Abstract
T-cell receptor-β (TCRβ) genes naturally acquire premature termination codons (PTCs) as a result of programmed gene rearrangements. PTC-bearing TCRβ transcripts are dramatically down-regulated to protect T-cells from the deleterious effects of the truncated proteins that would otherwise be produced. Here we provide evidence that two responses collaborate to elicit this dramatic down-regulation. One is rapid mRNA decay triggered by the nonsense-mediated decay (NMD) RNA surveillance pathway. We demonstrate that this occurs in highly purified nuclei lacking detectable levels of three different cytoplasmic markers, but containing an outer nuclear membrane marker, suggesting that decay occurs either in the nucleoplasm or at the outer nuclear membrane. The second response is a dramatic partitioning shift in the nuclear fraction-to-cytoplasmic fraction mRNA ratio that results in few TCRβ transcripts escaping to the cytoplasmic fraction of cells. Analysis of TCRβ mRNA kinetics after either transcriptional repression or induction suggested that this nonsense codon-induced partitioning shift (NIPS) response is not the result of cytoplasmic NMD but instead reflects retention of PTC+ TCRβ mRNA in the nuclear fraction of cells. We identified TCRβ sequences crucial for NIPS but found that NIPS is not exclusively a property of TCRβ transcripts, and we identified non-TCRβ sequences that elicit NIPS. RNA interference experiments indicated that NIPS depends on the NMD factors UPF1 and eIF4AIII but not the NMD factor UPF3B. We propose that NIPS collaborates with NMD to retain and degrade a subset of PTC+ transcripts at the outer nuclear membrane and/or within the nucleoplasm.
Approximately one-third of inherited genetic disorders are caused by nonsense or frameshift mutations, both of which generate premature termination codons (PTCs)5 (1). PTCs also arise from biosynthetic errors, including mistakes during both transcription and mRNA splicing (1, 2). PTC-bearing aberrant mRNAs are typically rapidly degraded by a quality control mechanism called nonsense-mediated mRNA decay (NMD) (3–6). By rapidly degrading these aberrant mRNAs, NMD reduces the translation of C-terminally truncated proteins encoded by PTC-bearing transcripts. This is important, as C-terminally truncated proteins often possess dominant-negative or deleterious gain-of-function activity (7, 8). The NMD response is conserved across the phylogentic scale and requires trans-acting factors that have been defined in Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, and mammals (3, 9, 10). Mammalian NMD requires recognition of the stop codon by the translation machinery (11–14) and also typically requires a spliceable intron downstream of the stop codon (15–17). The latter requirement derives from the fact that the splicing machinery deposits a dynamic assembly of proteins, known as the exon junction complex (EJC), which acts as a second signal for NMD (18–23).
Transcripts encoded by the T-cell receptor (TCR) and immunoglobulin (Ig) genes are a unique class of NMD substrates because they acquire PTCs at an extremely high frequency as a result of error-prone programmed gene rearrangements that increase immune receptor diversity (24). This frequent acquisition of PTCs may have led to strong selection pressure to efficiently eliminate PTC-bearing TCRβ transcripts. Consistent with this hypothesis, we previously showed that TCRβ transcripts harboring PTCs are down-regulated more dramatically (to ∼1–5% of normal levels) than are transcripts from most nonrearranging genes that have been tested (to ∼10–30% of normal levels) (3, 24, 25). We have previously shown that this robust down-regulation is neither specific to T-cells, nor does it require a TCRβ promoter; rather it is elicited by TCRβ sequences that promote efficient RNA splicing (25, 26). Recent evidence from mice harboring NMD-deficient T-cells with or without PTC-bearing TCRβ genes indicates that the dramatic down-regulation of aberrant TCR transcripts is essential for the survival of T-cells (27).
Here, we examined the underlying mechanism responsible for the dramatic down-regulation of aberrant PTC-bearing TCRβ transcripts. We provide evidence that PTCs elicit two responses that collaborate to dramatically reduce the level of PTC-containing TCRβ transcripts in the cytoplasmic fraction of cells. The first response is rapid decay of TCRβ transcripts in the nuclear fraction of cells. This is consistent with previous findings from several other groups who found that the introduction of PTCs in mammalian transcripts often triggers their down-regulation (at the steady-state level) in the nuclear fraction of cells (28–32). Our mRNA half-life analysis provided direct evidence that it is the result of more rapid mRNA decay. Analysis of three cytoplasmic markers indicated that the procedure we used for nuclear isolation yielded nuclei with <1% contamination of these particular markers. This, combined with our finding that the isolated nuclei retained an outer nuclear membrane marker, indicated that TCRβ NMD most likely occurs either at the outer nuclear membrane or in the nucleus itself. The second response is a dramatic partitioning shift in the nuclear fraction-to-cytoplasmic fraction mRNA ratio that results in few TCRβ transcripts escaping to the cytoplasmic fraction of cells. This nonsense codon-induced partitioning shift (NIPS) appeared to not be the result of cytoplasmic NMD, as PTCs did not elicit more rapid cytoplasmic mRNA decay when measured by three independent approaches. Instead, our analysis suggested that NIPS is the result of TCRβ mRNA retention in the nuclear fraction of cells. To begin to understand the underlying mechanism for NIPS, we defined cis-acting sequences and trans-acting factors required for it. We also examined the generality of the NIPS response and identified an instance when NIPS and nucleus-associated NMD are separable. Together, our data suggested that NIPS collaborates with nucleus-associated NMD to dramatically reduce the levels of PTC-bearing transcripts in the cytoplasmic fraction of cells. We propose that NIPS serves as a quality control mechanism that reduces the translation of truncated proteins that would otherwise cause deleterious gain-of-function or dominant-negative effects in mammalian cells.
EXPERIMENTAL PROCEDURES
Plasmids—Plasmid constructs A– (β-290) and A+ (PTC at codon 68; β-367) are pIF and pFS1, respectively, as described in Carter et al. (33). Constructs A+ (PTC at codon 51; β-583), Am (codon 51; β-584), and Am (codon 68; β-368) are constructs A′, B′, and D′, respectively in Wang et al. (34). Constructs B– (β-901) and B+ (β-902) were generated in two steps. First, a 3.2-kb SalI/BamHI fragment containing the entire TCRβ gene from construct A was subcloned into the XhoI and BamHI sites of the pREP9Δ B→ X vector (EV-137d; obtained from Dr. Alan Cochrane) to generate the plasmid β-841. The tet promoter amplified by PCR from B1-CMVT (EV-137a) using the primers MDA-1009 (5′-GGTGGCGGCCGCTCTAGAACTAG-3′) and MDA-1010 (5′-GACTCGGGGGGGGGGGGCTAGCGGGCGAATTGGGTACCG-3′) was then inserted into the NheI and NotI sites of β-841 to generate construct B–. Construct B+ (β-902) is identical to B– except it contains a nonsense mutation (UAG) at codon 51 in the VDJβ exon. Constructs C– (β-617) and C+ (β-1001) are described in Wang et al. (13). Constructs A+ (β-1025; harbors a PTC at codon 150, which is in the Cβ2.1 exon), D– (β-780), D+ (β-1026), E– (β-1116), E+ (β-1117), F– (β-974), and F+ (β-1038) are described in Chan et al. (35). Constructs G– (β-955) and G+ (β-1037) are described in Gudikote et al. (26). The PTC– (G-266) and PTC+ (G-267) versions of TPI are constructs O– and O+, respectively, in Gudikote et al. (26). The PTC– (G-435) and PTC+ (G-436) versions of human β-globin are a generous gift from Dr. Kulozik, University of Heidelberg, Germany.
Cell Culture, Transfection, and RNAi—HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. HeLa cells cultured to 50–70% confluency in 10-cm plates were transiently transfected with plasmid DNA (1–2 μg) using Lipofectamine (Invitrogen) according to the manufacturer's instructions. To deplete UPF1 and eIF4AIII, HeLa cells were transiently transfected with a UPF1- or eIF4AIII-specific siRNAs, respectively (Ambion), for 48 h as described previously (20, 23, 36). To deplete UPF3B, stable clones expressing UPF3B shRNA were generated as described in Chan et al. (35). UPF3B expression was further reduced in these cell clones by transiently transfecting 1 μg of shRNA expression plasmid (G-479) against UPF3B. A firefly luciferase-specific siRNA (Ambion) was used as a negative control (35). To generate stable cell lines expressing constructs A– and A+, HeLa cells cultured to 50% confluency in 60-mm plates were transfected with 1 μg of plasmid using Lipofectamine (Invitrogen). Clones resistant to G-418 antibiotic were selected using 700 μg/ml G-418.
RNA Isolation and Analysis—HeLa cells were resuspended in a buffer containing the detergent Nonidet P-40 (0.15 m NaCl, 0.6% Nonidet P-40, 10 mm Tris-HCl, 0.1 mm EDTA) and incubated for 10 min at 4 °C. The nuclei were pelleted by centrifugation at 1500 × g for 2 min, and the RNA in the supernatant (the cytoplasmic fraction) was purified by centrifugation over a 5.7 m CsCl cushion in guanidinium isothiocyanate lysis buffer, as described previously (37). The nuclear pellet was resuspended in Nonidet P-40 buffer containing 0.5% sodium deoxycholate, pelleted by centrifugation at 4500 × g for 2 min, and the RNA was purified from the nuclear pellet by centrifugation over a 5.7 m CsCl cushion in the same manner as the cytoplasmic fraction. RNase protection analysis was carried out on 5 μg of RNA using either probe a (VDJβ exon), probe b (Lβ exon) (26), probe c (71 nucleotides of the 3′ end of the L exon, the entire VDJ exon, and 60 nucleotides of the 5′ end of the Cβ2.1 exon), or probe d (probe j in Gudikote et al. (26)). The β-globin ribo-probe (probe e) was prepared as described in Wang et al. (13). Northern blotting analysis was carried out as described previously (38). Quantification of RNA levels was determined an Instant Imager (Packard Instrumentation Co.) or using a Storm PhosphorImager (Applied Biosystems). Real-time PCR analysis for analysis of UPF1, UPF3B, and eIF4AIII mRNA levels was performed as described previously (39).
RESULTS
PTCs Elicit Both Nuclear Fraction mRNA Decay and a Shift in Nuclear-to-Cytoplasmic Fraction mRNA Ratio—We previously demonstrated that PTCs result in the down-regulation (decrease in steady-state mRNA level) of TCRβ transcripts in the nuclear fraction of mammalian cells (15). To determine whether this down-regulation is the result of more rapid TCRβ mRNA decay in the nuclear fraction, we examined the half-life of PTC-containing (PTC+) and PTC-lacking (PTC–) TCRβ transcripts in the nuclear and cytoplasmic fractions of HeLa cells. Nuclear and cytoplasmic RNA was prepared as follows. The plasma membrane of HeLa cells was lysed by incubating cells in a buffer containing the detergent Nonidet P-40, and the nuclei freed by this procedure were pelleted. The cytoplasmic RNA fraction was made by centrifugation of the resulting supernatant over a cesium chloride cushion followed by extraction (see “Experimental Procedures” for more details). This fraction contained most of the endoplasmic reticulum (ER) in the cells, based on Western blot analysis with an antibody against calnexin, an abundant ER-associated protein (supplemental Fig. S1B). To obtain highly purified nuclei, the nuclear pellet was resuspended in a buffer containing Nonidet P-40 and the more stringent detergent sodium deoxycholate (DOC). The “washed” nuclei were gently centrifuged, and the nuclear pellet was resuspended and centrifuged over a cesium chloride cushion. This nuclear RNA fraction preparation was gauged to be highly pure, based on four criteria. First, 45 S and 32 S rRNA precursor transcripts were in the nuclear fraction but were undetectable in the cytoplasmic fraction (supplemental Fig. S1A). Second, the transcriptional regulator Sin3A, a nuclear marker, was present in the nuclear fraction, but it was not detectable in the cytoplasmic fraction (supplemental Fig. S1B). Third, the outer nuclear membrane marker Nesprin-1 was present in the nuclear fraction (supplemental Fig. S1C). Fourth, the nuclear fraction had undetectable levels of three cytoplasmic markers: calnexin, RCK, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 1B). The ER marker protein, calnexin, was present in crude nuclei (before DOC wash), but it was not detectable in highly purified nuclei (after a DOC wash) (Fig. 1B, –D and +D nuclei, respectively). Together with the finding that Nesprin-1 is present in DOC-washed nuclei, this indicates that DOC removes most of the ER but not the outer nuclear membrane. RCK is known to be present at high levels in P-bodies, which are cytoplasmic foci in which at least a subset of NMD is known to occur (40, 41). Like calnexin, RCK was present in the cytoplasmic and crude nuclei fractions, but not in the highly purified nuclei fraction (Fig. 1B). The absence of detectable RCK, calnexin, and GAPDH after the DOC wash indicates that the nuclear fraction preparation has negligible levels of cytoplasmic contamination (less than 1%, based on comparison of the nuclear fraction with dilutions of the cytoplasmic fraction).
FIGURE 1.
PTCs elicit nuclear fraction decay and NIPS. A, schematic diagram of TCRβ construct A, with or without a PTC (±) in the VDJ exon (codon 51 or 68), driven by the β-actin promoter (black box). Probe a (denoted by lowercase letter below the diagram) protects 72 nucleotides of mRNA. B, purity of the nuclear and cytoplasmic fractions from HeLa cells prepared as described under “Experimental Procedures,” as assessed by Western blotting. Equal cell volumes were loaded, or portions of the cell volume, as indicated. Calnexin, RCK, and GAPDH are cytoplasmic markers. β-Actin was used as a loading control. –D, before DOC wash; +D, after DOC wash. C and D, RNase protection analysis of the nuclear (C) and cytoplasmic (D) fraction RNA (5 μg) from HeLa cell lines stably expressing PTC– and PTC+ versions of construct A. Transcription was inhibited by adding actinomycin D (5 μg/ml), and nuclear and cytoplasmic fraction RNA was prepared at the indicated time points. TCRβ mRNA levels, measured using probe a, were normalized for loading against the level of endogenous β-actin. PTC+ TCRβ mRNA levels are relative to PTC– TCRβ mRNA levels, which are set to 100%. Graphs represent the average of at least three independent experiments. E, RNase protection analysis of nuclear fraction (N) and cytoplasmic fraction (C) RNA (5 μg) from HeLa cells transiently transfected with construct A harboring the nonsense mutations (A+) and missense mutations (Am) at the codons indicated, using probe a. TCRβ mRNA levels were normalized by measuring the level of human β-globin expressed from a vector cotransfected as a control for transfection efficiency. Similar results were obtained in at least two independent experiments.
We first examined the half-lives of PTC– and PTC+ TCRβ mRNA in nuclear and cytoplasmic fractions prepared as described above using the transcriptional inhibitor actinomycin D. PTC– and PTC+ TCRβ constructs (Fig. 1A) were stably transfected into HeLa cells; the cells were incubated with actinomycin D for various lengths of time; nuclear and cytoplasmic RNA was isolated, and the level of TCRβ mRNA was determined by ribonuclease protection analysis. This analysis showed that, compared with their PTC– counterparts, PTC+ TCRβ transcripts decreased in level more rapidly in the nuclear fraction (Fig. 1C). In contrast, the stability of both PTC+ and PTC– transcripts was virtually indistinguishable in the cytoplasmic fraction (Fig. 1D). These results suggested that PTCs cause TCRβ transcripts to selectively undergo NMD in the nuclear fraction. Further evidence for nuclear-fraction decay was the finding that PTCs at two different positions decreased the steady-state level of TCRβ mRNA in the nuclear fraction (Fig. 1E). We conclude from these results that PTCs cause TCRβ mRNA to decay in the nuclear fraction but not in the cytoplasmic fraction.
As a second approach to measure the half-lives of PTC+ and PTC– TCRβ mRNA in the nuclear and cytoplasmic fractions, we used the tetracycline (tet)-regulated promoter system. We generated PTC– and PTC+ TCRβ constructs driven by the tet promoter (supplemental Fig. S2A) and transiently transfected them, along with the tet promoter activator tTA, into HeLa cells. To measure the rate of mRNA decay, TCRβ transcription was blocked for different lengths of time by adding tetracycline to the medium. Analysis of nuclear and cytoplasmic fraction RNA from these cells indicated that TCRβ transcripts derived from the tet promoter exhibited rapid decay in response to a PTC in the nuclear fraction but not in the cytoplasmic fraction (supplemental Fig. S2, B and C). These results provided additional evidence that PTC-containing TCRβ transcripts undergo NMD in the nuclear fraction and not in the cytoplasmic fraction. Whether this decay occurs in the nucleoplasm, the outer nuclear membrane, or another site copurifying with the nucleus is addressed under the “Discussion.”
If nuclear fraction NMD was the only mechanism acting to decrease the level of PTC+ TCRβ transcripts, then PTCs should decrease TCRβ mRNA level by the same magnitude in both the nuclear and cytoplasmic fractions. However, contrary to this prediction, we found that PTCs decreased the level of TCRβ transcripts by a much greater magnitude in the cytoplasmic fraction than in the nuclear fraction (Fig. 1E). Quantification of the nuclear-to-cytoplasmic fraction mRNA ratio revealed that it was dramatically higher for PTC+ transcripts than it was for PTC– transcripts (∼12.5- and ∼36-fold higher for PTCs at codons 51 and 68, respectively). Comparison of highly purified nuclei (generated by washing with DOC) with the DOC wash itself indicated that this nonsense codon-induced partitioning shift (NIPS) response primarily resulted from the accumulation of PTC+ TCRβ transcripts in the highly purified nuclear fraction (supplemental Fig. S2D). However, we also observed that PTCs elicited a modest increase in the nuclear wash/cytoplasmic fraction ratio (supplemental Fig. S2D), indicating that a minor proportion of PTC+ TCRβ transcripts are also retained at a subcellular site removed by the DOC wash. To determine whether NIPS is a nonsense codon-specific response, we determined the nuclear-to-cytoplasmic ratio for TCRβ transcripts harboring missense mutations at the same codons as the nonsense mutations we tested (codon 51 and codon 68). We found that the nuclear-to-cytoplasmic ratio for transcripts harboring missense mutations was comparable with that of wild-type transcripts (Fig. 1E).
Evidence That NIPS Is Caused by Nuclear Retention—One explanation for NIPS is that PTCs cause retention or accumulation of TCRβ mRNA in the nuclear fraction of HeLa cells (for simplicity, we will use the term “retention” in this paper). To test this possibility, we performed “approach-to-steady-state” mRNA analysis. In this approach, tet promoter-driven PTC– and PTC+ TCRβ constructs (supplemental Fig. S2A) were transfected into HeLa cells in the presence of a low dose of tetracycline (50 ng/ml) just sufficient to inhibit transcription and optimal for rapid induction upon removal of tetracycline. Approximately 16 h later, transcription was initiated by removing tetracycline from the medium, and the nuclear and cytoplasmic fraction RNA were prepared at different time points. Analysis of nuclear fraction RNA revealed that, as expected, the level of TCRβ transcripts increased after removal of tetracycline (Fig. 2A). The upward slopes for PTC– and PTC+ transcripts in the nuclear fraction were similar, consistent with previous studies showing that introduction of a PTC does not affect the rate of TCRβ transcription (42, 43). In contrast, the cytoplasmic fraction had a much shallower upward slope for PTC+ transcripts than for PTC– transcripts, consistent with inhibited entry into the cytoplasmic fraction (Fig. 2B). This was supported by comparison of the slopes in the nuclear and cytoplasmic fractions. The slope for PTC– transcripts was almost identical in the cytoplasmic and nuclear fractions, indicating that PTC– transcripts are efficiently exported (Fig. 2, A and B). In contrast, PTC+ TCRβ transcripts only minimally increased in level in the cytoplasmic fraction (over a 26-h period) after removal of tetracycline (Fig. 2B). As we found that PTC+ TCRβ transcripts are not degraded in the cytoplasmic fraction (Fig. 1D and supplemental Fig. S2C), we believe that the simplest interpretation of these data is that PTCs inhibit TCRβ mRNA from leaving the nuclear fraction and entering the cytoplasmic fraction.
FIGURE 2.
Kinetic analysis of TCRβ transcripts. A and B, RNase protection analysis, using probe a, of nuclear fraction (A) and cytoplasmic fraction (B) RNA (5 μg) from HeLa cells transiently cotransfected with the tet activator plasmid EV-137d/TTA and construct B (with or without a PTC at codon 51 in the VDJ exon; see supplemental Fig. 2A for schematic). Nuclear and cytoplasmic fraction RNA was prepared at the indicated time points. Similar results were obtained in at least four independent experiments. C, nuclear-to-cytoplasmic (N/C) ratio at the indicated time points, using the data from A and B. Quantification was done as described in Fig. 1E.
This is depicted in an alternative manner by plotting the nuclear-to-cytoplasmic ratio at different time points in Fig. 2C. The nuclear-to-cytoplasmic ratio for PTC– transcripts decreased over time, consistent with rapid export of these transcripts and their accumulation in the cytoplasm. In contrast, the nuclear-to-cytoplasmic ratio for PTC+ transcripts increased over time, indicative of their accumulation in the nuclear fraction. Collectively, these data suggest that PTCs cause the retention or accumulation of TCRβ transcripts in the nucleus-associated fraction of cells. As with nucleus-associated NMD, this retention may occur either in the nucleoplasm or at the outer nuclear membrane (see the “Discussion”). An apparent contradiction with the notion that PTC+ transcripts accumulate in the nuclear fraction is that PTC+ and PTC– transcripts exhibited similar upward slopes in the nuclear fraction after transcriptional induction (Fig. 2A). We suggest two non-mutually exclusive explanations for this. First, the rate of TCRβ transcription may greatly exceed that of TCRβ mRNA export, thereby masking an accumulation of PTC+ TCRβ transcripts in the nuclear fraction. Second, our half-life analysis (described above) indicated that the retained PTC+ TCRβ transcripts are also rapidly degraded in the nuclear fraction, which would be expected to lead to little net change in the nuclear level of newly synthesized TCRβ transcripts.
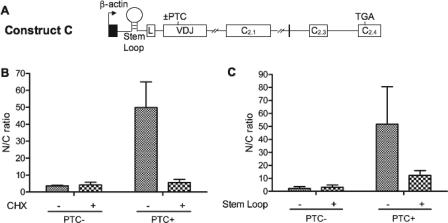
NIPS Depends on Translation—Because we found that NIPS is triggered specifically by nonsense mutations, this predicts that it depends on translation. To test this, we used two independent approaches to inhibit translation: 1) treatment with the protein synthesis inhibitor cycloheximide (CHX) (33), and 2) insertion of a translation-inhibiting stem-loop upstream of the AUG start codon (Fig. 3A). In the first approach, HeLa cells were transiently transfected with PTC– and PTC+ TCRβ constructs and incubated with or without CHX. Nuclear and cytoplasmic fraction RNA was harvested and analyzed by ribonuclease protection analysis. This analysis showed that CHX treatment reduced the nuclear-to-cytoplasmic ratio of PTC+ transcripts to a value similar to that for PTC– transcripts, indicating that CHX reversed the NIPS response (Fig. 3B). Specificity was demonstrated by the observation that CHX did not have a significant effect on the nuclear-to-cytoplasmic ratio of PTC– transcripts.
FIGURE 3.
Evidence that NIPS depends on translation. A, schematic diagram of TCRβ constructs containing a stem-loop 42 nucleotides upstream of the start AUG (with or without a PTC (±) at codon 68 in the VDJ exon). B, RNase protection analysis, using probe a, of nuclear fraction (N) and cytoplasmic fraction (C) RNA (5 μg) from HeLa cells transiently transfected with 1 μg of construct A (with or without a PTC at codon 68 in the VDJ exon) and treated with CHX (100 μg/ml) 6 h before harvesting. C, RNase protection analysis of nuclear fraction (N) and cytoplasmic fraction (C) RNA (5 μg) from HeLa cells transiently transfected with 1 μg of construct A or C (with or without a PTC at codon 68 in the VDJ exon), using probe a. The data in B and C were quantified as described in Fig. 1E. Similar results were obtained in at least three independent experiments.
For the stem-loop translation-inhibition experiments, we introduced a stem-loop previously shown to inhibit translation (13) into the 5′-untranslated region of PTC– and PTC+ TCRβ constructs (Fig. 3A). These two constructs, along with control constructs lacking the stem-loop, were transiently transfected into HeLa cells, and cytoplasmic and nuclear fraction RNA was harvested and analyzed by ribonuclease protection analysis. This analysis demonstrated that the NIPS response of PTC+ transcripts was decreased by the stem-loop (Fig. 3C). Specificity was demonstrated by the finding that the stem-loop did not significantly affect the nuclear-to-cytoplasmic ratio of PTC– transcripts. Although the reversal of NIPS by PTCs was incomplete (the nuclear-to-cytoplasmic ratio of stem-loop PTC+ transcripts was not as low as that for PTC– transcripts), this was expected given that the stem-loop is known to not completely inhibit translation (11, 44). Together, the stem-loop and CHX experiments provide strong evidence that NIPS depends on translation.
NIPS Depends on NMD Factors—We next examined whether NIPS depends on NMD factors. We first determined the role of UPF1, an NMD factor that several groups, including our own, have shown is crucial for TCRβ NMD (35, 45, 46). Using an siRNA previously shown to specifically knock down UPF1 (35, 36), we depleted the level of UPF1 RNA (supplemental Fig. S3A) and found that this selectively decreased the nuclear-to-cytoplasmic ratio of PTC+ transcripts, not PTC– transcripts (Fig. 4, A and B). Although depletion of UPF1 strongly decreased NIPS of PTC+ transcripts (by ∼5-fold), it was not as low as that for PTC– transcripts (Fig. 4, A and B) consistent with the fact that TCRβ NMD is only partially reversed by the degree of UPF1 depletion achieved by RNAi (Fig. 4A) (35, 36).
FIGURE 4.
Depletion of UPF1 and eIF4AIII, but not UPF3B, inhibits NIPS. A, C, and E, RNase protection analysis of nuclear fraction (N) and cytoplasmic fraction (C) RNA (5 μg) from HeLa cells transfected with the indicated short interfering RNAs (siRNA) or shRNAs and construct A (with or without a PTC at codon 68 in the VDJ exon). Quantification was performed as in Fig. 1E. The data in E are representative of at least two independent experiments. B and D, quantification of data obtained from experiments performed as described in A and C, respectively, from at least three independent experiments.
We next examined the role of eIF4AIII, a core component of the EJC necessary for maximal TCRβ NMD (23, 35, 47, 48). We efficiently knocked down eIF4AIII using a previously described siRNA (23, 35, 47, 48) (supplemental Fig. S3B) and found that this modestly, but significantly, decreased the nuclear-to-cytoplasmic ratio for PTC+ transcripts (Fig. 4, C and D), indicating that NIPS at least partially depends on eIF4AIII. The effect was specific, as depletion of eIF4AIII reduced the nuclear-to-cytoplasmic ratio of transcripts harboring PTCs at two independent positions, but it did not significantly affect the nuclear-to-cytoplasmic ratio of the PTC– transcript (data not shown). The modest decrease in the nuclear-to-cytoplasmic ratio of the PTC+ transcripts could be due to either a partial eIF4AIII requirement or insufficient eIF4AIII depletion to observe a strong dependence. We suspect the former, as the level of eIF4AIII protein was strongly reduced (by 90%, data not shown) in response to siRNA treatment.
Finally, we examined whether NIPS requires the EJC factor UPF3B (Upf3x). Although required for the down-regulation of some NMD substrates, we recently reported that strong depletion of this EJC factor has no effect on either PTC+ TCRβ transcripts or a subset of endogenous human NMD substrates, suggesting that a subset of PTC-bearing transcripts are degraded by an alternative UPF3B-independent branch of the NMD pathway (35). To test the role of UPF3B in NIPS, we used a “super RNAi” approach in which UPF3B levels were depleted by stably transfecting a UPF3B shRNA construct into HeLa cells, selecting a cell clone with depleted UPF3B levels, and then transiently transfecting the UPF3B shRNA construct into this cell clone to further reduce the level of UPF3B (35). This approach allowed us to reduce the UPF3B mRNA level to ∼20% of normal (supplemental Fig. S3C). This depletion of UPF3B did not significantly change the nuclear-to-cytoplasmic ratio of transcripts harboring PTCs in two different exons (Fig. 4E, and data not shown). It also did not significantly inhibit TCRβ NMD, in agreement with our previous report (35). We conclude that neither the decay of PTC+ TCRβ transcripts nor their NIPS response is affected by UPF3B depletion, suggesting that PTCs affect the fate of TCRβ transcripts by mechanisms independent of this EJC-associated factor.
NIPS Cis Elements—We previously showed that the robust down-regulation of TCRβ transcripts in response to PTCs requires a region encompassing the VDJ exon and its flanking intronic sequences (25, 26). However, in these past studies, we only measured the level of TCRβ transcripts in total cellular mRNA. Thus we did not determine the cellular compartment in which this TCRβ regulatory region acts to confer robust down-regulation. Our fractionation studies herein established that TCRβ transcripts do not undergo NMD in the cytoplasmic fraction and are only modestly decreased in level in the nuclear fraction (Fig. 1E), suggesting the hypothesis that the TCRβ regulatory region acts by conferring nuclear fraction retention (the NIPS response). To test this hypothesis, we transiently transfected HeLa cells with constructs lacking the VDJ exon and its flanking intronic sequences (Fig. 5A, construct D) and examined whether this abolished the NIPS response. Ribonuclease protection analysis revealed that the nuclear-to-cytoplasmic ratio of PTC+ transcripts derived from this construct was dramatically less than the parental construct and was similar to that of PTC– counterparts (Fig. 5, B and E, construct D). These results indicated that the cis element(s) responsible for NIPS reside within the VDJ exon and its flanking intronic sequences.
FIGURE 5.
TCRβ and TPI sequences that trigger NIPS. A, schematic diagram of TCRβ constructs with or without a PTC (±) at codon 150 in the Cβ2.1 exon. Construct E has intron 1 (rIVS1) and intron 2 (rIVS2) from rabbit β-globin (indicated in boldface); constructs F and G have TPI exons 2 and 4, respectively, in place of the VDJ exon. Probe b (denoted by lowercase letter below the diagram) protects 73 nucleotides of mRNA. Probe c is described under “Experimental Procedures.” B–D, RNase protection analysis using probe b (B and D) or probe c (C) of nuclear fraction (N) and cytoplasmic fraction (C) RNA (5 μg) from HeLa cells transiently transfected with the constructs shown. E, quantification of the nuclear-to-cytoplasmic (N/C) ratio of the transcripts derived from the constructs shown. Quantification was done as described in Fig. 1E. Similar results were obtained in at least three independent experiments.
To determine whether the intronic sequences flanking the VDJ exon are required for NIPS, we replaced TCRβ IVS1 and IVS2 with heterologous introns (Fig. 5A, construct E). Transient transfection followed by ribonuclease protection analysis revealed that transcripts derived from these “intron substitution” constructs maintained the ability to undergo NIPS (Fig. 5C). These data indicated that the intronic sequences in the TCRβ regulatory region are not required for the NIPS response.
To assess whether the VDJ exon is essential for NIPS, we substituted the VDJ exon with exon 2 from the triose-phosphate isomerase (TPI) gene (Fig. 5A, construct F). We hypothesized that TPI exon 2 would confer the ability to undergo NIPS, as we previously showed that this exon drives the robust down-regulation of PTC+ transcripts as well as does the VDJ exon (26). Transient transfection and analysis of the resulting chimeric transcripts by ribonuclease protection assay indicated that transcripts containing TPI exon 2 exhibited a strong NIPS response (Fig. 5, D and E, construct F). To examine the specificity of the response, we replaced the VDJ exon with TPI exon 4 (Fig. 5A, construct G). We previously showed that this exon does not confer the ability to undergo robust down-regulation (26), and thus we hypothesized it also would not confer NIPS. Indeed, we found that, unlike TPI exon 2-containing transcripts, exon 4-containing transcripts did not exhibit NIPS (Fig. 5, D and E, construct G). TPI exons 2 and 4 have greatly different densities of exonic splicing enhancers, which may be responsible for their difference in activity. Collectively, our results indicated that a region containing the VDJ exon and adjacent intron sequences is required for the NIPS response of PTC+ TCRβ transcripts, but that neither the VDJ exon nor the adjacent introns is absolutely essential for this response.
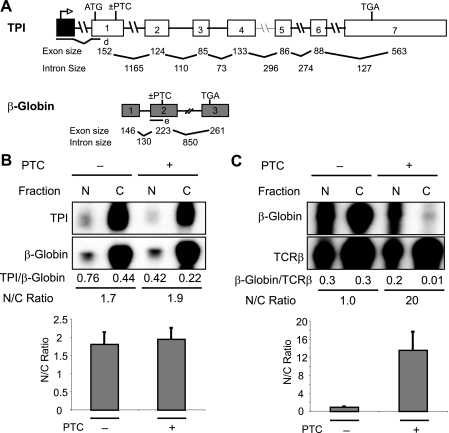
Generality of NIPS—To test the generality of the NIPS response, we examined two mammalian transcripts that have been well studied for their NMD responses: β-globin and TPI (16, 29, 32, 49–51). Both of these transcripts have been shown to be down-regulated by PTCs in the nuclear fraction of cells (29, 32). Analysis of the nuclear and cytoplasmic fraction RNA from HeLa cells transfected with PTC+ and PTC– versions of β-globin and TPI constructs (Fig. 6A) showed that β-globin transcripts underwent the NIPS response, but TPI transcripts did not (Fig. 6, B and C). The ability of β-globin, but not TPI transcripts, to undergo NIPS explains why PTCs down-regulate β-globin mRNA much more strongly (to ∼5% of the normal level, Fig. 6C) than TPI mRNA (to ∼50% of the normal level, Fig. 6B) in the cytoplasm. Collectively, these data suggest that NIPS acts on a specific subset of transcripts to confer reduced mRNA levels in the cytoplasm.
FIGURE 6.
Generality of NIPS. A, schematic diagram of TPI and human β-globin constructs with or without a PTC. B and C, RNase protection analysis of nuclear (N) and cytoplasmic (C) fraction RNA prepared from HeLa cells transiently transfected with either TPI (B) or β-globin (C) constructs with or without a PTC. Human β-globin (B) or TCRβ construct A (C) were cotransfected as normalizing controls for transfection efficiency (the values below the gels indicate values from the gels shown). The histograms indicate the average values from at least three independent experiments, quantified as in Fig. 1E.
DISCUSSION
In this study, we show by mRNA half-life analysis that PTCs cause TCRβ transcripts to undergo rapid decay in a nucleus-associated fraction containing <1% cytoplasmic contamination (Fig. 1, B–D). To our knowledge, the only other transcript experimentally demonstrated by RNA half-life analysis to undergo more rapid decay in the nuclear fraction in response to a PTC is TPI (29). However, it is likely that nuclear-fraction NMD is a general phenomenon, as several other mammalian transcripts harboring PTCs have been shown to exhibit reduced steady-state levels in the nuclear fraction (28–32). We provide evidence that in addition to nuclear-fraction decay, PTCs trigger a novel response not previously reported, i.e. a dramatic increase in the nuclear-to-cytoplasmic mRNA ratio (Fig. 1E). Only nonsense mutations, not missense mutations, triggered this NIPS response (Fig. 1E). This NIPS response was not the result of a measurable increase of mRNA decay in the cytoplasm (Fig. 1, C and D; supplemental Fig. S2, B and C). Instead, NIPS appears to be the result of retention in the nuclear fraction, based on our “approach-to-steady-state” analysis, which showed that the presence of a PTC caused newly synthesized TCRβ transcripts to accumulate over time in the nuclear fraction (Fig. 2, A–C). Although the dramatic increase in nuclear-to-cytoplasmic mRNA ratio that we observed in response to nonsense codons is consistent with their triggering mRNA retention in the nuclear fraction of cells, we cannot exclude that NIPS is instead caused by rapid cytoplasmic decay not measurable by standard methods.
We found that NIPS was inhibited by perturbations in translation, and it depended on the NMD factors UPF1 and eIF4AIII (Figs. 3 and 4). These data, along with our finding that NIPS was elicited specifically by nonsense, not missense, mutations (Fig. 1E), led us to conclude that NIPS is a translation-dependent event that may serve as an RNA surveillance mechanism to recognize deleterious translation signals. Although the NIPS response depended on UPF1 and eIF4AIII, we found it was not perturbed by knockdown of the NMD factor UPF3B (Fig. 4). This insensitivity to UPF3B depletion agrees with our recent discovery that the down-regulation of PTC+ TCRβ transcripts is not affected by depletion of UPF3B (35). It remains for future experiments to determine whether UPF3B independence is an intrinsic feature of the NIPS response. We recently identified two classes of NMD substrates; one class was up-regulated by UPF3B depletion, but the other class was not (35). Analysis of the nuclear-to-cytoplasmic ratio of these two classes of transcripts will begin to address whether UPF3B independence is a general feature of NIPS.
Our mapping experiments suggested that NIPS is conferred by cis elements in the VDJ exon (Fig. 5). This is intriguing, as this exon is uniquely generated by programmed rearrangements (24). However, despite the role of the VDJ exon in increasing nuclear-to-cytoplasmic ratio, TCRβ transcripts are not alone in having this property. First, we found that β-globin transcripts also undergo the NIPS response (Fig. 6). Second, we identified an exon from another gene that was also able to confer the NIPS response (Fig. 5, D and E). It remains for future studies to determine the underlying mechanism responsible for NIPS. We previously showed that one parameter that increases the magnitude of PTC+ mRNA down-regulation is efficient splicing (26), suggesting the possibility that efficient splicing might also trigger the NIPS response. Although an attractive hypothesis, our analysis of a series of mutant TCRβ constructs possessing different splicing efficiencies does not support this hypothesis.6 It also remains for future studies to understand why PTCs elicit NIPS in some transcripts but not in others. Because the NIPS response leads to dramatically reduced concentrations of the mRNA in the cytoplasmic fraction, one possibility is that NIPS is a property of transcripts that, like TCR, are under strong selection pressure to undergo robust down-regulation, e.g. those that commonly acquire PTCs and/or those that, if translated, give rise to truncated proteins with highly deleterious effects.
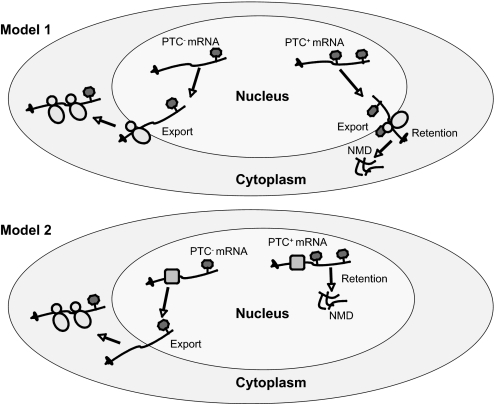
Nucleus-associated mRNA retention and rapid decay are paradoxical, as they are only known to be triggered by a signal (a nonsense codon) read outside of the nucleus. We suggest two solutions as follows: (i) stop codon recognition, mRNA retention, and rapid mRNA decay are triggered by cytoplasmic ribosomes associated with the outer nuclear membrane, or (ii) stop codons are read by nuclear ribosomes or a novel codon scanner in the nucleus, which results in their retention and decay there (Fig. 7). In the first model, ribosomes associated with the outer nuclear membrane proofread transcripts. If a PTC is recognized in this “pioneer round” of translation, the mRNA is retained and rapidly degraded at that site (Fig. 7, model 1). An attractive possibility is that such nuclear membrane-associated ribosomes proofread mRNAs soon after they emerge from the nuclear pore. After this pioneer round of translation, mRNAs deemed as lacking PTCs would be released to undergo bulk translation in other regions of the cytoplasm. The notion that the site of mRNA retention and rapid mRNA decay occurs at the outer nuclear envelope is consistent with our finding that our nuclear purification procedure generated nuclei that retain the outer nuclear membrane (supplemental Fig. S1C). Also consistent with this model is the finding that S. cerevisiae transcripts harboring PTCs are bound to polysomes (52, 53). Additional support for this model comes from the fact that outer nuclear membrane-associated ribosomes are in the cytoplasmic milieu, where translation is known to occur. Although we consider the outer nuclear membrane the most likely site of retention and rapid mRNA decay, it is possible that these events occur in another highly specialized portion of the cytoplasm that cofractionates with the nucleus. For example, these events could occur in a subset of the ER that remains bound to the nucleus after our purification procedure. We think this is unlikely, as our purification procedure generated nuclei containing undetectable levels of the ER marker calnexin (Fig. 1B). Another site where retention and mRNA decay might occur is in P-bodies associated with the nucleus. This is an attractive idea, as P-bodies are cytoplasmic foci harboring high concentrations of mRNA decay factors that have been shown to be a major site of NMD (40, 41). However, our nuclear fraction preparations had an undetectable amount of the P-body marker RCK (less than 1% of the cytoplasmic level, Fig. 1B), indicating that either P-bodies are not involved or that a subset of P-bodies lacking the RCK marker is where these events occur.
FIGURE 7.
Models. Model 1, retention and decay of PTC+ transcripts occurs at the outer nuclear membrane. Nuclear membrane-associated cytoplasmic ribosomes read transcripts after they emerge from the nuclear pore. Transcripts deemed normal during this pioneer round of translation are released into the cytoplasm for bulk translation. Model 2, stop codon recognition, retention, and decay of PTC+ transcripts occur in the nucleoplasm.
The second model posits that there is a ribosome or ribosome-like entity that “proofreads” mRNAs in the nucleus proper. Once this nuclear scanner identifies a PTC-containing mRNA, the aberrant mRNA is held in the nucleus and degraded. Consistent with this model, mutations that generate PTCs elicit a frame-dependent up-regulation of precursor and alternatively spliced TCRβ mRNAs in the nucleus and the nuclear fraction, respectively, of human cell lines (43, 45, 54–56). Further support for this model comes from a study showing that TCRβ transcripts trapped in the nucleus as a result of incubation with nuclear-export blockers are not perturbed in their ability to be down-regulated in response to PTCs (57). The nucleus has at least some of the factors required for translation, including charged tRNAs and some translation factors (58, 59). There is also evidence that a small but significant fraction of translation occurs in nuclei in D. melanogaster (60), Dictyostelium discoideum (61), and human cell lines (62). However, there is also considerable evidence against the notion that translation occurs in the nucleus (63), including the inability of highly purified nuclei to engage in translation (64) and the fact that some translation factors are present at extremely low concentrations in the nucleus (65). Furthermore, there has been evidence that nonsense and frameshift mutations do not elicit nuclear effects as a result of disruption of reading frame, but rather because of disruption of exonic splicing enhancers (66, 67).
To begin to distinguish between these models, it will be necessary to determine the precise cellular location of nucleus-associated mRNA retention and NMD. Imaging techniques will probably be required to resolve this. Although several laboratories have developed imaging techniques to localize specific transcripts (68), no laboratory has been able to identify the location of NMD in mammalian cells. New, more advanced techniques will probably be needed to address this, as the current imaging technology cannot detect most processed mRNAs after they have left the site of transcription. Regardless of the location of nucleus-associated retention and NMD, it will be interesting in the future to determine whether the retention of PTC-bearing transcripts in this location makes these transcripts more susceptible to decay in that compartment. An alternative possibility is that NIPS is a fail-safe mechanism that serves to retain most of the PTC+ transcripts that escape decay.
If indeed PTC+ transcripts are selectively retained in the nuclear fraction of cells, this complicates the interpretation of studies in which only steady-state mRNA levels are examined. For example, if the amount of transcript retained in the nuclear fraction equals the amount that is degraded, retention will mask nuclear fraction NMD. Also, because nuclear fraction retention greatly reduces the steady-state level of PTC+ transcripts in the cytoplasmic fraction, its effects can be misinterpreted as cytoplasmic NMD. In summary, our study demonstrates that nonsense codons reduce the cytoplasmic levels of PTC-containing TCRβ mRNA by eliciting two responses, nuclear fraction NMD and NIPS. We propose that these two responses collaborate to almost completely block the entry of aberrant mRNAs into the translating pool of mRNAs in the cytoplasm. Future studies will be required to determine why only a subset of transcripts exhibits the NIPS response and the precise mechanism responsible for it.
Supplementary Material
Acknowledgments
We thank Jade Clement and John Hamilton for their contributions. We also thank Dr. Alan Cochrane (McGill University) for generously providing tet plasmids and Pierre McCrea and Humam Kadara for antisera.
This work was supported, in whole or in part, by National Institutes of Health Grant GM058595. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and additional references.
Footnotes
The abbreviations used are: PTC, premature termination codon; TCRβ, T-cell receptor-β; NMD, nonsense-mediated decay; EJC, exon junction complex; NIPS, nonsense codon-induced partitioning shift; RNAi, RNA interference; ER, endoplasmic reticulum; DOC, deoxycholate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CHX, cycloheximide; shRNA, short hairpin RNA; TPI, triose-phosphate isomerase; tet, tetracycline.
A. D. Bhalla, J. P. Gudikote, and M. F. Wilkinson, unpublished observations.
References
- 1.Culbertson, M. R. (1999) Trends Genet. 15 74–80 [DOI] [PubMed] [Google Scholar]
- 2.Mendell, J. T., and Dietz, H. C. (2001) Cell 107 411–414 [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y. F., Imam, J. S., and Wilkinson, M. F. (2007) Annu. Rev. Biochem. 76 51–74 [DOI] [PubMed] [Google Scholar]
- 4.Hentze, M. W., and Kulozik, A. E. (1999) Cell 96 307–310 [DOI] [PubMed] [Google Scholar]
- 5.Hilleren, P., and Parker, R. (1999) RNA (N. Y.) 5 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner, E., and Lykke-Andersen, J. (2002) J. Cell Sci. 115 3033–3038 [DOI] [PubMed] [Google Scholar]
- 7.Schell, T., Kulozik, A. E., and Hentze, M. W. (2002) Genome Biol. 3 REVIEWS1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson, M. F. (2005) Trends Genet. 21 143–148 [DOI] [PubMed] [Google Scholar]
- 9.Jacobson, A., and Peltz, S. W. (2000) in Translational Control of Gene Expression (Sonenberg, N., Hershey, J. W. B., and Mathews, M. B., eds) pp. 827–897, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 10.Wilusz, C. J., Wang, W., and Peltz, S. W. (2001) Genes Dev. 15 2781–2785 [DOI] [PubMed] [Google Scholar]
- 11.Belgrader, P., Cheng, J., and Maquat, L. E. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 482–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, S., Leonard, D., and Wilkinson, M. F. (1997) J. Exp. Med. 185 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, J., Vock, V. M., Li, S., Olivas, O. R., and Wilkinson, M. F. (2002) J. Biol. Chem. 277 18489–18493 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, J., and Maquat, L. E. (1997) EMBO J. 16 826–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter, M. S., Li, S., and Wilkinson, M. F. (1996) EMBO J. 15 5965–5975 [PMC free article] [PubMed] [Google Scholar]
- 16.Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M. W., and Kulozik, A. E. (1998) EMBO J. 17 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, J., Sun, X., Qian, Y., LaDuca, J. P., and Maquat, L. E. (1998) Mol. Cell. Biol. 18 5272–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehring, N. H., Kunz, J. B., Neu-Yilik, G., Breit, S., Viegas, M. H., Hentze, M. W., and Kulozik, A. E. (2005) Mol. Cell 20 65–75 [DOI] [PubMed] [Google Scholar]
- 19.Gehring, N. H., Neu-Yilik, G., Schell, T., Hentze, M. W., and Kulozik, A. E. (2003) Mol. Cell 11 939–949 [DOI] [PubMed] [Google Scholar]
- 20.Kim, Y. K., Furic, L., Desgroseillers, L., and Maquat, L. E. (2005) Cell 120 195–208 [DOI] [PubMed] [Google Scholar]
- 21.Le Hir, H., Izaurralde, E., Maquat, L. E., and Moore, M. J. (2000) EMBO J. 19 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L. E. (2002) EMBO J. 21 3536–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palacios, I. M., Gatfield, D., St Johnston, D., and Izaurralde, E. (2004) Nature 427 753–757 [DOI] [PubMed] [Google Scholar]
- 24.Gudikote, J. P., and Wilkinson, M. F. (2006) in NMD and the Immune System (Maquat, L. E., ed) pp. 71–83, R. G. Landes Co., Austin, TX
- 25.Gudikote, J. P., and Wilkinson, M. F. (2002) EMBO J. 21 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudikote, J. P., Imam, J. S., Garcia, R. F., and Wilkinson, M. F. (2005) Nat. Struct. Mol. Biol. 12 801–809 [DOI] [PubMed] [Google Scholar]
- 27.Weischenfeldt, J., Damgaard, I., Bryder, D., Theilgaard-Monch, K., Thoren, L. A., Nielsen, F. C., Jacobsen, S. E., Nerlov, C., and Porse, B. T. (2008) Genes Dev. 22 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belgrader, P., Cheng, J., Zhou, X., Stephenson, L. S., and Maquat, L. E. (1994) Mol. Cell. Biol. 14 8219–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng, J., and Maquat, L. E. (1993) Mol. Cell. Biol. 13 1892–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler, O., and Chasin, L. A. (1996) Mol. Cell. Biol. 16 4426–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozano, F., Maertzdorf, B., Pannell, R., and Milstein, C. (1994) EMBO J. 13 4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, J., Sun, X., Qian, Y., and Maquat, L. E. (1998) RNA (N. Y.) 4 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter, M. S., Doskow, J., Morris, P., Li, S., Nhim, R. P., Sandstedt, S., and Wilkinson, M. F. (1995) J. Biol. Chem. 270 28995–29003 [DOI] [PubMed] [Google Scholar]
- 34.Wang, J., Gudikote, J. P., Olivas, O. R., and Wilkinson, M. F. (2002) EMBO Rep. 3 274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan, W. K., Huang, L., Gudikote, J. P., Chang, Y. F., Imam, J. S., MacLean, J. A., II, and Wilkinson, M. F. (2007) EMBO J. 26 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell, J. T., Sharifi, N. A., Meyers, J. L., Martinez-Murillo, F., and Dietz, H. C. (2004) Nat. Genet. 36 1073–1078 [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson, M. F. (2000) in Essential Molecular Biology (Brown, T. A., ed) pp. 69–88, Oxford University Press, Oxford, UK
- 38.Clement, J. Q., Maiti, S., and Wilkinson, M. F. (2001) J. Biol. Chem. 276 16919–16930 [DOI] [PubMed] [Google Scholar]
- 39.Maclean, J. A., II, Chen, M. A., Wayne, C. M., Bruce, S. R., Rao, M., Meistrich, M. L., Macleod, C., and Wilkinson, M. F. (2005) Cell 120 369–382 [DOI] [PubMed] [Google Scholar]
- 40.Durand, S., Cougot, N., Mahuteau-Betzer, F., Nguyen, C. H., Grierson, D. S., Bertrand, E., Tazi, J., and Lejeune, F. (2007) J. Cell Biol. 178 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheth, U., and Parker, R. (2006) Cell 125 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buhler, M., Mohn, F., Stalder, L., and Muhlemann, O. (2005) Mol. Cell 18 307–317 [DOI] [PubMed] [Google Scholar]
- 43.Muhlemann, O., Mock-Casagrande, C. S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M. F., and Moore, M. J. (2001) Mol. Cell 8 33–43 [DOI] [PubMed] [Google Scholar]
- 44.Kozak, M. (1989) Mol. Cell. Biol. 9 5073–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendell, J. T., ap Rhys, C. M., and Dietz, H. C. (2002) Science 298 419–422 [DOI] [PubMed] [Google Scholar]
- 46.Paillusson, A., Hirschi, N., Vallan, C., Azzalin, C. M., and Muhlemann, O. (2005) Nucleic Acids Res. 33 e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferraiuolo, M. A., Lee, C. S., Ler, L. W., Hsu, J. L., Costa-Mattioli, M., Luo, M. J., Reed, R., and Sonenberg, N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4118–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibuya, T., Tange, T. O., Sonenberg, N., and Moore, M. J. (2004) Nat. Struct. Mol. Biol. 11 346–351 [DOI] [PubMed] [Google Scholar]
- 49.Cheng, J., Fogel-Petrovic, M., and Maquat, L. E. (1990) Mol. Cell. Biol. 10 5215–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daar, I. O., and Maquat, L. E. (1988) Mol. Cell. Biol. 8 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baserga, S. J., and Benz, E. J., Jr. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 2056–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He, F., Peltz, S. W., Donahue, J. L., Rosbash, M., and Jacobson, A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, S., Welch, E. M., Hogan, K., Brown, A. H., Peltz, S. W., and Jacobson, A. (1997) RNA (N. Y.) 3 234–244 [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, J., Hamilton, J. I., Carter, M. S., Li, S., and Wilkinson, M. F. (2002) Science 297 108–110 [DOI] [PubMed] [Google Scholar]
- 55.Chang, Y. F., Chan, W. K., Imam, J. S., and Wilkinson, M. F. (2007) J. Biol. Chem. 282 29738–29747 [DOI] [PubMed] [Google Scholar]
- 56.Wang, J., Chang, Y. F., Hamilton, J. I., and Wilkinson, M. F. (2002) Mol. Cell 10 951–957 [DOI] [PubMed] [Google Scholar]
- 57.Buhler, M., Wilkinson, M. F., and Muhlemann, O. (2002) EMBO Rep. 3 646–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lund, E., and Dahlberg, J. E. (1998) Science 282 2082–2085 [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson, M. F., and Shyu, A. B. (2002) Nat. Cell Biol. 4 E144–E147 [DOI] [PubMed] [Google Scholar]
- 60.Brogna, S., Sato, T. A., and Rosbash, M. (2002) Mol. Cell 10 93–104 [PubMed] [Google Scholar]
- 61.Mangiarotti, G. (1999) Biochemistry 38 3996–4000 [DOI] [PubMed] [Google Scholar]
- 62.Iborra, F. J., Jackson, D. A., and Cook, P. R. (2001) Science 293 1139–1142 [DOI] [PubMed] [Google Scholar]
- 63.Dahlberg, J. E., Lund, E., and Goodwin, E. B. (2003) RNA (N. Y.) 9 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nathanson, L., Xia, T., and Deutscher, M. P. (2003) RNA (N. Y.) 9 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bohnsack, M. T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Gorlich, D. (2002) EMBO J. 21 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caputi, M., Kendzior, R. J., Jr., and Beemon, K. L. (2002) Genes Dev. 16 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lytle, J. R., and Steitz, J. A. (2004) RNA (N. Y.) 10 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez, A. J., Condeelis, J., Singer, R. H., and Dictenberg, J. B. (2007) Semin. Cell Dev. Biol. 18 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.