstract
Copper is an essential yet toxic trace element. The Ctr1 family of proteins plays a critical role for copper uptake in eukaryotes. However, the mechanisms of action of Ctr1 are largely unknown. Our previous data demonstrated that copper transport induces conformational changes in the cytosolic C terminus of the yeast Saccharomyces cerevisiae Ctr1. To define the physiological significance of this molecular event and gain better insights into the mechanism of Ctr1-mediated copper uptake, we have characterized the functional roles of the Ctr1 C terminus. A Ctr1 mutant lacking the entire C-terminal cytosolic tail is functional in high affinity copper uptake; however, yeast cells expressing this mutant are extremely sensitive to excess copper. Toxic copper uptake is not attributed to elevated expression or distinct subcellular localization of this mutant as compared with wild type Ctr1. Further characterization of the function of Ctr1 containing deletions or site-directed mutations at the C terminus indicates a structural role for the C terminus in controlling Ctr1 activities. In response to excess copper, Ctr1-mediated copper transport is rapidly blocked in a C terminus-dependent mechanism associated with direct binding of copper. We propose that conformational changes in the cytosolic tail of yeast Ctr1 by copper sensing within this domain lead to the inhibition of Ctr1-mediated copper transport. These data suggest a new regulatory mechanism by which yeast cells maintain homeostatic copper acquisition.
Copper functions in electron transfer reactions by virtue of its ability to donate and accept electrons through redox reactions, which enables copper to act as a cofactor for vital enzymes (1–4). Serious defects in growth and development from nutritional or genetic copper deficiencies indicate the vital roles that copper plays in various physiological processes (5–7). However, excess accumulation of copper is highly toxic (8). Individuals diagnosed with Wilson disease experience abnormal copper accumulation in the liver due to a defect in its excretion, which in turn leads to hepatic dysfunction and neurological symptoms (6, 7).
Identification and characterization of the molecular mechanisms involved in transport, distribution, and excretion of copper and also the regulation of these processes have revealed how cells maintain homeostatic copper metabolism (3, 4). Ctr1 (copper transporter 1) serves as a high affinity copper transporter in the yeast Saccharomyces cerevisiae (9). This yeast also expresses two additional Ctr1 family members, Ctr2 and Ctr3. Ctr3 is functionally redundant to Ctr1; however, Ctr3 expression in many yeast strains is blocked by a transposon element (10). Ctr2 appears to mobilize copper from the vacuole (11). The Ctr1 family of copper transporters is conserved in eukaryotes, including humans (3, 4). Ctr1 transports Cu+, which is reduced by cell surface metalloreductases (12, 13), with a Km of 1∼5 μm copper (9, 14). After copper is transported across the cell membrane, it is incorporated into copper-requiring proteins via copper-delivery molecules and assembly factors (15–17). Cytosolic copper chelators, such as metallothionein and phytochelatin, sequester excess copper (18, 19). P-type ATPases play a role in copper detoxification by extruding cellular copper (6, 7).
Given that copper levels in the environment likely fluctuate and excess copper is toxic, delicate control of copper uptake, distribution, and detoxification is necessary for copper homeostasis. Transcriptional regulation of genes encoding components of copper transport and detoxification via Mac1 and Ace1 has been characterized in yeast (3, 22, 23). Moreover, excess copper rapidly induces degradation of yeast Ctr1 (t½ < 30 min) in the apparent absence of endocytosis (24). However, in contrast to these data, another group reported that when excess copper is added to culture media, yeast Ctr1 is accumulated at the vacuole in an Rsp5 ubiquitin ligase and endocytosis-dependent mechanism (25). Ctr1 that accumulates within the vacuole is stable after 2 h. Two lysine residues (Lys-340 and Lys-345) at the C terminus are necessary for copper-dependent ubiquitination followed by the internalization of Ctr1 (25). This mode of post-translational regulation of yeast Ctr1 appears to occur in mammalian Ctr1 as well (26, 27), although other research groups have reported contradictory data (28, 29). Collectively, the nature and mechanisms of copper-dependent post-translational regulations of Ctr1 are not firmly established yet in both yeast and mammals.
Ctr1 family members are integral membrane proteins possessing three predicted transmembrane domains (3, 4). Conserved methionine residues, one at the extracellular N terminus and two that face into the periplasmic space in the predicted second trans-membrane domain, are proposed to be involved in copper translocation (30). However, overall sequence identity among Ctr1 family of proteins is minimal. In particular, the N-terminal extracellular domain and C-terminal cytosolic tail are distinct in length and amino acid sequence. The predicted C terminus of yeast Ctr1 and human Ctr1 encompass125 and 12 amino acids, respectively (3, 4). However, despite of this difference in length, many of Ctr1 family members possess potential metal binding residues at the N and C termini. For instance, the C-terminal cytosolic tail of yeast Ctr1 contains six Cys residues, and human Ctr1 possesses His-Cys-His residues at the C terminus (3, 4). Several lines of evidence indicate that Ctr1 forms a multimer complex (30–32), and indeed a projection structure of the human Ctr1 confirmed that Ctr1 forms a compact trimer with a novel channel-like architecture and no structural occlusion of the predicted pore for copper (33). However, it is largely unknown how Ctr1 transports copper. Our previous data demonstrate that copper induces conformational changes in the C terminus of yeast Ctr1, which is coupled with copper transport (34). Co-expression of Ctr1 monomers fused with either cyan or yellow fluorescent protein resulted in fluorescence resonance energy transfer, which is consistent with multimer assembly of Ctr1 in situ (34). Copper near the Km value of Ctr1 enhanced fluorescence resonance energy transfer in a manner that correlated with cellular copper uptake, which revealed an implication of structural remodeling of the C terminus in function and/or regulation of Ctr1 in yeast (34). Copper translocation through the transmembrane pore within a homotrimer complex could culminate with a motion of the C terminus when copper is released into the cytoplasm. Alternatively, structural remodeling of Ctr1 could be implicated in regulation of Ctr1. Given that Ctr1 in yeast and mammalian cells might undergo copper-induced endocytosis and degradation (24–27), a specific conformation induced in response to excess copper may recruit the machinery for internalization and degradation of Ctr1. It is also possible that the conformational change in the cytosolic C terminus controls the copper translocation across the transmembrane pore when cells accumulate sufficient copper.
To gain better insights into the mechanisms of action and regulation of Ctr1 and the physiological significances of the conformational changes in the C terminus (34), we have characterized the functional roles for the C terminus of Ctr1 in yeast S. cerevisiae. Cells expressing Ctr1 lacking the entire C-terminal tail are active in high affinity copper uptake but extremely sensitive to excess copper. The yeast Ctr1 with serial deletion or site-directed mutations of residues within the C terminus indicates that in response to excess copper, the C terminus rapidly blocks Ctr1-mediated copper uptake with no significant changes in expression levels and subcellular distribution of Ctr1. Our data suggest a new regulatory mechanism of Ctr1 by which yeast cells prevent toxic accumulation of copper.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions—A BY4741 haploid S. cerevisiae strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), in which the Ctr1 gene is deleted (ctr1::kanMX4) and the Ctr3 gene is silent (Open Biosystems), was used in this study. Drs. Bruno André (Université Libre de Bruxelles), Caroline Philpott (National Institutes of Health), and David Eide (University of Wisconsin-Madison) kindly provided rsp5/npi1 (35), end4-1 (36), and chc1-1 (37) strains and their isogenic wild type strains. In the rsp5/npi1 strain, expression of Rsp5/Npi1 is reduced >10-fold compared with isogenic wild type cells as a result of the insertion of a Ty1 element in its 5′ region (35). end4-1 and chc1-1 are temperature-sensitive mutant alleles (36, 37). Yeast cells were cultured with synthetic complete (SC)2 media (2% dextrose, 0.2% amino acid mixture, 0.67% yeast nitrogen base) lacking uracil for plasmid selection (SC-ura) or YPD (2% dextrose, 2% Bacto-peptone, 1% yeast extract) media. Solid media was prepared with the supplementation of 1.5% agar. Cell growth on non-fermentable carbon sources was tested using YPEG media (2% Bacto-peptone, 1% yeast extract, 2% ethanol, 3% glycerol, 1.5% agar). Yeast cells were cultured in SC-ura media at 30 °C if no media or temperature is specified.
Plasmid Construction and Manipulation—If no expression vector is indicated, single copy yeast vectors (p416-TEF) (38) were used for a TEF2 gene promoter-driven constitutive expression. For triple hemagglutinin (HA) epitope-tagging at the N-terminal extracellular domain, a NotI restriction enzyme site was created between the 119th and 120th amino acid. Site-directed mutagenesis was conducted by the primer overlap extension method (39). Serial truncation of the C terminus was made by the PCR method. Common molecular biology techniques, including plasmid amplification and purification using Escherichia coli, followed previously established methods (40). All constructs were sequenced to confirm mutations and inframe fusion. Plasmid transformation into yeast was performed using the lithium acetate method (41).
Cell Growth Assays—The functionality of Ctr1 mutant alleles was determined by assessing the complementation of the growth defect of Δctr1 cells on YPEG media. Cells transformed with Ctr1 plasmid were cultured to mid-log phase in SC-ura media. Cells (5 μl diluted at 0.1 optical density at a wavelength of 600 nm (A600 = 0.1)) were spotted on solid YPEG media. To further deplete copper in this media, we supplemented copper chelator bathocuproine disulfonate (BCS) (20 or 50 μm). Plates were incubated at 30 °C for 3 days before photography. To determine copper toxicity conferred by the Ctr1 mutant alleles, cells were spotted on SC-ura media supplemented with copper (CuCl2) at concentrations indicated in figures. Cell growth was assessed after 1 day. Each assay was repeated at least two times to confirm results.
Western Blot Analysis—Total protein extracts were prepared by breaking cells with glass beads in phosphate-buffered saline (PBS) containing protease inhibitor mixture (Complete Mini, Roche Applied Science) and 1% Triton X-100. Protein concentrations were measured using a BCA kit (Pierce) according to the manufacturer's specifications. Cell lysates were denatured in a SDS sample buffer containing dithiothreitol (100 mm) for 15 min at 60 °C, resolved by SDS-PAGE, and then transferred to a nitrocellulose membrane. HA epitope-tagged Ctr1 was detected by chemiluminescence using anti-HA antibodies (Rockland) and horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Santa Cruz). Phosphoglycerate kinase (PGK) was probed as a loading control using anti-PGK antibodies (Molecular Probes) and horseradish peroxidase-conjugated anti-mouse IgG antibodies (GE Healthcare).
64Cu Uptake Assays—Radioactive 64Cu (specific activity of 15–30 mCi/μg of CuCl2) (Isotrace Technique) was added to exponentially growing cells to the indicated concentrations, and then the cells were incubated for 10 min (for determining copper transport kinetics) or 5 min (for other copper uptake assays) at room temperature. 64Cu uptake into cells was quenched with ice-cold PBS containing 10 mm EDTA. These cells were filtered using nitrocellulose membranes (0.22 μm, Millipore) and washed twice with PBS containing 10 mm EDTA. Cell-associated 64Cu was quantified using a γ-counter (PerkinElmer Life Sciences 1470 WIZARD). 64Cu levels in Δctr1 cells expressing an empty vector (Ctr1-independend background uptake) were subtracted. Copper uptake was calculated using a standard curve and then normalized to the cell number. Apparent Vmax and Km were determined via extrapolation to zero in the reciprocal plot for velocity versus copper concentration using GraphPad PRISM™ software.
Subcellular Fractionation—Subcellular fractionation was performed essentially as described previously (42) with the following modification. Exponentially growing cells were precultured with cycloheximide (100 μg/ml) for 30 min and then cultured with or without copper supplementation (0.1 mm CuCl2, 30 min). Metabolic inhibitors (10 mm sodium azide and potassium fluoride) were added to cells, and then cells were collected by centrifugation. Cells were washed 1 time with a breaking buffer (10 mm Tris-HCl (pH 7.6), 10% sucrose, 10 mm EDTA, and protease inhibitor mixture (Complete Mini, Roche Applied Science)). Cells were then vortexed with glass beads. After centrifugation for 3 min at 300 × g, the supernatant (0.5 ml) was loaded onto a linear sucrose gradient prepared by subsequently layering 10, 20, 35, or 60% sucrose to form an 9-ml step gradient. Samples were subjected to centrifugation (at 4 °C at 50,000 rpm for 14 h). Fractions (500 μl each) were collected from the top of the gradient. Aliquots of each fraction were denatured in an SDS sample buffer containing dithiothreitol (100 mm) at 60 °C for 15 min. HA-tagged Ctr1 in these fractions was detected by Western blotting using anti-HA antibodies.
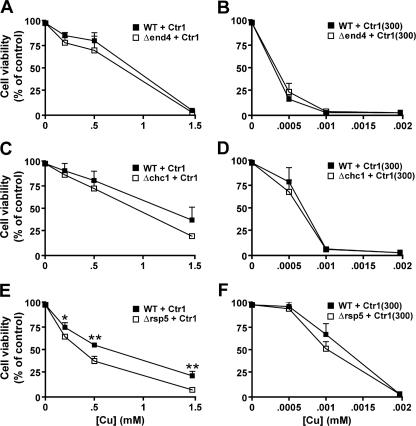
Lethality Assays—rsp5/npi1 (35), end4-1 (temperature-sensitive mutation of end4) (36), and chc1-1 (temperature-sensitive mutation of chc1) (37) strains and their isogenic wild type strains were transformed with expression constructs of wild type Ctr1 or Ctr1 with C-terminal 106 amino acid deletion (Ctr1(300)). rsp5/npi1 cells grown to mid-log phase were cultured with and without supplementation of copper (CuCl2) at concentrations indicated in the figures (see Fig. 5, E and F) for 1 h at 30 °C. For experiments using temperature-sensitive end4-1 and chc1-1 mutant strains, cells were cultured at 25 °C and then shifted to a restrictive temperature (37 °C) for 30 min before adding copper to culture media. After culturing the cells with copper for 1 h, they were washed twice and diluted to the same cell density (A600 = 0.001), and then 100 μl were plated on solid YPD media. Our assumption is that cells accumulate toxic copper and/or are severely damaged during the culture in media supplemented with copper and are not able to grow on YPD plates. After 2 days, the colonies that had grown were counted, and colony numbers were then compared with those of control plates.
FIGURE 5.
Copper sensitivity of cells defective in endocytosis or ubiquitination pathways. Wild type Ctr1 or Ctr1 with C-terminal truncation of the last 106 amino acids (Ctr1(1–300)) were expressed in end4-1 strain (A and B), chc1-1 strain (C and D), Δrsp5 strain (E and F), and isogenic wild type (WT) control strains. Temperature-sensitive end4-1 and chc1-1 cells and their isogenic control cells were cultured at 25 °C and then shifted to a restrictive temperature (37 °C) to block endocytosis. Δrsp5 and isogenic control strains were cultured at 30 °C to mid-log phase. Cells were cultured with supplementation of copper (CuCl2) in media at the indicated concentrations for 1 h than washed twice, and then 100 μl of diluted (A600 = 0.001) cells were plated on solid YPD media. After incubating plates at 30 °C for 2 days, growing colonies on plates were counted, and then the colony numbers were compared with those on control plates where cells precultured without copper were plated. Each data point indicates the average ± S.D. of at least four independent assays. *, p < 0.05; **, p < 0.01, Student's t test.
Superoxide Dismutase Activity Assays—Cell lysates were prepared by breaking cells with glass beads in PBS containing protease inhibitor mixture (Complete Mini, Roche Applied Science). We first inactivated manganese-containing superoxide dismutase as described previously (43) and then measured copper and zinc-containing superoxide dismutase (SOD1) activities by determining the inhibition of nitrite formation from hydroxylamine in the presence of superoxide anion generator (44). SOD1 activities were normalized by protein concentrations and then converted to units using a standard curve of bovine liver SOD1 (Sigma).
RESULTS
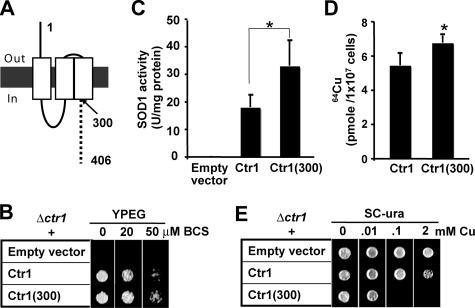
C-terminal Cytosolic Tail of Ctr1 Is Dispensable for Copper Transport—To determine the roles for the cytosolic C-terminal tail of Ctr1 in copper transport, we examined functionality of Ctr1 with deletion of the C terminus encompassing the last 106 amino acids (Ctr1(300)) (Fig. 1A). Δctr1 cells were not able to grow on the YPEG media containing solely non-fermentable carbon sources because of the defect in copper-requiring cytochrome c oxidase (Fig. 1B). However, the expression of either full-length Ctr1 or Ctr1(300) complemented the growth defect of Δctr1 cells on YPEG media and the same media supplemented with BCS copper chelator that reduces bio-available copper (Fig. 1B). No difference in growth was observed between cells expressing either Ctr1 or Ctr1(300), which suggests that the C terminus of Ctr1 does not play a critical role in copper transport followed by its insertion into cytochrome c oxidase. Next, we measured the activities of copper-containing SOD1 in these cells. Although SOD1 activities in Δctr1 cells were hardly detectable, the expression of either Ctr1 or Ctr1(300) dramatically enhanced SOD1 activities (Fig. 1C). Interestingly, cells expressing Ctr1(300) exhibited higher SOD1 activities when compared with cells expressing the full-length Ctr1 (Fig. 1C). To directly monitor Ctr1 and Ctr1(300)-mediated cellular copper acquisition, we measured radioactive 64Cu uptake into cells. Cells were cultured to exponential growth phase in SC media supplemented with copper chelator BCS (10 μm) for 12 h, and then 64Cu (0.1 μm, 64CuCl2) uptake was measured for 5 min. Cellular 64Cu uptake was also higher in cells expressing Ctr1(300) when compared with cells expressing Ctr1 (Fig. 1D). Collectively, these data suggest that the C-terminal tail of Ctr1 is dispensable for high affinity uptake and subsequent distribution of copper to copper-requiring proteins.
FIGURE 1.
Functionality of Ctr1 lacking the C-terminal cytosolic tail. A, schematic depiction of Ctr1 in yeast S. cerevisiae. The dotted line indicates the predicted C-terminal tail encompassing amino acids 301–406. White rectangles indicate three transmembrane domains. B–E, empty vector, full-length Ctr1, or Ctr1 lacking its last 106 amino acids (Ctr1(300)) were expressed in a Δctr1 strain, and then cellular copper metabolism was examined. B, Ctr1(300) functionally complements the growth defect of Δctr1 cells on non-fermentable (YPEG) media. Exponentially growing cells (5 μl, diluted to A600 = 0.1) were spotted on YPEG media supplemented with BCS copper chelator. Cell growth was assessed after 3 days. C, cytosolic fractions were subjected to activity assays for copper- and zinc-containing SOD1. D, 64Cu uptake assays. Cells cultured in SC-ura and supplemented with copper chelator BCS (10 μm for 12 h) were washed twice with prewarmed SC-ura media and re-suspended in the same media. Cells were incubated with 64Cu (0.1 μm 64CuCl2) for 5 min. Cell-associated 64Cu was measured using a γ-counter. E, cells expressing Ctr1(300) are hypersensitive to copper. Exponentially growing cells (5μl of cells diluted to A600 = 0.1) were spotted on SC-ura media and supplemented with indicated concentrations of copper (CuCl2)(Cu). Plates were incubated at 30 °C for 1 day, and cell growth was assessed. Each bar in C and D represents the average ± S.D. of at least four independent experiments. *, p < 0.01, Student's t test.
Cells Expressing Ctr1 with a C Terminus Deletion Are Hypersensitive to Copper—Given that Ctr1(300) transports more copper as compared with Ctr1, we hypothesized that the C terminus may play a regulatory role in copper transport. We tested this hypothesis by monitoring cell growth on media containing toxic copper. Δctr1 cells transformed with an empty vector, Ctr1, or Ctr1(300) were cultured to mid-log phase, and then cells (5 μl cells diluted to A600 = 0.1) were spotted on SC media supplemented with excess copper. Indeed, cells expressing Ctr1(300) are extremely sensitive to copper. Although cells expressing empty vector or Ctr1 grow on media containing 2 mm copper, cells expressing Ctr1(300) could not grow even on media containing 0.1 mm copper (Fig. 1E). These cells were also hypersensitive to silver but not other metals tested, including zinc, iron, or cadmium (data not shown). Given that both Cu+ and Ag+ have been proposed as substrates of Ctr1 (14), these data suggest that Ctr1(300) transport excess copper when cells grow in high copper concentration media and that C terminus may contain a regulatory domain preventing excess copper uptake.
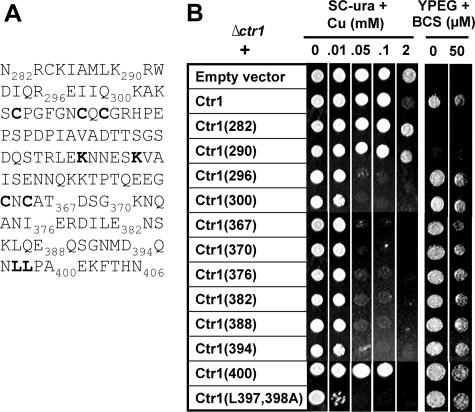
Mapping of the C Terminus to Identify Residues That Play Regulatory Roles—To identify residues within the Ctr1 C terminus that play inhibitory roles in copper transport in response to excess copper, we mapped the C terminus of Ctr1. Expression vectors of Ctr1 mutants with serial deletion of the C terminus were constructed (Fig. 2A) and then transformed into a Δctr1 strain. Cell growth on both SC media containing excess copper and copper-requiring YPEG media was then assessed (Fig. 2B). BCS copper chelator was supplemented to YPEG media to further deplete bio-available copper. Growth defects on the YPEG media of cells expressing Ctr1 with the deletion of the last 116 or 124 amino acids (Ctr1(290) or Ctr1(282)) suggested the non-functionality of these Ctr1 alleles in high affinity copper transport. Cells that express Ctr1 with the deletion of the last 6 amino acids (Ctr1(400)) showed slight but significant copper sensitivity on SC media containing excess copper when compared with cells expressing the wild type Ctr1. Cells expressing Ctr1 with deletion of the last 12 amino acids (Ctr1(394)) or additional amino acids up to 110 amino acids (Ctr1(296)) are extremely sensitive to copper. Complementation of growth defects of Δctr1 cells on copper-requiring YPEG media indicates that these Ctr1 alleles are fully functional in copper uptake.
FIGURE 2.
Growth assays of cells expressing Ctr1 with deletions or site-directed mutations within its C terminus. A, C-terminal (amino acids 282–406) sequence of yeast Ctr1. Truncations are indicated by the number of amino acid residues. Amino acid residues that were subjected to site-directed mutagenesis in this and subsequent figures are in bold type. B, empty vector, full-length Ctr1, and Ctr1 with serial deletions of the C terminus or substitutions of Leu-397 and Leu-398 residues to Ala (Ctr1(L397A,L398A)) were expressed in a Δctr1 strain. Exponentially growing cells were diluted (A600 = 0.1), and then cells (5 μl) were spotted on non-fermentable (YPEG) or uracil-deleted synthetic complete (SC-ura) media supplemented with indicated concentrations of copper (CuCl2) or copper chelator BCS. Cell growth was assessed after 1 and 3 days for SC-ura and YPEG media, respectively.
Mapping of the C terminus suggests that the amino acids between 395 and 400 may contain important residues for the regulation of homeostatic acquisition of copper. This domain contains a dileucine motif (Fig. 2A) that is known to play a role in internalization of membrane protein in mammalian cells (31). Although the function of this motif in yeast S. cerevisiae has not been established, a regulatory role of a dileucine motif in the yeast Gap1 amino acid permease was suggested (35). To examine the roles for this motif in Ctr1 function and regulation, we substituted dileucine residues to alanine (Ctr1(L397A,L398A)), expressed this mutant in a Δctr1 strain, and then tested cell growth on copper-requiring or toxic copper media. This Ctr1 allele complemented growth of Δctr1 cells on YPEG media; however, cells expressing this mutant are extremely sensitive to copper toxicity (Fig. 2B). This suggests an important role for these leucine residues in the regulation of Ctr1-mediated copper transport. When cellular copper is in excess, the C terminus might regulate subcellular localization and/or expression of Ctr1 or play a structural role for controlling copper transport activities of Ctr1 multimer complexes.
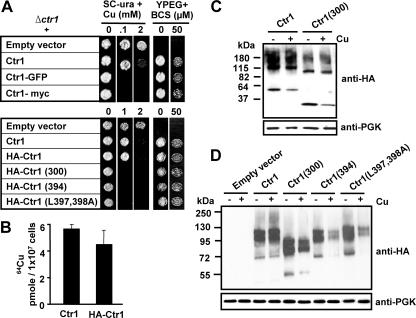
Epitope Tagging at the N or C Terminus Further Suggested Critical Roles for the C Terminus in Preventing Copper Toxicity—To determine the role of the C terminus in expression levels and subcellular localization of yeast Ctr1, we have made epitope-tagged Ctr1 constructs. Given that previous reports used the Ctr1 allele fused with either single c-Myc at an EcoR1 site encompassing amino acids 382, 383, and 384 at the C terminus (Ctr1-Myc) (24) or GFP at the end of the C terminus (Ctr1-GFP) (25), we expressed these constructs in Δctr1 yeast cells and then tested cell growth on non-fermentable YPEG media and synthetic complete SC media. Both Ctr1-Myc and Ctr1-GFP functionally complement growth defect of Δctr1 cells on YPEG media (Fig. 3A, upper panel), suggesting similar copper transport activities of these alleles to that of non-tagged Ctr1. However, cells expressing Ctr1-GFP or Ctr1-Myc are extremely sensitive to copper when grown on SC media (Fig. 3A, upper panel). This data suggests that structural perturbation of the C terminus of yeast Ctr1 confers copper sensitivity when expressed in yeast cells.
FIGURE 3.
Expression levels of Ctr1 possessing C-terminal deletion or mutation. A, functionality of Ctr1 fused with epitopes or GFP. GFP coding sequence was inserted into a NotI site generated by PCR at the end of C terminus. Single c-Myc epitope was inserted into the EcoRI site within the C-terminal tail. Triple HA epitopes were inserted into a NotI site generated by PCR after the 119th amino acid residue of wild type Ctr1, Ctr1(300), Ctr1(394), and Ctr1(L397A,L398A). An empty vector, intact Ctr1, and epitope or GFP-fused Ctr1 were expressed in a Δctr1 strain. Exponentially growing cells in synthetic complete media omitting uracil (SC-ura) were diluted (A600 = 0.1), and then cells (5 μl) were spotted on solid non-fermentable (YPEG) or SC-ura media supplemented with indicated concentrations of copper (CuCl2) or copper chelator BCS. Cell growth was assessed after 1 and 3 days for SC-ura and YPEG media, respectively. B, 64Cu uptake assays. Cells cultured in SC-ura media containing copper chelator BCS (10 μm for 12 h) were washed twice with prewarmed SC-ura media and resuspended in the same media. Cells were incubated with 64Cu (0.1 μm 64CuCl2) for 5 min. Cell-associated 64Cu was measured using a γ counter. Data represent the average ± S.D. of three independent experiments. C, HA-epitope tagged full-length Ctr1 (Ctr1–3HA) or Ctr1 with a truncation of the last 106 amino acids at the C terminus (HA-Ctr1(300)) were expressed in a Δctr1 strain. Exponentially growing cells were cultured with or without supplementation of copper (CuCl2, 100 μm) for 30 min. Cycloheximide (100 μg/ml) was added to the cell culture 30 min prior to copper supplementation. Total protein extracts were subjected to Western blot analysis using anti-HA antibodies. PGK, phosphoglycerate kinase. D, Δctr1 cells expressing HA-tagged Ctr1, Ctr1(300), Ctr1(394), and Ctr1(L397A,L398A) were cultured in media supplemented with (+) or without (–) copper (CuCl2, 25 μm) for 12 h. Total protein extracts were subjected to Western blot analysis using anti-HA antibodies. Experiments were conducted three times, and a representative figure is presented. Expression levels were quantified using phosphoglycerate kinase loading control and then normalized to wild type Ctr1.
For epitope tagging at the N terminus, a NotI restriction enzyme site was artificially generated directly after the start codon or at several locations within the extracellular N terminus. Triple HA epitope was inserted into the NotI site, and then the functionality of the Ctr1 fused with HA was tested. Among these fusion proteins, Ctr1 with HA inserted after the 119th amino acid (Ctr1–3HA) was fully functional in both complementation of the growth defects of Δctr1 cells on YPEG media (Fig. 3A, lower panel) and 64Cu uptake (Fig. 3B) and easily detectable by Western blotting using anti-HA antibodies (Fig. 3, C and D). No significant difference in growth was observed on SC media supplemented with toxic copper either (Fig. 3A, lower panel). Ctr1(300), Ctr1(394), and Ctr1(L397A,L398A) tagged with HA at the same position were also fully functional in complementation of cell growth and conferred copper sensitivity (Fig. 3A, lower panel). These data indicate that HA epitope in the N terminus does not perturb the function and regulation of Ctr1.
Expression Levels of Ctr1 Possessing Deletion or Site-directed Mutations at the C Terminus—Copper toxicity in cells expressing Ctr1 possessing a deletion or site-directed mutations in the C terminus may be attributed to the higher expression levels of Ctr1 mutants relative to wild type Ctr1. Δctr1 cells expressing HA epitope-tagged Ctr1 or Ctr1(300) were cultured to mid-log phase. Cycloheximide (100 μg/ml) was added 30 min before copper supplementation to the culture (0.1 mm CuCl2 for 30 min). Total protein extracts of collected cells were subjected to Western blot analyses of Ctr1 using anti-HA antibodies. Predicted monomer, dimer, and trimer species were detected, and expression levels of Ctr1 and Ctr1(300) were similar to each other (Fig. 3C). Copper supplementation in the culture media slightly enhanced Ctr1 turnover especially monomer species; however, no significant difference in protein stability was observed between Ctr1 and Ctr1(300).
Second, we determined whether the C terminus affects Ctr1 expression levels in cells cultured long term in media containing excess copper. Δctr1 cells expressing empty vector, HA-tagged Ctr1, Ctr1(300), Ctr1(394), or Ctr1(L397A,L398A) were cultured without or with copper supplementation (25 μm CuCl2) in liquid SC media for 12 h. Copper supplementation results in slow growth of cells expressing Ctr1(300), Ctr1(394), or Ctr1(L397A,L398A) compared with cells expressing wild type Ctr1 (data not shown), which is consistent with growth defects on copper-supplemented solid media. Western blot analyses of Ctr1 using anti-HA antibodies demonstrated no significant difference in expression levels among these Ctr1 alleles when cells were cultured in media without copper supplementation (Fig. 3D). However, when cells were cultured in the media supplemented with copper, expression of Ctr1(300), Ctr1(394), or Ctr1(L397A,L398A) are lower than wild type Ctr1 (Fig. 3D). Collectively, the differences in the expression levels do not explain why cells expressing Ctr1 possessing deletion or mutation at the C terminus are hypersensitive to copper when compared with cells expressing wild type Ctr1.
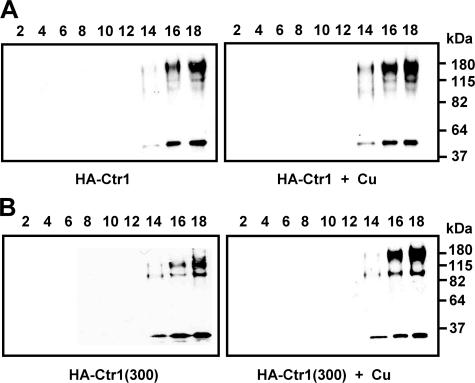
C-terminal Truncation Does Not Affect Subcellular Distribution of Ctr1—Although the expression levels of Ctr1 and Ctr1(300) are similar, the cellular location of Ctr1(300) could be distinct from Ctr1. To determine subcellular location of HA-tagged Ctr1 and Ctr1(300), lysates of cells cultured with or without supplementation of copper (0.1 mm for 30 min) were fractionated by a sucrose gradient. Western blot analysis of the collected fractions showed that Ctr1 is detectable mainly at high density fractions which were predicted to contain the plasma membrane (Fig. 4A, left panel). However, copper supplementation to the culture media did not alter subcellular distribution of Ctr1 (Fig. 4A, right panel). No significant difference in the distribution pattern between Ctr1 (Fig. 4A) and Ctr1(300) was observed (Fig. 4B) either. These data indicate that the copper hypersensitivity of cells expressing Ctr1(300) is not attributed to a different pattern of subcellular distribution of Ctr1(300) when compared with that of Ctr1.
FIGURE 4.
Subcellular distribution of Ctr1 was not altered by C-terminal deletion or copper in culture media. Triple HA epitopes were inserted into a NotI site generated by PCR after the 119th amino acid residue of wild type Ctr1 and Ctr1(300). Cells expressing HA-tagged Ctr1 (HA-Ctr1)(A) or Ctr1(300) (HA-Ctr1(300))(B) were cultured without (left panel) or with (right panel) copper supplementation (CuCl2, 0.1 mm) in the growth media for 30 min. Cell lysates prepared using glass beads were separated on sucrose gradients, and then portions of collected fractions (0.5 ml each, fractions 1–18 from the top) were subjected to Western blotting using anti-HA antibodies. Fractions used for Western blotting are indicated in each figure.
Cells Defective in Endocytosis Pathway Are Not Hypersensitive to Copper—Because our data were inconsistent with previous reports (24, 25) suggesting copper-responsive internalization and degradation of yeast Ctr1 and the proposed role for dileucine motif in regulation of subcellular trafficking of plasma membrane proteins of mammalian cells (31), we further investigated the roles for the endocytosis pathways in regulation of Ctr1-mediated copper transport. If this molecular event plays a significant role in preventing excess copper transport, expression of Ctr1 in cells defective of endocytosis pathways would lead to hypersensitivity to copper in these cells as compared with isogenic wild type control cells. Consistently, if Ctr1 endocytosis is dependent on the C terminus of Ctr1, wild type strains expressing C terminus-deleted Ctr1(300) would manifest the similar copper sensitivity to the endocytosis-defective strains expressing full-length Ctr1. Yeast strains possessing a temperature-sensitive allele of End4 or Chc1 (end4-1 or chc1-1) are defective in the internalization of cell surface proteins when cells are cultured at a restrictive temperature (36, 37). These strains were transformed with Ctr1 and Ctr1(300) expression constructs, cultured in permissive temperature (25 °C), and then shifted to a restrictive temperature (37 °C) to inactivate End4 or Chc1. Copper (CuCl2) was supplemented into the culture media at concentrations indicated in Fig. 5 for 1 h. Cells were washed twice, and then diluted cells (100 μl, A600 = 0.001) were plated on YPD media. Colonies that had grown were counted after 2 days. Δend4 or Δchc1 cells expressing wild type control Ctr1 have slightly higher sensitivity to copper when compared with their isogenic wild type strains (no statistically significant difference) (Fig. 5, A and C). However, the wild type cells and Δend4 or Δchc1 cells expressing Ctr1(300) were extremely sensitive to copper. Cells expressing Ctr1(300) were not able to form colonies after culture in media containing 2 μm CuCl2 (Fig. 5, B and D); however, ∼50% cells expressing Ctr1 were able to grow after culture in media containing 500 μm CuCl2 (Fig. 5, A and C). These data suggest that the endocytosis pathway does not play a significant role in the regulation of Ctr1-mediated copper uptake and that copper sensitivity conferred by Ctr1(300) is not associated with a defect of its internalization in response to excess copper.
Cells Defective in Rsp5/Npi1 Ubiquitin-protein Ligase Are Not Hypersensitive to Copper—A recent paper showed that Ctr1 undergoes Rsp5 ubiquitin-protein ligase-dependent ubiquitination followed by internalization (25). To determine the roles for the Rsp5/Npi1 ubiquitin ligase in Ctr1-mediated copper transport, we transformed the expression constructs of Ctr1 and Ctr1(300) into rsp5/npi1 (35) and isogenic wild type strains. Exponentially growing cells were supplemented with copper (CuCl2) in culture media at the concentrations indicated in Fig. 5, E and F, for 1 h. Cells were washed twice, and then diluted cells (100 μl, A600 = 0.001) were plated on YPD media. Colonies that had grown were counted after 2 days. Colony numbers were lower in the rsp5/npi1 strain (Δrsp5) when compared with wild type control cells transformed with the expression construct of full-length Ctr1, suggesting Rsp5/Npi1-mediated ubiquitination might play a protective role in copper toxicity (Fig. 5E). However, both wild type and rsp5/npi1 cells expressing Ctr1(300) were again extremely sensitive to copper (Fig. 5F) when compared with the same strains expressing the full-length wild type Ctr1 (Fig. 5E). These data suggest that copper sensitivity conferred by Ctr1(300) does not rely on the defect in Rsp5-mediated ubiquitination of Ctr1 in a Ctr1 C terminus-dependent manner.
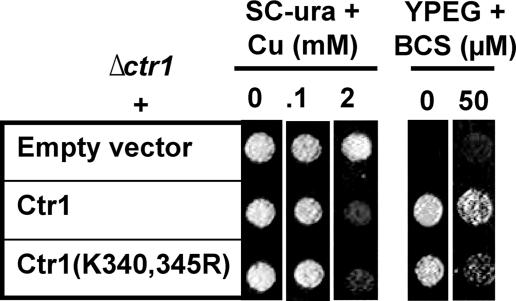
Given a previous report suggesting that Rsp5/Npi1 ubiquitinates two lysine residues in the C terminus (Lys-340 and -345) (Fig. 2A) (25), we substituted these residues to arginine and then tested the function and regulation of this Ctr1 allele. Δctr1 cells expressing Ctr1(K340R,K345R) could complement the Ctr1 deletion on copper requiring YPEG media (Fig. 6), and this Ctr1 allele did not confer copper hypersensitivity to cells growing on SC media containing excess copper (Fig. 6), which further suggests that defects in ubiquitination of the Lys-340 and -345 residues are not a reason for copper sensitivity of cells expressing Ctr1 with a C-terminal deletion. Copper sensitivity of rsp5/npi1 strain relative to its isogenic wild type strain when Ctr1 is expressed (Fig. 5E) might reflect an overall poor health of rsp5/npi1 strain.
FIGURE 6.
Determination of the roles for lysine residues (Lys-340 and -345) within the C terminus of Ctr1 in copper uptake. Full-length Ctr1 or Ctr1 possessing site-directed mutation of Lys-340 and -345 to arginine (Ctr1(K340R,K345R) were expressed in a Δctr1 strain. Exponentially growing cells were diluted (A600 = 0.1), and then cells (5 μl) were spotted on non-fermentable (YPEG) and SC-ura media supplemented with indicated concentrations of copper (CuCl2) or copper chelator BCS. Cell growth was assessed after 1 and 3 days for SC-ura and YPEG media, respectively.
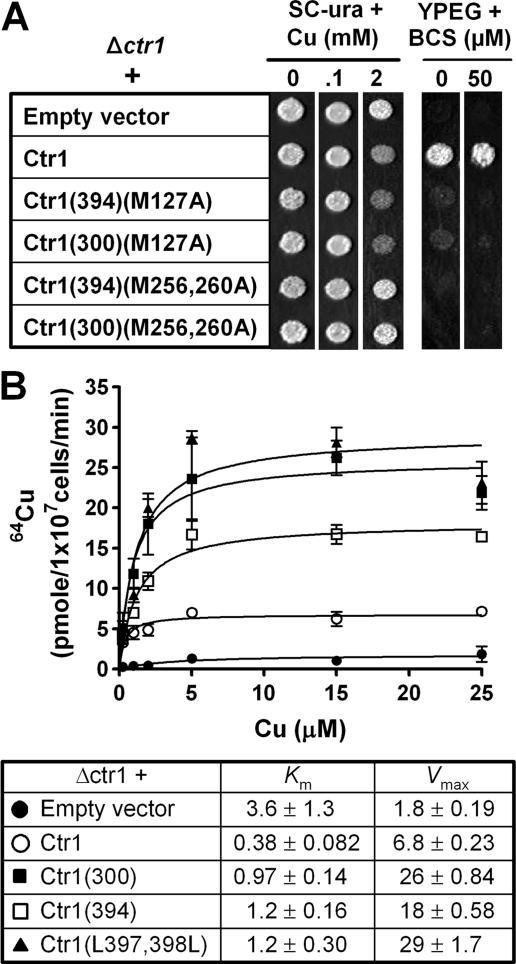
Copper Sensitivity Conferred by Ctr1 C-terminal Mutants Is Dependent on Their Functionality—We next determined whether hypersensitivity of cells expressing Ctr1 possessing a deletion or site-directed mutations is associated with excess copper transport by these mutants. We hypothesized that deletion of the C terminus in a non-functional Ctr1 does not confer copper sensitivity if copper transport activities are necessary for copper sensitivity of cells expressing these Ctr1 mutants. Ctr1 possessing site-directed mutations of conserved Met-127, Met-256, and/or Met-260 to alanine (Ctr1(M127A) or Ctr1(M256A,M260A)) is non-functional in copper transport (35). Expression constructs of these Ctr1 mutants with a deletion of the last 106 or 12 amino acids, Ctr1(300)(M127A), Ctr1(394)(M127A), Ctr1(300)(M256A, M260A), and Ctr1(394)(M256A,M260A), were transformed into a Δctr1 strain. Cell growth on copper-requiring YPEG media and SC media containing excess copper was examined. The data presented in Fig. 7A suggest that hypersensitivity to copper conferred by Ctr1(300) and Ctr1(394) is dependent on their copper transport activities.
FIGURE 7.
Ctr1 mutants possessing C-terminal deletion transport excess copper. A, non-functional Ctr1 alleles possessing a C-terminal deletion did not confer copper sensitivity. Conserved methionine residues (Met-127, Met-256, and Met-260) in Ctr1 (30) were substituted with alanine by site-directed mutagenesis. The last 106 or 12 amino acids in the C terminus were deleted in these Ctr1 mutants (Ctr1(300) and Ctr1(394), respectively). These mutant Ctr1 alleles were expressed in a Δctr1 strain. Exponentially growing cells were diluted (A600 = 0.1), and then cells (5 μl) were spotted on non-fermentable (YPEG) or SC-ura media supplemented with indicated concentrations of copper (CuCl2) or copper chelator BCS. Cell growth was assessed after 1 and 3 days for SC-ura and YPEG media, respectively. B, 64Cu uptake kinetics of cells expressing Ctr1 with a deletion or site-directed mutations within its C terminus. An empty vector, full-length Ctr1 (Ctr1), or Ctr1 with a deletion of the last 106 or 12 amino acids (Ctr1(300) or Ctr1(394), respectively) or site-directed mutations of Leu-397 and Leu-398 (Ctr1(L397A,L398A)) within the C terminus were expressed in a Δctr1 strain. Cells exponentially growing in SC-ura media were incubated with 64Cu (0.4, 1, 2, 5, 15, or 25 μm) for 10 min, and then cell-associated 64Cu was measured using a γ counter. 64Cu uptake was normalized to cell numbers. Each data point represents the average ± S.D. of four independent assays. Apparent Km (μm) and Vmax (pmol/1 × 107 cells/min) for copper were calculated.
We predicted that Ctr1 containing deletion or site-directed mutations at the C terminus would transport excess copper. This hypothesis was tested by determining the 64Cu uptake kinetics of cells expressing Ctr1(300), Ctr1(394), or Ctr1(L397A,L398A). Δctr1 cells expressing empty vector, Ctr1, or Ctr1 mutant alleles were subjected to 64Cu uptake assays (Fig. 7B). Copper uptake at the concentration of 0.4 μm (the lowest copper concentration) was similar between cells expressing Ctr1 or Ctr1 mutants; however, the uptake at more than 2 μm copper concentrations was significantly higher in cells expressing Ctr1 mutants (Fig. 7B, upper panel). For instance, apparent Vmax of Ctr1(300) for copper was ∼4-fold higher than that of full-length Ctr1 (Fig. 7B, lower panel).
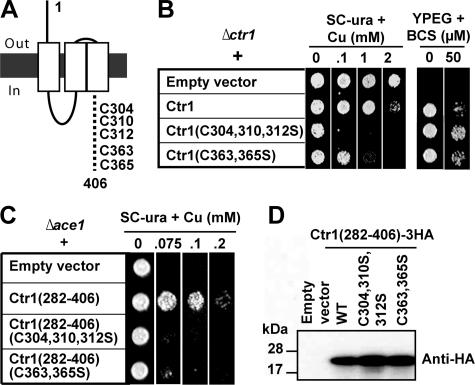
Regulatory Roles for the Cysteine Residues in the C Terminus of Ctr1—We investigated whether the C terminus plays a role in copper-responsive control of Ctr1 activities. The conformational changes in the Ctr1 C terminus that are associated with copper transport (34) might reflect the sensing of intracellular copper followed by the inhibition of copper translocation. The C terminus of Ctr1 encompassing the last 106 amino acids possesses five cysteine residues (Cys-304, Cys-310, Cys-312, Cys-363, and Cys-365) (Figs. 2A and 8A). To determine the roles of these cysteine residues in the function and regulation of Ctr1, we substituted these cysteine residues to serine. These mutants were expressed in a Δctr1 strain. Cell growth on YPEG media and SC media supplemented with toxic copper was examined. Expression of Ctr1 possessing these cysteine mutations complemented the growth defect of Δctr1 cells on YPEG media, suggesting no significant role of the cysteine residues in Ctr1-mediated copper transport (Fig. 8B). However, cells expressing Ctr1(C304S,C310S,C312S) or Ctr1(C363S, C365S) were hypersensitive to copper (Fig. 8B) on SC-ura media, suggesting vital roles of these cysteine residues in blocking excess copper transport.
FIGURE 8.
Determination of the roles of cysteine residues in the C terminus of Ctr1. A, five cysteine residues within the C terminus of Ctr1 encompassing amino acids 301–406 are indicated. B, full-length Ctr1 or Ctr1 possessing site-directed mutation of cysteine residues within the C terminus were expressed in a Δctr1 strains. Exponentially growing cells were diluted (A600 = 0.1), and then cells (5 μl) were spotted on non-fermentable (YPEG) or SC media supplemented with indicated concentrations of copper (CuCl2) or copper chelator BCS. Cell growth was assessed after 1 day for SC-ura media and 3 days for YPEG media. C, C-terminal tail of Ctr1 encompassing the last 124 amino acids (Ctr1(282–406)) without or with site-directed mutation of cysteine residues to serine as indicated was expressed in copper-sensitive Δace1 strain. Exponentially growing cells were diluted (A600 = 0.1), and then cells (5 μl) were spotted on SC media supplemented with indicated concentrations of copper. Cell growth was assessed after 1 day. D, total protein extracts of cells transformed with expression constructs of the C-terminal peptides fused with triple HA epitopes in frame were subjected to Western blotting using anti-HA antibodies.
These Cys residues at the C terminus might bind with copper when cellular copper is in excess, which could trigger conformational changes followed by inhibition of Ctr1 copper transport activities. To probe the copper binding to the C terminus of Ctr1, we examined whether expression of C terminus itself in the cytosol can sequester copper like metallothionein. If copper ion(s) binds to the C terminus, overexpression of the C termini in the cytosol would lead to copper resistance in copper sensitive Δace1 cells. Ace1 is a transcription factor that induces expression of Cup1 and Crs5 metallothionein in response to excess copper (23). Thus, Δace1 cells are sensitive to copper toxicity (23). Peptide encompassing the last 125 amino acids of Ctr1 C terminus (Ctr1(282–406)) was expressed in a Δace1 strain, and then the cell growth on excess copper media was monitored. Indeed, expression of the C terminus allowed Δace1 cells to grow on media containing toxic copper (Fig. 8C). However, the same peptide in which cysteine residues are substituted by serine failed to confer copper resistance (Fig. 8C). Expression levels of these peptides bearing mutations of cysteine residues were similar to those of the wild type control peptide (Fig. 8D). These data suggest copper binding to the C terminus of Ctr1 in a cysteine-dependent mechanism.
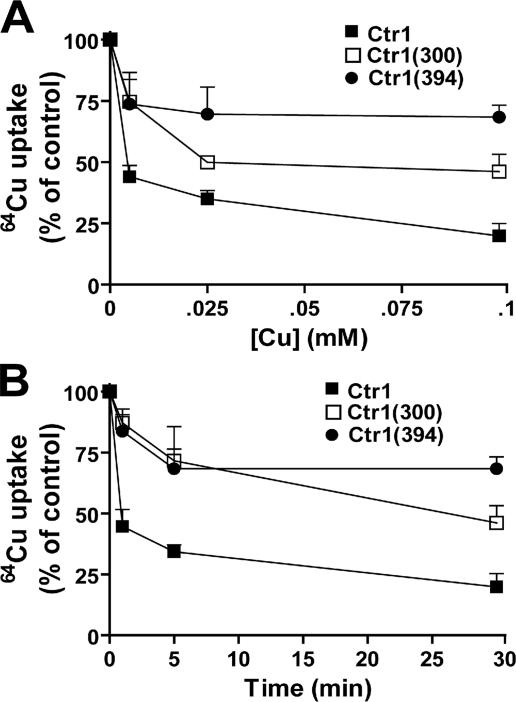
Excess Copper in Culture Media Rapidly Inactivates Ctr1-mediated Copper Transport in a C Terminus-dependent Mechanism—We hypothesized that excess copper in media inhibits Ctr1-mediated copper transport in a C terminus-dependent manner. To test this, we cultured Δctr1 cells transformed with empty vector and expression constructs of Ctr1, Ctr1(300), or Ctr1(394) to mid-log phase with supplementation of cycloheximide (100 μg/ml) for 30 min. The same concentration of cycloheximide was maintained throughout the experiments to prevent new functional Ctr1 synthesis after inactivation of existing Ctr1. Cells were cultured in media supplemented with copper (CuCl2) at concentrations indicated in Fig. 9A for 30 min. To determine time-dependent responses to copper, cells were cultured with 100 μm CuCl2 for 1, 5, and 30 min (Fig. 9B). Cells were then washed twice with prewarmed fresh media, after which 64Cu uptake into cells was measured by adding 64Cu (0.1 μm 64CuCl2) for 5 min to the culture media. We predicted that if Ctr1 activities are inhibited in response to excess copper, cells precultured with copper would have a lower uptake of 64Cu than control cells precultured with no copper supplementation. Indeed, 64Cu uptake in Ctr1-expressing cells which were precultured with copper (0.1 mm CuCl2 for 30 min) was reduced to less than 20% that of 64Cu uptake in control cells which received no copper pretreatment (Fig. 9A). 5 or 25 μm copper pretreatment for 30 min reduced 64Cu uptake in cells expressing Ctr1 to 45 or 39% that in control cells which received no copper pretreatment, respectively (Fig. 9A). This indicates the copper concentration-dependent inhibition of Ctr1 activity. Copper (0.1 mm CuCl2) response is fast, as pretreatment for 5 min was sufficient for near maximum inhibition of 64Cu uptake (Fig. 9B). However, 64Cu uptake into cells expressing Ctr1(300) or Ctr1(394) and were precultured with copper (0.1 mm CuCl2 for 30 min) was ∼46 or 68% that in the control cells which received no copper pretreatment, respectively (Fig. 9A). Indeed, all data points of 64Cu uptake in cells expressing Ctr1(300) and Ctr1(394) are significantly higher than those of Ctr1 (p < 0.001, Student's t test) (Fig. 9, A and B), indicating the roles for the Ctr1 C terminus in control of Ctr1 activities in response to excess copper.
FIGURE 9.
Ctr1-mediated 64Cu uptake in cells precultured in media supplemented with copper. Empty vector, Ctr1, and Ctr1 with a C-terminal deletion of the last 106 or 12 amino acids (Ctr1(300) or Ctr1(394)) were expressed in a Δctr1 strain. Cells grown to exponential phase in SC-ura media were cultured with cycloheximide (100 mg/ml media) for 30 min, and the same concentration of cycloheximide was maintained throughout experiments to inhibit new Ctr1 synthesis. Copper (0, 5, 25, or 100 μm CuCl2) was added to culture media for 30 min (A), or copper (0 or 100 μm CuCl2) was added to culture media for 1, 5, or 30 min (B). Cells were washed twice using pre-warmed fresh SC media and then resuspended in the same media. To measure 64Cu uptake in these cells, cells were incubated with 64Cu (0.1 μm 64CuCl2) for 5 min and washed twice with PBS containing 10 mm EDTA. Cell-associated 64Cu was measured using a γ counter and normalized to cell numbers. 64Cu uptake in cells expressing an empty vector was subtracted as background uptake. Data were presented as percentage (%) of 64Cu uptake in control cells that were precultured without copper supplementation in media. Each data point represents the average ± S.D. of at least four independent assays.
DISCUSSION
Our data suggests that the C terminus of Ctr1 in yeast S. cerevisiae plays a regulatory role in preventing toxic copper transport. In the presence of excess copper, Ctr1 transport activity is rapidly inhibited. This response to copper is dependent on the C-terminal cytosolic tail of Ctr1 but not associated with apparent changes in expression levels and subcellular location of Ctr1. Given that copper is extremely toxic and its levels fluctuate in the environment, down-regulation of Ctr1 expression via the endocytosis pathway and/or transcriptional control may not be fast enough for the prevention of toxic accumulations of copper. Hence, control of Ctr1 activity via direct sensing of cellular copper provides a mechanism by which cells promptly inhibit excess copper uptake.
The data presented herein indicate that the C terminus is a regulatory domain that prevents the toxic accumulation of copper. We propose that when the C terminus senses excess copper through direct copper coordination by Cys residues, Ctr1-mediated copper transport is blocked. Thus, the C terminus of Ctr1 serves as a molecular switch that controls homeostatic copper uptake. Consistent to a report indicating that the purified C-terminal domain of yeast Ctr1 encompassing last 126 amino acids binds four Cu+ ions as a cuprous-thiolate polynuclear cluster (45), we were able to show that overexpression of the C terminus itself in the cytosol could suppress the copper sensitivity of Δace1 cells likely by sequestering copper. A caveat of this experiment was that copper binding at the C terminus might occur within a trimer complex (34); however, our data suggest that intermolecular coordination is not necessary for copper binding to the C terminus. Site-directed mutagenesis studies also suggest that cysteine residues at the C terminus serve as copper ligands. Hence, structural perturbation of the C terminus could result in ablation of the inhibitory role of the C terminus. This could be a reason why cells expressing Ctr1 with deletion, epitope-tagging, or site-directed mutation of either copper-sensing residues or any other residues involved in regulatory function of the C terminus transport excess copper and are sensitive to copper toxicity.
It is also interesting to note recent reports indicating a regulatory function of metal binding to cytosolic domains of other metal transporters. The x-ray structure of the Mg2+ importer CorA revealed that the cytoplasmic domain contains magnesium binding sites between monomers that do not appear to be necessary for a transport function (46, 47). The structures of another magnesium transporter, MgtA, solved with and without magnesium binding to the cytoplasmic domains indeed showed the Mg2+-dependent movement of the cytosolic domains, which reorganizes the transmembrane helices (48). Hence, MgtA-mediated magnesium transport could be controlled by intracellular magnesium concentration. Secondly, YiiP zinc exporter of E. coli is a homodimer that binds with four zinc ions per subunit (49). Among the four, two zinc ions bind in the cytoplasmic domain with a metallochaperone-like fold (49). It was suggested that zinc binding to the cytosolic domain of YiiP is necessary for the formation of a functional dimeric complex, which was proposed as a mechanism by which cells activate this exporter only when cellular zinc levels are in excess (20). Our data presented herein and reported previously (34) suggest that a similar mechanism controls Ctr1-mediated copper uptake in yeast.
We could not observe significant changes in cell surface expression of Ctr1 in cells cultured in media supplemented with excess copper, which is contradictory to other reports (24, 25). The underlying reasons of these discrepancies among experimental data are not known. However, we observed that Ctr1 in lysates prepared from cells cultured in copper-supplemented media migrates higher than trimer species on SDS-PAGE if samples are not thoroughly denatured (data not shown). Liu et al. (25) also commented on these copper-dependent high molecular weight complexes. This could reduce Ctr1 species corresponding to trimer, dimer, or monomer that are detected by immunoblotting, resulting in a misinterpretation of these data as a down-regulation of Ctr1. It is also necessary to note that, whereas we fused triple HA epitopes at the N terminus of Ctr1, others used Ctr1 fused with GFP or c-Myc epitope at the C terminus (24, 25). Because this domain undergoes conformational changes when cells are cultured with copper supplementation (34), the detection efficiency of GFP or c-Myc epitope tagged at the C terminus under incomplete protein denaturation conditions could be different depending on copper levels in culture media. Hence, the post-translational regulations determined by monitoring expression levels and subcellular location of Ctr1 tagged with GFP or c-Myc at the C terminus might reflect copper-dependent conformational changes in Ctr1.
Our genetic analyses did not confirm any significant role for the endocytosis and degradation pathway in homeostatic copper acquisition. We predicted that cells defective in Rsp5 ubiquitin ligase (21, 35) or molecular factors for endocytosis such as End4 and Chc1 (36, 37) will be hypersensitive to copper if these factors are necessary for the down-regulation of Ctr1 as suggested previously (25). Second, if copper hypersensitivity of cells expressing Ctr1 with deletion or site-directed mutations at its C terminus is attributed to defects in copper-responsive down-regulation of Ctr1, cells defective in endocytosis pathway would manifest a similar sensitivity when wild type Ctr1 or Ctr1 with C-terminal deletion is expressed in these cells. However, in contrast to this prediction, End4- or Chc1-defective cells are not more sensitive to copper when compared with their isogenic wild type cells, and these strains expressing Ctr1 with C-terminal deletion are hypersensitive to copper as compared with cells where the full-length Ctr1 is expressed. Site-directed mutation of Lys-340 and Lys-345 to arginine, which abolishes copper-induced internalization of Ctr1 (25), did not lead to a significant increase in copper sensitivity to cells expressing this mutant allele. Collectively, our data suggest that ubiquitination and endocytosis of Ctr1, if any, play a minor role in the prevention of excess uptake of copper as compared with C terminus-mediated control of Ctr1 activity.
Ctr1 family members share similar structural features, including three transmembrane domains and several potential copper binding residues (3, 4). Site-directed mutagenesis of conserved methionine residues of Ctr1 in yeast and humans suggest that the mechanisms of action among Ctr1 family members are similar (30). However, the C-terminal cytosolic tail is distinct in length and amino acid sequence. For example, the predicted C terminus of yeast Ctr1 and human Ctr1 encompasses the last 125 and 12 amino acid residues of each polypeptide, respectively (3, 30). Despite this difference in length, many of the family members possess predicted metal binding residues at the C terminus. For instance, the C-terminal cytosolic tail of yeast Ctr1 contains five cysteine residues. Our mutational analysis of these residues revealed that they are not necessary for high affinity copper uptake but play a vital role in the prevention of copper toxicity when cells grow in media containing excess copper. The presence of potential metal binding residues at the C terminus of other Ctr1 family members suggests that the regulatory roles of the C terminus characterized in yeast may be conserved in this family of proteins. We are currently testing this hypothesis.
Several amino acid residues within the C terminus play critical roles in the control of Ctr1-mediated copper transport. Our in vivo data suggest that cysteine residues in the C terminus bind with copper, which could be a copper-sensing mechanism necessary for subsequent inactivation of Ctr1. However, the roles for the dileucine motif located within the C terminus in the prevention of toxic copper transport are not clear yet. Although a sorting signal in mammalian cells that contains the dileucine motif has been relatively well characterized (31), function of this motif in yeast is not elucidated yet. Given that turnover rates and subcellular distribution of Ctr1 are not significantly altered by copper, copper sensitivity in cells expressing Ctr1 where the dileucine residues are substituted to alanine is unlikely attributed to the role of this motif in endocytosis and degradation.
Taken together, our data provide an initial glimpse into how copper regulates the activities of its own transporter. Characterization of high resolution structures of Ctr1 would provide direct evidences of structural changes that are associated with copper sensing by the C terminus followed by inactivation of Ctr1.
Acknowledgments
We thank Drs. Bruno André, Caroline Philpott, and David Eide for providing the rsp5/npi1, end4-1 and chc1-1 yeast strains and isogenic wild type strains, respectively. We also thank Lee laboratory members for technical assistance and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK79209 (to J. L.) and P20-RR-17675 (to the Nebraska Redox Biology Center). This work was also supported by a Nutricia Foundation Fellowship (to D. S.), and funds provided through the Hatch Act in the University of Nebraska Agricultural Research Division. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SC, synthetic complete; BCS, bathocuproine disulfonic acid; GFP, green fluorescent protein; HA, hemagglutinin; PBS, phosphate-buffered saline; SOD1, Cu,Zn superoxide dismutase.
References
- 1.Linder, M. C. (1991) Biochemistry of Copper, Plenum Press, New York
- 2.Uauy, R., Olivares, M., and Gonzalez, M. (1998) Am. J. Clin. Nutr. 67 Suppl. 5, 952–959 [DOI] [PubMed] [Google Scholar]
- 3.Lee, J., Adle, D., and Kim, H. (2006) in Molecular Biology of Metal Homeostasis and Detoxification (Tamás, M. J., and Martinoia, E., eds) pp. 1–36, Springer-Verlag, Berlin Heidelberg, Germany
- 4.Kim, B. E., Nevitt, T., and Thiele, D. J. (2008) Nat. Chem. Biol. 4 176–185 [DOI] [PubMed] [Google Scholar]
- 5.Prohaska, J. R. (2000) Nutrition 16 502–504 [DOI] [PubMed] [Google Scholar]
- 6.Gitlin, J. D. (2003) Gastroenterology 125 1868–1877 [DOI] [PubMed] [Google Scholar]
- 7.Lutsenko, S., Barnes, N. L., Bartee, M. Y., and Dmitriev, O. Y. (2007) Physiol. Rev. 87 1011–1046 [DOI] [PubMed] [Google Scholar]
- 8.Halliwell, B., and Gutteridge, J. M. (1990) Methods Enzymol. 186 1–85 [DOI] [PubMed] [Google Scholar]
- 9.Dancis, A., Yuan, D. S., Haile, D., Askwith, C., Eide, D., Moehle, C., Kaplan, J., and Klausner, R. D. (1994) Cell 76 393–402 [DOI] [PubMed] [Google Scholar]
- 10.Knight, S. A., Labbe, S., Kwon, L. F., Kosman, D. J., and Thiele, D. J. (1996) Genes Dev. 10 1917–1929 [DOI] [PubMed] [Google Scholar]
- 11.Rees, E. M., Lee, J., and Thiele, D. J. (2004) J. Biol. Chem. 279 54221–54229 [DOI] [PubMed] [Google Scholar]
- 12.Hassett, R., and Kosman, D. J. (1995) J. Biol. Chem. 270 128–134 [DOI] [PubMed] [Google Scholar]
- 13.Georgatsou, E., Mavrogiannis, L. A., Fragiadakis, G. S., and Alexandraki, D. (1997) J. Biol. Chem. 272 13786–13792 [DOI] [PubMed] [Google Scholar]
- 14.Lee, J., Pena, M. M., Nose, Y., and Thiele, D. J. (2002) J. Biol. Chem. 277 4380–4387 [DOI] [PubMed] [Google Scholar]
- 15.Pufahl, R. A., Singer, C. P., Peariso, K. L., Lin, S.-J., Schmidt, P. J., Fahrni, C. J., Culotta, V. C., Penner-Hahn, J. E., and O'Halloran, T. V. (1997) Science 278 853–856 [DOI] [PubMed] [Google Scholar]
- 16.Cobine, P. A., Pierrel, F., and Winge, D. R. (2006) Biochim. Biophys. Acta 1763 759–772 [DOI] [PubMed] [Google Scholar]
- 17.Culotta, V. C., Klomp, L. W. J., Strain, J., Casareno, R. L. B., Krems, B., and Gitlin, J. D. (1997) J. Biol. Chem. 272 23469–23472 [DOI] [PubMed] [Google Scholar]
- 18.Klaassen, C. D., Liu, J., and Choudhuri, S. (1999) Annu. Rev. Pharmacol. Toxicol. 39 267–294 [DOI] [PubMed] [Google Scholar]
- 19.Cobbett, C., and Goldsbrough, P. (2002) Annu. Rev. Plant Biol. 53 159–182 [DOI] [PubMed] [Google Scholar]
- 20.Deleted in proof
- 21.Deleted in proof
- 22.Yamaguchi-Iwai, Y., Serpe, M., Haile, D., Yang, W., Kosman, D. J., Klausner, R. D., and Dancis, A. (1997) J. Biol. Chem. 272 17711–17718 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, P., and Thiele, D. J. (1993) Biofactors 4 105–115 [PubMed] [Google Scholar]
- 24.Ooi, C. E., Rabinovich, E., Dancis, A., Bonifacino, J. S., and Klausner, R. D. (1996) EMBO J. 15 3515–3523 [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Sitaram, A., and Burd, C. G. (2007) Traffic 8 1375–1384 [DOI] [PubMed] [Google Scholar]
- 26.Petris, M. J., Smith, K., Lee, J., and Thiele, D. J. (2003) J. Biol. Chem. 278 9639–9646 [DOI] [PubMed] [Google Scholar]
- 27.Guo, Y., Smith, K., Lee, J., Thiele, D. J., and Petris, M. J. (2004) J. Biol. Chem. 279 17428–17433 [DOI] [PubMed] [Google Scholar]
- 28.Klomp, A. E., Tops, B. B., Van Denberg, I. E., Berger, R., and Klomp, L. W. (2002) Biochem. J. 364 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisses, J. F., Chi, Y., and Kaplan, J. H. (2005) J. Biol. Chem. 280 9635–9639 [DOI] [PubMed] [Google Scholar]
- 30.Puig, S., Lee, J., Lau, M., and Thiele, D. J. (2002) J. Biol. Chem. 277 26021–26030 [DOI] [PubMed] [Google Scholar]
- 31.Sandoval, I. V., and Bakke, O. (1994) Trends Cell Biol. 4 292–297 [DOI] [PubMed] [Google Scholar]
- 32.Dancis, A., Haile, D., Yuan, D. S., and Klausner, R. D. (1994) J. Biol. Chem. 269 25660–25667 [PubMed] [Google Scholar]
- 33.Aller, S. G., and Unger, V. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinani, D., Adle, D. J., Kim, H., and Lee, J. (2007) J. Biol. Chem. 282 26775–26785 [DOI] [PubMed] [Google Scholar]
- 35.Hein, C., Springael, J. Y., Volland, C., Haguenauer-Tsapis, R., and André, B. (1995) Mol. Microbiol. 18 77–87 [DOI] [PubMed] [Google Scholar]
- 36.Raths, S., Rohrer, J., Crausaz, F., and Riezman, H. (1993) J. Cell Biol. 120 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, P. K., Davis, N. G., Sprague, G. F., and Payne, G. S. (1993) J. Cell Biol. 123 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mumberg, D., Muller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 39.Overton, M. C., and Blumer, K. J. (2002) J. Biol. Chem. 277 41463–41472 [DOI] [PubMed] [Google Scholar]
- 40.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman J. G., Smith, J. A., and Struhl, K. (1987) Current Protocol in Molecular Biology, John Wiley & Sons, Inc., New York
- 41.Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995) Yeast 11 355–360 [DOI] [PubMed] [Google Scholar]
- 42.Katzmann, D. J., Epping, E. A., and Moye-Rowley, W. S. (1999) Mol. Cell. Biol. 19 2998–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nebot, C., Moutet, M., Huet, P., Xu, J. Z., Yadan, J. C., and Chaudiere, J. (1993) Anal. Biochem. 214 442–451 [DOI] [PubMed] [Google Scholar]
- 44.Oyanagui, Y. (1984) Anal. Biochem. 142 290–296 [DOI] [PubMed] [Google Scholar]
- 45.Xiao, Z., Loughlin, F., George, G. N., Howlett, G. J., and Wedd, A. G. (2004) J. Am. Chem. Soc. 126 3081–3090 [DOI] [PubMed] [Google Scholar]
- 46.Lunin, V. V., Dobrovetsky, E., Khutoreskaya, G., Zhang, R., Joachimiak, A., Doyle, D. A., Bochkarev, A., Maguire, M. E., Edwards, A. M., and Koth, C. M. (2006) Nature 440 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eshaghi, S., Niegowski, D., Kohl, A., Martinez Molina, D., Lesley, S. A., and Nordlund, P. (2007) Science 313 354–357 [DOI] [PubMed] [Google Scholar]
- 48.Hattori, M., Tanaka, Y., Fukai, S., Ishitani, R., and Nureki, O. (2007) Nature 448 1072–1075 [DOI] [PubMed] [Google Scholar]
- 49.Lu, M., and Fu, D. (2007) Science 317 1746–1748 [DOI] [PubMed] [Google Scholar]
- 50.Nies, D. H. (2007) Science 317 1695–1696 [DOI] [PubMed] [Google Scholar]
- 51.Horák, J. (2003) Biochim. Biophys. Acta 1614 139–155 [DOI] [PubMed] [Google Scholar]