Abstract
A signaling pathway involving ZAP-70, LAT, and SLP76 has been regarded as essential for receptor-driven T cell development and activation. Consistent with this model, mice deficient in SLP76 have a complete block at the double negative 3 stage of T cell development. Recently, however, it has been reported that inactivation of Cbl, a ubiquitin-protein isopeptide ligase, partially rescues T cell development in SLP76-deficient mice. To probe the influence of Cbl on domain-specific SLP76 functions, we reconstituted SLP76-/- Cbl-/- mice with Slp76 transgenes bearing mutations in each of three functional domains of SLP76 as follows: Y3F, in which the amino-terminal tyrosine residues of SLP76 were mutated, eliminating sites of SLP76 interaction with Vav, Nck, and Itk; Δ20, in which 20 amino acids in the proline-rich region of SLP76 were deleted, removing a binding site for Gads; and RK, in which arginine 448 of SLP76 was replaced by lysine, abolishing function of the Src homology 2 domain. Although each of these transgenes has been shown to partially rescue T cell development in SLP76-/- mice, we report here that Cbl inactivation completely reverses the severe double negative 3 developmental block that occurs in SLP76-deficient mice expressing the Y3F transgene (Y3F mice) and partially rescues the defect in positive selection in T cell receptor transgenic Y3F mice, but in contrast fails to rescue thymic development of SLP76-deficient mice expressing the Δ20 or RK transgene. Rescue in SLP76-/-Cbl-/-Y3F double-positive thymocytes is associated with enhanced tyrosine phosphorylation of signaling molecules, including Lck, Vav, PLC-γ1, and ERKs, but not Itk, in response to T cell receptor stimulation. Thus, our data demonstrate that Cbl suppresses activation of a bypass signaling pathway and thereby enforces SLP76 dependence of early T cell development.
T cell development proceeds through multiple stages that regulate the generation and selection of T cells whose T cell receptors (TCR)2 have an appropriate range of affinity for peptides presented by major histocompatibility complex (MHC) molecules (1). Precursors give rise to immature CD4-CD8- double negative (DN) cells that can be further divided into DN1, DN2, DN3, and DN4 stages, distinguished by cell surface phenotype as well as by critical events, including expansion of DN3 cells that have successfully rearranged TCRβ and have expressed and signaled through the pre-TCR complex (2). DN3 cells differentiate to the DN4 and then CD4+CD8+ double-positive (DP) stage following pre-TCR signaling. DP thymocytes rearrange TCRα, express a mature TCRαβ receptor, and develop into mature CD4+CD8- or CD4-CD8+ single-positive (SP) cells through a process of positive and negative selection that is based on signaling through this mature TCR and selection of a T cell repertoire that is tolerant to self but capable of responding to foreign-peptide-MHC (pMHC) complexes (1, 3, 4). Finally, SP cells exit from the thymus as mature T cells capable of recognizing and responding to foreign antigens.
The signals from pre-TCR and TCR, which determine the fate of developing thymocytes, have been intensely studied. Ligation of the TCR by pMHC complexes results in activation of a signaling cascade initiated by phosphorylation and activation of TCR-ζ, Lck, and ZAP-70, which in turn phosphorylate downstream targets, including LAT and SLP76. ZAP-70, LAT and SLP76 proteins (3) have been shown to be essential for thymocyte development by studies, including genetic manipulation in mice (5–8). There are essentially no detectable DP or SP thymocytes or peripheral T cells in LAT-/- or SLP76-/- mice, in which thymocyte development is blocked at the DN3 stage (5, 7). ZAP70-/- thymocytes are blocked at the DP stage of T cell development, and ZAP70-/- mice have very few SP thymocytes or peripheral T cells (6). These studies suggest that signal transduction required for early T cell development proceeds through a pathway that involves critical roles of multiple molecules, including ZAP-70, LAT, and SLP76.
SLP76 consists of three functional domains as follows: an amino-terminal domain containing targets for tyrosine phosphorylation, a proline-rich region, and a carboxyl-terminal SH2 domain (9). The amino-terminal tyrosine residues (Tyr-112, Tyr-128, and Tyr-145) are phosphorylated by tyrosine kinases following TCR engagement, enabling SLP76 to interact with Vav, a Rho guanine nucleotide exchange factor, Nck, an adaptor protein, and Itk, a member of Tec family PTK. The proline-rich region of SLP76 has the capacity to bind Gads, a Grb2 homolog, which results in the recruitment of SLP76 to cell surface membrane lipid rafts through binding to LAT following TCR engagement. The carboxyl-terminal SH2 domain of SLP76 interacts with ADAP (adhesion and degranulation-promoting protein) (10) an adaptor protein, and HPK-1, a serine kinase (9). Reconstitution of SLP76-deficient mice with transgenes containing mutations in each of these domains has demonstrated that each region is required for normal thymocyte development (5, 8). Two groups have reconstituted SLP76-deficient mice with T cell-specific expression of wild-type and mutant SLP76 transgenes, including a mutant in which three tyrosine residues (Tyr-112, Tyr-128, and Tyr-145) in the amino-terminal domain of SLP76 were substituted by phenylalanines (Y3F); a mutant in which 20 amino acids (amino acids 224–244) in the proline-rich region of SLP76 were deleted (Δ20); and a mutant in which arginine 448 of SLP76 was replaced by lysine (RK) (11, 12). The profound defects in T cell development and activation that are observed in SLP76 knock-out mice are completely reversed by reconstitution with a wild-type SLP76 transgene. In contrast, however, reconstitution with SLP76 that has been mutated in any of its three functional domains only partially rescues T cell development in SLP76 knock-out mice.
c-Cbl (Cbl) is a ubiquitin ligase and adaptor protein (regulator) with multiple domains that associate with multiple molecules involved in signal transduction (13). Thymocytes from Cbl knock-out mice have enhanced cell surface expression of TCR and CD3 in comparison with control mice (14, 15). In addition, it has been observed that phosphorylation of ZAP-70, LAT, and SLP76 is increased in Cbl-/- mouse thymocytes (14, 15). Recently, we reported that inactivation of Cbl partially rescues T cell development in LAT and SLP76-deficient mice (16), and Myers et al. (17) reported that inactivation of Cbl partially rescues T cell development in ZAP-70-deficient mice. These observations indicate that Cbl mediates requirements for LAT, SLP76, and ZAP-70 by preventing signaling that is capable of supporting T cell differentiation independent of LAT, SLP76, or ZAP-70. However, the rescue of T cell development in these model systems is strikingly incomplete, failing to substantially reconstitute development through the pre-TCR-dependent DN3-DN4 transition and thus failing to generate normal numbers of DP or functionally mature SP thymocytes. These findings suggest that Cbl inactivation functions to enable pathways that are capable of bypassing some but not all of the requirements for ZAP-70, LAT, and SLP76 during T cell development. To define these signaling pathways, normally suppressed by Cbl, that can support T cell development, we assessed the ability of Cbl inactivation to rescue T cell development in the presence of Y3F, Δ20, or RK SLP76 mutant transgenes. In this study, we report that Cbl inactivation completely reverses the DN3-DN4 developmental defect and partially reverses alterations in positive selection in thymocytes of SLP76 knock-out mice reconstituted with the SLP76 mutant Y3F, which lacks amino-terminal phosphotyrosine residues. In contrast, Cbl inactivation has no effect on the thymic developmental defects observed in SLP76 knock-out mice reconstituted with Slp76 transgenes mutated in the proline-rich Gads-binding region (Δ20) or the carboxyl-terminal SH2 domain (RK). Biochemical studies revealed that rescue of development in SLP76-/-Y3F thymocytes by inactivation of Cbl was marked by reversal of defects in tyrosine phosphorylation of multiple molecules, including Lck, Vav, PLC-γ1, and ERKs in response to TCR stimulation of DP thymocytes. Thus, Cbl normally enforces SLP76 dependence of T cell development by inhibiting an alternative pathway that may be independent of SLP76 association with Vav, Nck, and Itk (18).
MATERIALS AND METHODS
Mice—c-Cbl knock-out (Cbl-/-) (14), SLP76 knock-out (SLP76-/-) (5), and SLP76 transgenic (Y3F, Δ20 and RK) mice (12) were described previously. AND TCR transgenic mice (19) were bred to Cbl-/-, SLP76-/-, and Y3F transgenic mice on a B6 background (H-2b). All animals were housed at BIOQUAL, Inc. (Rockville, MD).
Antibodies—Anti-mouse CD3ε (2C11), anti-CD28 (4F10), anti-ZAP-70, anti-TCR-ζ, anti-CD4-PE, anti-CD8-FITC, anti-TCR-Vβ3-biotin, anti-TCR-Vα11-biotin, anti-CD25-biotin, anti-leu4-biotin, anti-CD62L-biotin, anti-CD5-biotin, anti-CD69-biotin, anti-HSA-biotin, and anti-TCR-β (H57)-biotin monoclonal antibodies were purchased from Pharmingen. Anti-phosphotyrosine (4G10), anti-PLC-γ1, and anti-Itk monoclonal antibodies were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). Anti-ERKs, anti-pY(416)Lck, anti-Lck, and anti-phospho-ERKs polyclonal antibodies were purchased from Cell Signaling (Beverly, MA). Anti-Vav1 polyclonal rabbit antibody was purchased from Santa Cruz Biotechnology (San Diego, CA).
Western Blot Analysis—Thymocytes were incubated with 5 μg/ml biotin-conjugated anti-CD3 for 30 min and then stimulated with 20 μg/ml streptavidin for the indicated time. The stimulated cells were lysed in buffer containing 50 μm Tris (pH 7.4), 150 μm NaCl, 1 mm Na2VO4, 1% Nonidet P-40, and protease inhibitor mixture. Protein lysates were used for biochemical analysis.
RESULTS
To assess the function of Cbl in regulating T cell differentiation, we determined the effect of Cbl inactivation in mice expressing SLP76 mutants, each of which is associated with defective T cell development. SLP76-/-Cbl-/- mice were bred with each of three mutant SLP76 transgenic strains (12) as follows: Y3F, in which three tyrosine residues in the amino-terminal domain of SLP76 were replaced by phenylalanines; Δ20, in which 20 amino acids in the proline-rich region of SLP76 were deleted; and RK, in which arginine 448 of SLP76 was replaced by lysine. The resulting mouse strains were designated SLP76-/-Cbl-/-Y3F, SLP76-/-Cbl-/-Δ20, and SLP76-/-Cbl-/-RK.
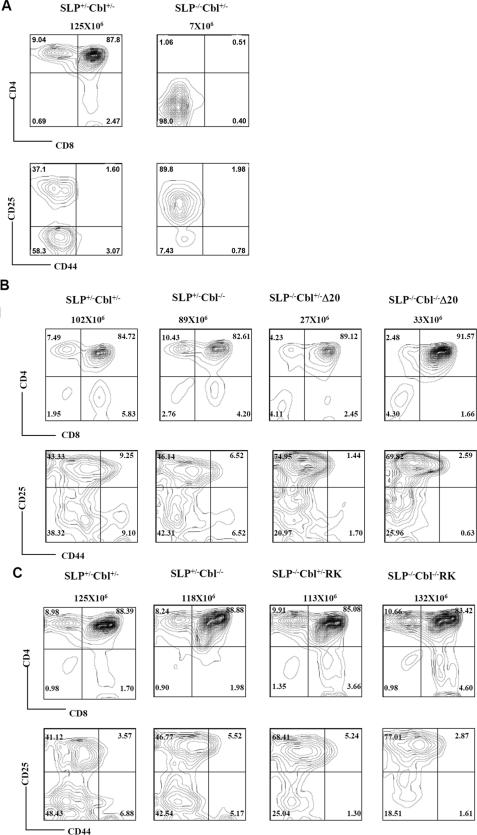
Cbl Inactivation Fails to Rescue the Defects in Thymic T Cell Development in SLP76-/-Δ20 and SLP76-/-RK Mice—Consistent with previous reports, reconstitution of SLP76-/- mice with the Δ20 transgene partially rescues the thymocyte developmental defect of SLP76-/- mice (5, 12). In contrast to the absence of detectable DP and SP cells in SLP76-/- mice (Fig. 1A) (5), the DN3 (CD25+CD44-) block observed in SLP76-/- mice is partially relieved in SLP76-/-Δ20 mice, and DP and SP thymocytes are generated in the presence of this transgene (Fig. 1B). However, the total cellularity of thymocytes in SLP76-/-Δ20 mice is only 20% that of wild type (Fig. 1B).
FIGURE 1.
Cbl inactivation fails to rescue the defects of T cell development in SLP76-/-Δ20 and SLP76-/-RK mice. A, thymocytes from SLP76+/-Cbl+/- (wild type) and SLP76-/-Cbl+/- (SLP76-/-) mice were stained with antibodies specific for CD4 and CD8 (top row) or with antibodies specific for CD25 and CD44 after excluding CD4+/CD8+/NK1.1+/Mac-1+/Gr1+/CD3+/B220+ thymocytes (bottom row). Genotypes of mice and total numbers of thymocytes for each mouse are shown above the plots. Numbers in quadrants indicate the frequency of DN (CD4-CD8-), DP (CD4+CD8+), CD4(CD4+CD8-), CD8(CD4-CD8+), DN1 (CD25-CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44-), and DN4 (CD25-CD44-) thymocytes. B, thymocytes from SLP76+/-Cbl+/- (wild-type), SLP76+/-Cbl-/- (Cbl-/-), SLP76-/-Cbl+/-Δ20 (SLP76-/-Δ20) and SLP76-/-Cbl-/-Δ20 mice were stained with CD4 and CD8 (top row); or with CD25 and CD44 after excluding CD4+/CD8+/NK1.1+/Mac-1+/Gr1+/CD3+/B220+ thymocytes (bottom row). Genotypes of mice used in the experiment and total thymocytes of each mouse are shown above plots. Numbers in quadrants indicate the frequency of DN (CD4-CD8-), DP (CD4+CD8+), CD4 (CD4+CD8-), CD8 (CD4-CD8+), DN1 (CD25-CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44-), and DN4 (CD25-CD44-) thymocytes. Results shown are representative of three independent experiments. C, thymocytes from SLP76+/-Cbl+/- (wild type), SLP76+/-Cbl-/- (Cbl-/-), SLP76-/-Cbl+/-RK (SLP76-/-RK), and SLP76-/- Cbl-/-RK mice were stained with CD4 and CD8 (top row) or with CD25 and CD44 excluding CD4+/CD8+/NK1.1+/Mac-1+/Gr1+/CD3+/B220+ thymocytes (bottom row). Genotypes of mice used in the experiment and total thymocytes of each mouse are shown above the plots. Numbers in quadrants indicate the frequency of DN (CD4-CD8-), DP (CD4+CD8+), CD4 (CD4+CD8-), CD8 (CD4-CD8+), DN1 (CD25-CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44-), and DN4 (CD25-CD44-) thymocytes. Results shown are representative of three or more independent experiments.
Inactivation of Cbl had no significant effect on the defects in thymocyte development observed in SLP76-/-Δ20 mice. Both SLP76-/-Δ20 and SLP76-/-Cbl-/-Δ20 mice had much reduced total thymic cellularity, as well as reduced proportions of SP cells compared with wild-type or Cbl-/- mice. DN development in SLP76-/-Cbl-/-Δ20 mice was partially blocked at the DN3 (CD25+CD44-) stage, again similar to SLP76-/-Δ20 mice with intact Cbl expression (Fig. 1B). These data indicate that Cbl inactivation does not overcome or bypass the requirement for the proline-rich Gads-binding domain that is mutated in SLP76-/-Δ20 mice.
In SLP76-/- mice carrying the SLP76 RK transgene, overall thymic cellularity is near-normal, as are proportions of DP and SP cells; however, the DN development of thymocytes is still partially blocked in SLP76-/-RK mice (Fig. 1C) (12). Again, Cbl inactivation failed to alter the phenotype of SLP76-/-RK mice, with SLP76-/-RK and SLP76-/-Cbl-/-RK thymuses having similarly decreased CD4/CD8 ratios and partial blocks in DN3 to DN4 transition (Fig. 1C).
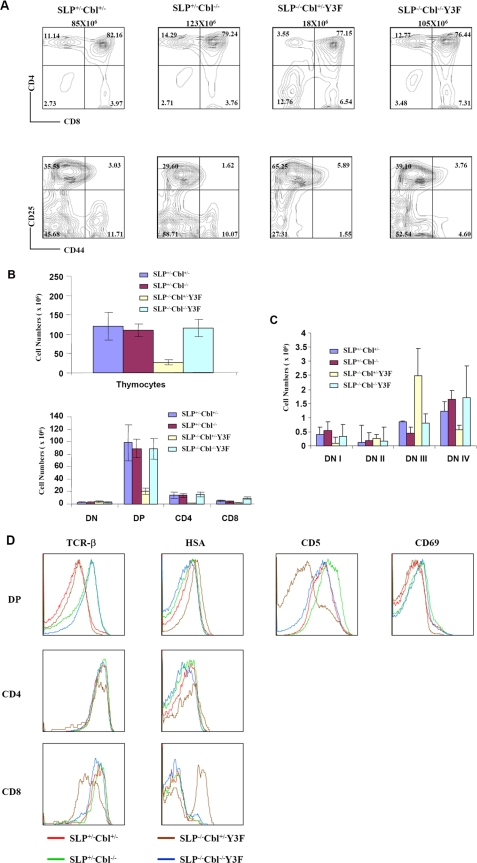
Cbl Inactivation Rescues Defects in Thymic T Cell Development in SLP76-/-Y3F Mice—The amino-terminal tyrosine residues of SLP76 are phosphorylated by tyrosine kinases following TCR engagement, which enables SLP76 to interact with Vav, a Rho guanine nucleotide exchange factor, Nck, an adaptor protein, and Itk, a member of Tec family protein-tyrosine kinase, each through association with a distinct phosphotyrosine on SLP76 (9). The SLP76 mutant Y3F, in which three tyrosine residues (Tyr-112, Tyr-128, and Tyr-145) in the amino-terminal domain of SLP76 were substituted by phenylalanines, is thus unable to interact with Vav, Nck, and Itk (9). Consistent with previously reported results (12), the Y3F transgene only partially rescued thymocyte development of an SLP76-/- mouse. SLP76-/-Y3F transgenics had significantly decreased total thymic cellularity and numbers of DP cells compared with wild-type or Cbl-/- mice (Fig. 2, A and B). The percentage of CD4 cells was decreased relative to controls, whereas the percentage of DN was dramatically increased in SLP76-/-Y3F thymus compared with control mice (Fig. 2A). The DN development of SLP76-/-Y3F was severely impaired at the DN3 (CD25+CD44-) to DN4 (CD25-CD44-) transition relative to wild-type or Cbl-/- mice (Fig. 2A). In contrast, SLP76-/-Cbl-/-Y3F mice had normal thymic cellularity, with normal proportions and numbers of CD4+ SP and DP cells, as well as normalized proportions of DN3 and DN4 subsets (Fig. 2, A and C). Thus, Cbl inactivation completely rescues the defect of DN development in SLP76-/-Y3F mice.
FIGURE 2.
Cbl inactivation rescues defects in T cell development in SLP76-/-Y3F mice. A, thymocytes from SLP76+/-Cbl+/- (wild type), SLP76+/-Cbl-/- (Cbl-/-), SLP76-/-Cbl+/-Y3F (SLP76-/-Y3F), and SLP76-/-Cbl-/-Y3F mice were stained with CD4 and CD8 (top row) or with CD25 and CD44 excluding CD4+/CD8+/NK1.1+/Mac-1+/Gr1+/CD3+/B220+ thymocytes (bottom row). Genotypes of mice used in the experiment and total thymocytes of each mouse are shown above the plots. Numbers in quadrants indicate the frequency of DN (CD4-CD8-), DP (CD4+CD8+), CD4 (CD4+CD8-), CD8 (CD4-CD8+), DN1 (CD25-CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44-), and DN4 (CD25-CD44-) thymocytes. Results shown are representative of four or more experiments. B, bar graph representing the mean total cellularity of thymocytes in wild type (n = 5), Cbl-/-(n = 5), SLP76-/-Y3F (n = 4), and SLP76-/-Cbl-/-Y3F (n = 5) mice (upper panel), and the mean numbers of DN, DP, CD4+CD8- (CD4), and CD4-CD8+ (CD8) thymocytes in wild-type (n = 5), Cbl-/-(n = 5), SLP76-/-Y3F (n = 4), and SLP76-/-Cbl-/-Y3F (n = 5) mice (lower panel). C, bar graph representing the mean numbers of DN1, DN2, DN3, and DN4 thymocytes in wild-type (n = 5), Cbl-/-(n = 5), SLP76-/-Y3F (n = 4), and SLP76-/-Cbl-/-Y3F (n = 5) mice. D, expression of cell surface molecules (H57, CD5, CD69, and HSA indicated at the top of histograms) on DP, CD4, and CD8 cells of SLP76+/-Cbl+/- (wild type, red), SLP76+/-Cbl-/- (Cbl-/-, green), SLP76-/-Cbl+/-Y3F (SLP76-/-Y3F, brown), and SLP76-/-Cbl-/-Y3F (blue) thymocytes were assessed by flow cytometry.
The effect of Cbl inactivation on thymic development was further analyzed by characterizing the phenotype of cells that develop through this pathway. TCR expression was up-regulated in DP thymocytes of Cbl-/- mice relative to wild-type controls, as described previously (12). Cell surface TCRβ expression on SLP76-/-Y3F DP cells was similarly elevated by Cbl inactivation (Fig. 2D). Levels of cell surface TCRβ expressed on SLP76-/-Cbl-/-Y3F DP thymocytes are equivalent to those on Cbl-/- DP cells, and significantly increased in comparison with wild-type and SLP76-/-Y3F mice. In contrast, TCRβ expression on CD4 single-positive cells was similar for wild-type, Cbl-/-, SLP76-/-Y3F, and SLP76-/-Cbl-/- Y3F mice. SLP76-/-Y3F had dramatically decreased expression of TCRβ on CD8 single-positive cells, and this decrease was completely normalized by Cbl inactivation (Fig. 2D). It is possible that SLP76-/-Y3F CD8 cells expressing low density of TCR represent, at least in part, an increased population of immature SP cells and that this population is substantially reduced by Cbl inactivation. Patterns of CD3 expression were similar to those of TCRβ on wild-type, Cbl-/-, SLP76-/-Y3F, and SLP76-/-Cbl-/-Y3F thymocytes (data not shown).
The level of HSA expression has been shown to be a marker for the maturational state of SP thymocytes, with less mature cells expressing higher levels of HSA, and levels decreasing progressively with maturation (20). SLP76-/-Y3F DP and CD8+ thymocytes had increased expression of HSA, whereas SLP76-/-Cbl-/-Y3F DP and CD8+ thymocytes had normal expression of HSA, indicating that they had progressed to a more mature state (Fig. 2D). CD5 is a negative regulator of signal transduction, and its level of cell surface expression has been shown to be proportional to the strength of TCR signaling of DP thymocytes. CD69 is an early marker of TCR signaling and in DP thymocytes is an indication that cells have been positively selected by recognition of a selecting ligand. Expression of both CD5 and CD69 was decreased on SLP76-/-Y3F DP thymocytes, indicating that these cells had received diminished signaling through the TCR (Fig. 2D). Notably, expression of both CD5 and CD69 was increased in SLP76-/-Cbl-/-Y3F DP cells to levels comparable with wild-type DP cells (Fig. 2D).
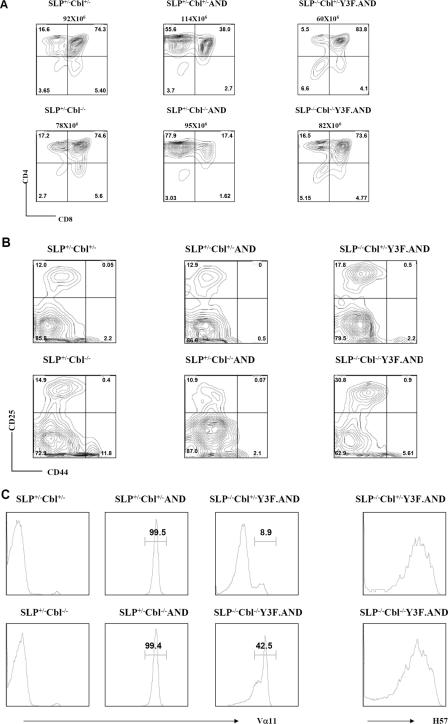
Expression of a TCR Transgene Rescues Defective DN3-DN4 Development in SLP76-/-Y3F Mice—As shown above, thymic development in SLP76-/-Y3F mice is marked by a defect in maturation at the DN3-DN4 transition or β checkpoint, a transition that is mediated by signaling through the pre-TCR. It has been observed that expression of a transgenic αβ TCR can replace the pre-TCR in mediating DN3-DN4 differentiation. To first determine whether DN3-DN4 differentiation mediated by a transgenic TCR is also defective in the SLP76-/-Y3F mice, the AND transgenic TCR (19) was expressed in SLP76-/-Y3F mice. Expression of the AND transgene in wild-type mice results in positive selection that is highly skewed toward development of CD4 SP cells, as reported previously for I-Ek-expressing mice (19), and is shown here to also be the case for wild-type H-2b mice (Fig. 3A). Notably, total thymocyte numbers were increased in SLP76-/-Y3F.AND mice (78 ± 8 × 106 for 3 mice) in comparison with SLP76-/-Y3F mice (21 ± 7 × 106 for 5 mice), whereas the percentage of DN was reduced in SLP76-/-Y3F.AND mice in comparison with SLP76-/-Y3F mice (representative data in Fig. 2A and Fig. 3A). Analysis of DN development showed that the defect in DN3-DN4 transition observed in thymocytes of SLP76-/-Y3F mice (Fig. 2B) was normalized in SLP76-/- Y3F.AND mice (Fig. 3B). Thus, mutation of amino-terminal tyrosines in the Y3F mutant results in deficient DN3-DN4 transition mediated by the pre-TCR, but it does not compromise development mediated by a transgenic mature αβ TCR.
FIGURE 3.
TCR AND transgene rescues the defect in DN development in SLP76-/-Y3F mice, and Cbl inactivation partially rescues positive selection in SLP76-/-Y3F.AND mice. Thymocytes from SLP76+/- Cbl+/- (wild type), SLP76+/-Cbl-/- (Cbl-/-), SLP76+/-Cbl+/-AND, SLP76+/-Cbl-/-AND, SLP76-/- Cbl+/-Y3F.AND, and SLP76-/-Cbl-/-Y3F.AND mice were stained with CD4 and CD8 (A) or with CD25 and CD44 (B) after excluding CD4+/CD8+/NK1.1+/Mac-1+/Gr1+/CD3+/B220+ thymocytes. Genotypes of mice used in the experiment and total thymocytes of each mouse are shown above the plots. Numbers in quadrants indicate the frequency of DN (CD4-CD8-), DP (CD4+CD8+), CD4 (CD4+CD8-), CD8 (CD4-CD8+), DN1 (CD25-CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44-), and DN4 (CD25-CD44-) thymocytes. C, thymocytes were stained with anti-CD4, anti-CD8, and anti-Vα11. Histograms represent expression of Vα11 on gated CD4 SP thymocytes. Results shown are representative of three independent experiments.
Cbl Inactivation Modulates Positive Selection of TCR Transgenic T Cells in SLP76-/-Y3F Mice—Expression of TCR transgenes has been a valuable approach to assessing T cell repertoire selection. The AND transgene is composed of Vα11 and Vβ3 chains and is specific for cytochrome c (19). Although expression of the AND transgene substantially reversed the DN3-DN4 block in thymic development in SLP76-/-Y3F mice, as reflected in increased numbers of DP and SP cells, the repertoire selected in the SLP76-/-Y3F.AND mice differed substantially from that selected in SLP76+/-.AND mice. Both CD4 and CD8 SP thymocytes were generated, but there was not a strong preponderance of CD4 cells in SLP76-/-Y3F.AND mice (Fig. 3A). When TCR expression was analyzed, CD4 SP thymocytes expressed homogeneously high levels of TCR Vα11 (Fig. 3C) as well as Vβ3 (data not shown) in both SLP76+/-Cbl+/-AND and SLP76+/- Cbl-/-AND mice, indicating that selection was highly efficient for cells expressing transgenic α and β chains. However, the TCR repertoire in SLP76-/-Cbl+/-Y3F.AND thymuses was markedly different. Vα11 expression in these mice was bimodal, with the majority (84 ± 14%, n = 3) of thymocytes expressing low levels of Vα11 and a smaller population expressing high levels of Vα11 comparable with those expressed in SLP76 wild-type AND transgenics (Fig. 3C). SLP76-/- Cbl-/-Y3F.AND mice were analyzed to determine whether Cbl inactivation would affect the altered TCR selection observed in SLP76-/- Y3F.AND mice. Inactivation of Cbl in SLP76-/-Cbl-/- Y3F.AND mice resulted in a substantial increase in the total number of CD4 SP cells (21 ± 6.8 × 106, n = 3 versus 5.3 ± 2.3 × 106, n = 3) (Fig. 3A) and in the percentage of Vα11-high CD4 SP cells (53 ± 9%, n = 3 versus 17 ± 10%, n = 3) (Fig. 3C) over levels observed in SLP76-/-Y3F.AND mice, although these values did not reach the levels in SLP76+/-Cbl+/-AND mice (65 ± 7 × 106 CD4 SP cells, 100% Vα11-high) (Fig. 3, A and C). This difference in Vα11 expression resulting from Cbl inactivation was not the result of differences in overall expression of TCRαβ as measured by Cβ staining (Fig. 3C), and thus reflects a shift from selection of cells expressing low levels of transgenic Vα11 and high levels of endogenous nontransgenic Vα to predominant selection of cells expressing Vα11-high transgenic receptors (Fig. 3C). Thus, inactivation of Cbl both enhanced overall CD4 SP selection and altered specific TCR repertoire selection in the presence of the AND transgene in SLP76-/-Y3F mice.
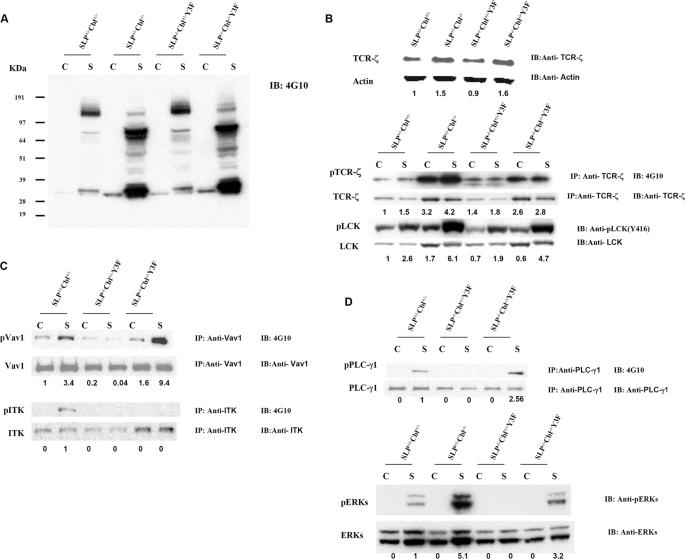
Phosphorylation of Multiple Signaling Proteins in Response to TCR Signals Is Increased by Cbl Inactivation in SLP76-/-Cbl-/- Y3F Thymocytes—The findings presented above indicate that Cbl inactivation rescues the defects in early T cell development observed in SLP76-/-Y3F mice. To understand the effect of Cbl inactivation on TCR signaling, thymocytes from wild-type, Cbl-/-, SLP76-/-Y3F, and SLP76-/-Cbl-/-Y3F mice were subjected to biochemical analysis. Fig. 4A shows total protein tyrosine phosphorylation of DP thymocytes from wild-type, Cbl-/-, SLP76-/-Y3F, and SLP76-/-Cbl-/-Y3F mice in response to TCR stimulation with anti-CD3 antibody. Hyper-phosphorylation of multiple proteins was observed in stimulated Cbl-/- thymocytes relative to wild-type controls, consistent with previous reports (14, 15). Tyrosine phosphorylation in SLP76-/-Y3F thymocytes was similar to that observed in wild-type cells and was similarly enhanced by Cbl inactivation in SLP76-/-Cbl-/-Y3F thymocytes (Fig. 4A).
FIGURE 4.
Cbl inactivation reverses selective signaling defects in SLP76-/-Y3F thymocytes. DP thymocytes were sorted from SLP76+/-Cbl+/- (wild type), SLP76+/-Cbl-/- (Cbl-/-), SLP76-/-Cbl+/-Y3F (SLP76-/-Y3F), and SLP76-/-Cbl-/-Y3F mice by flow cytometry and were unstimulated or stimulated with anti-CD3 Abs for 5 min at 37 °C. Protein lysates were prepared from DP thymocytes. A, protein lysates were immunoblotted (IB) with anti-phosphotyrosine Ab (4G10). B, protein lysates were immunoblotted (IB) with anti-TCR-ζ, anti-actin, anti-Lck, or anti-phospho(P)-lck antibody or were immunoprecipitated (IP) with anti-TCR-ζ and immunoblotted with anti-phosphotyrosine Ab (4G10) and anti-TCR-ζ,(upper panels). C, protein lysates were immunoprecipitated with anti-Vav1 and immunoblotted with anti-phosphotyrosine Ab (4G10) and anti-Vav1 (upper panels) or were immunoprecipitated with anti-ITK and immunoblotted with 4G10 and anti-ITK (lower panels). D, protein lysates were immunoprecipitated with anti-PLCγ1 and immunoblotted with anti-phosphotyrosine (4G10) or anti-PLCγ1(upper panels), or lysates were immunoblotted with anti-phospho-ERKs or anti-ERK (lower panels). Lane C indicates control cells without stimulation; lane S indicates cells were stimulated with 10 μg/ml of anti-mouse CD3 antibodies for 5 min.
We next evaluated the status of specific signaling proteins. It has been reported that TCR-ζ chain is degraded by the Cbl-SLAP complex and that levels of total TCR-ζ protein are therefore increased in Cbl-deficient thymocytes (17). The level of expression of the TCR-ζ chain was analyzed in lysates of thymocytes from WT, Cbl-/-, SLP76-/-Y3F, and SLP76-/- Cbl-/-Y3F mice by immunoblotting with anti-TCR-ζ antibody. The level of total TCR-ζ protein in Cbl-/- or SLP76-/- Cbl-/-Y3F thymocytes was greater than that in wild-type or SLP76-/-Y3F thymocytes, consistent with the reported role of Cbl in TCR-ζ degradation (Fig. 4B). It has been demonstrated that phosphorylation of TCR-ζ in DP thymocytes occurs in vivo as a result of TCR engagement (21). When the effect of Cbl inactivation on this in vivo phosphorylation was assessed, it was found that TCR-ζ phosphorylation, in the absence of in vitro stimulation, was substantially greater in Cbl-/- thymocytes than in WT cells (Fig. 4B). TCR-ζ phosphorylation in SLP76-/- Y3F thymocytes was not substantially different from WT levels, and notably, Cbl inactivation in SLP76-/-Cbl-/-Y3F thymocytes resulted in increased TCR-ζ phosphorylation in SLP76-/- Cbl-/-Y3F comparable with that found in Cbl-/- cells. In vitro stimulation with anti-CD3 did not enhance phosphorylation substantially above the levels observed ex vivo. Thus, in addition to the effect of Cbl on total TCR-ζ protein levels, in vivo phosphorylation of TCR-ζ is markedly enhanced by Cbl inactivation, and this effect is intact in SLP76-/-Cbl-/- Y3F thymocytes.
Phosphorylation and consequent activation of Lck have been well characterized as critical proximal events in TCR-mediated signaling. We assessed phosphorylation of Lck in the response of DP thymocytes to anti-CD3 stimulation, using antibody specific for phosphorylation of Tyr-416 of Lck, a modification that is specifically related to activation of Lck kinase activity (22, 23). SLP76-/-Y3F thymocytes had levels of Lck Tyr-416 phosphorylation similar to those of wild-type cells upon CD3 stimulation (Fig. 4B). Tyr-416 phosphorylation of Lck in response to anti-CD3 was substantially increased in Cbl-/- thymocytes relative to wild-type controls. Similarly, Tyr-416 phosphorylation of SLP76-/-Cbl-/-Y3F thymocytes in response to anti-CD3 was dramatically increased in comparison with responses of wild type or SLP76-/-Y3F (Fig. 4B). Thus, Lck phosphorylation and activation, an early step in TCR signaling and proximal to SLP76 in the signaling cascade, is intact in SLP76-/-Y3F thymocytes, as is the enhancement of this response by Cbl inactivation.
The amino-terminal tyrosine residues of SLP76 are phosphorylated by tyrosine kinases following TCR engagement, which enables SLP76 to interact with Vav and Itk (9). The Y3F mutation eliminates the capacity of SLP76 to interact with Vav and Itk, whose activities or phosphorylation might therefore be affected in SLP76-/-Y3F thymocytes. Protein lysates from wild-type, SLP76-/-Y3F, and SLP76-/-Cbl-/-Y3F DP thymocytes were immunoprecipitated with antibodies against Vav1 and Itk, and the immunoprecipitated products were immunoblotted with antibodies against phosphotyrosine, Vav1, and Itk. In wild-type DP thymocytes, anti-CD3 stimulation resulted in phosphorylation of both Vav1 and Itk (Fig. 4C). In striking contrast, no phosphorylation of Vav1 or Itk was detected in stimulated SLP76-/-Y3F DP thymocytes (Fig. 4C). Thus, disruption of Vav- and Itk-binding sites on the SLP76 Y3F mutant molecule completely abrogates tyrosine phosphorylation of Vav and Itk in response to TCR cross-linking. Strikingly, however, Vav1 tyrosine phosphorylation in response to TCR signaling was completely restored in SLP76-/- Cbl-/-Y3F DP thymocytes, in fact to levels substantially greater than those in wild-type mice in response to TCR stimulation (Fig. 4C). In contrast, no detectable phosphorylation of Itk occurred in SLP76-/- Cbl-/-Y3F DP thymocytes (Fig. 4C), indicating that Cbl inactivation rescued the defect in Vav signaling but not the defect in Itk phosphorylation in SLP76-/-Y3F thymocytes. To further study the Vav signaling pathway in SLP76-/-Cbl-/-Y3F thymocytes, we next analyzed phosphorylation of ERKs and PLC-γ1, proteins that are downstream in the Vav signaling pathway (24). No phosphorylation of ERKs or PLC-γ1 was detected in SLP76-/-Y3F DP thymocytes in response to TCR stimulation, indicating that these events are normally highly dependent upon intact sites mutated in the SLP76 Y3F transgene (Fig. 4D). However, Cbl inactivation reversed this defect, and SLP76-/-Cbl-/-Y3F DP thymocytes generated substantial tyrosine phosphorylation of both PLC-γ1 and ERKs in response to TCR stimulation (Fig. 4D). Thus, the Vav signaling rescued by Cbl inactivation in SLP76-/-Y3F thymocytes appears to be functional as reflected in these downstream events.
DISCUSSION
A widely accepted model for signal transduction during T cell development, as well as for TCR-mediated activation of mature T cells, involves critical early roles for Lck, ZAP-70, LAT, and SLP76, which in turn transduce signals to molecules, including PLC-γ1, Vav, Itk, Gads, Grb2, and ERKs (9). Thus, deletion of ZAP-70, LAT, or SLP76 results in a profound arrest in T cell development (5, 7, 8). However, it has been demonstrated that inactivation of Cbl, a ubiquitin ligase with negative regulatory activity, can partially rescue the defects in T cell development observed in ZAP-70-, LAT-, and SLP76-deficient mice (16, 17), indicating the existence of signaling pathways, normally inhibited by Cbl, that can function in thymic T cell differentiation. To elucidate the role normally mediated by Cbl in regulating such pathways, we have utilized a series of transgenes with mutations in specific domains of SLP76. Each of these SLP76 mutants, Y3F, Δ20, and RK SLP76, partially rescues the defects in thymocyte development that occur in SLP76-/- mice (12), but mice expressing these SLP76 mutants have residual defects in thymic development. We have thus been able to study the role of Cbl by analyzing the effects of Cbl inactivation on the T cell developmental and signaling defects that result from discrete molecular lesions in the SLP76 pathway. We found that Cbl inactivation reverses the defect in thymic development in amino-terminal tyrosine mutant SLP76-/-Y3F mice but not the defects resulting from SLP76-/-Δ20 and SLP76-/-RK mice.
The SLP76-/-Δ20 mutant lacks 20 amino acids in the proline-rich region and has lost the capacity to bind Gads (9). Cbl inactivation failed to rescue the defect in T cell development of SLP76-/-Δ20 mice (Fig. 1B), indicating that Cbl inactivation does not enable an alternative pathway capable of bypassing the requirement for Gads. Gads appears to function in binding to both SLP76 and LAT and thus in targeting SLP76-containing signaling complexes to LAT at the cell membrane and in proximity to the TCR (25, 26). The failure of Cbl inactivation to overcome the requirement for Gads binding to SLP76 suggests that there may be a relatively stringent requirement for the function of Gads in targeting SLP76-containing complexes to the cell membrane. Similarly, the signaling pathway that is defective in the RK mutant, which has lost the capacity to interact with ADAP and HPK-1, was not rescued by Cbl inactivation (Fig. 1C), indicating that no mechanisms were activated that are capable of compensating for this SLP76 mutation.
The amino-terminal tyrosine residues (Tyr-112, Tyr-128, and Tyr-145) of SLP76 are phosphorylated by ZAP-70 following TCR engagement, enabling SLP76 to interact with molecules, including Vav (at the Tyr-112 site), Nck (at the Tyr-128 site), and Itk (at the Tyr-145 site) (9). The interactions of SLP76 with these signaling molecules have been shown to be essential events for thymic development. The SLP76 mutant Y3F, in which three tyrosine residues (Tyr-112, Tyr-128, and Tyr-145) were substituted by phenylalanines, was introduced as a transgene into SLP76-deficient mice. Mutation of the three tyrosine residues of SLP76 results in multiple defects of T cell development, including decreased thymic size, a block in development at the DN3 stage, reduced numbers of DP and SP thymocytes, and reduced expression of TCRβ, CD5, and CD69 on DP thymocytes (12) (Fig. 2, A–D). It is notable that the defects in T cell development in SLP76-/-Y3F mice are similar to those observed in the Vav1 knock-out mouse (24). Our data showed that the substantial defects in T cell development caused by the mutation of Tyr-112, Tyr-128, and Tyr-145 residues of SLP76 could be effectively rescued by introducing the Cbl mutation (Fig. 2, A–D). It is thus possible that rescue of defective T cell development by Cbl inactivation in Y3F mutant mice is mediated by rescue of the defect in Vav signaling in SLP76-/-Y3F thymocytes. Indeed, Cbl inactivation completely rescued the profound defect in Vav1 phosphorylation seen in SLP76-/- Y3F thymocytes in response to TCR signaling (Fig. 4C). This rescued Vav signaling by Cbl mutation might play a role in the reversal of defects involving potential downstream molecules, such as ERKs and PLC-γ1, in response of SLP76-/-Y3F thymocytes to TCR signaling (Fig. 4D). However, it is also possible that ERK and PLC-γ1 phosphorylation may be rescued by Vav-independent pathways that are also affected by Cbl, such as those downstream of Lck. Phosphorylation of Lck is an early event in TCR signaling and appears to be upstream of SLP76 in this cascade. Consistent with this, there was no defect in Lck phosphorylation in SLP76-/-Y3F mutant thymocytes (Fig. 4B). Inactivation of ubiquitin ligase Cbl, which results in increased levels of TCR-ζ and increased phosphorylation (and presumably increased phosphorylation-dependent activity) of Lck in Cbl-/- mice, similarly increased these responses in cells expressing the mutant SLP76-/-Y3F (Fig. 4B).
Our findings suggest that activation of Vav may be a critical pathway in the mechanism by which Cbl inactivation permits normalization of T cell development in the presence of the Y3F mutation that, in Jurkat cells, has been shown to disrupt the ability of SLP76 to interact with Vav (27). Consistent with the functional importance of normalized Vav phosphorylation in SLP76-/-Cbl-/-Y3F thymocytes is the accompanying restoration of phosphorylation of putative downstream mediators, including PLC-γ1 and ERKs. The mechanism by which Vav is phosphorylated in the presence of Y3F mutant SLP76 is not yet established, but intact proximal events involving TCR-ζ and Lck may play a role. The interaction of Itk with SLP76 is also abrogated by the Y3F mutation in Jurkat cells. However, in contrast to the ability of Cbl inactivation to restore defective phosphorylation of Vav in response to TCR engagement, phosphorylation of Itk remained undetectable in SLP76-/-Cbl-/-Y3F thymocytes. This finding may reflect a strict requirement for the interaction of Itk with SLP76 for Itk activation (28). The Itk knock-out mouse in fact expresses less severe reductions in thymocyte numbers (60–70% of WT cellularity) and partial defects in thymocyte selection that lead to increased numbers of “innate-type” CD8+ T lymphocytes (29). Preliminary studies indicate that Cbl inactivation does not rescue these phenotypes in Itk knock-out mice (supplemental Fig. 1). It therefore appears that Cbl inactivation facilitates activation of Vav, but not Itk, in the absence of interaction of these molecules with SLP76. These results may reflect activation of Vav via multiple pathways in contrast to a strict requirement for Itk interaction with SLP76 for activation of this kinase. A potential mechanism for Vav activation in SLP76-/- Cbl-/-Y3F mice is suggested by the reported ability of Lck to directly phosphorylate and activate Vav (30). Cbl binds directly to Lck and negatively regulates Lck localization to lipid rafts, as well as mediating Lck ubiquitination and degradation independent of ZAP-70 (31, 32). Inactivation of Cbl in SLP76-/- Cbl-/-Y3F mice resulted in elevated Lck activation, which therefore might be sufficient to mediate Vav activation directly, independent of ZAP-70 or Vav interaction with SLP76.
In addition to the effects of Cbl inactivation on early events in T cell development, we also assessed the effect of SLP76 mutation and Cbl inactivation on selection of mature SP cells, using the AND TCR transgenic model. Expression of the MHC class II-restricted AND transgene was found to overcome the defect in DN3-DN4 differentiation in SLP76-/-Y3F mice, in contrast to the failure of pre-TCR signaling to induce DN3-DN4 differentiation in these mice in the absence of a TCR transgene. These results demonstrate the difference, either quantitative or qualitative, in SLP76-dependent signaling requirements downstream of pre-TCR and transgenic TCR. Although the AND transgene allowed differentiation of DN4, DP, and SP cells in SLP76-/- Y3F mice, positive selection was notably different in these mice than in those expressing AND and wild-type SLP76. The strong skewing to differentiation of CD4 SP thymocytes that is observed in wild-type AND transgenics was absent in SLP76-/-Y3F.AND mice. In addition, the strong predominance of transgenic TCR Vα11 and Vβ3 observed in wild-type AND mice was altered in SLP76-/- Y3F.AND mice, where the majority of CD4 SP cells selected in these mice expressed low levels of transgenic Vα11 and substantial levels of nontransgenic (non-Vα11) TCR, indicating a marked skewing in positive selection in the presence of mutant SLP76. Cbl inactivation had substantial effects on both of these aspects of positive selection, resulting in skewing to differentiation of CD4 SP cells and to the predominant expression of transgenic TCR Vα11/Vβ3 receptors. Thus, in addition to its effects on early pre-TCR-mediated T cell development, Cbl expression substantially affects later stages of TCR-mediated positive selection in the presence of mutated SLP76.
Because Cbl inactivation allows efficient generation of mature SP thymocytes in SLP76-/-Cbl-/-Y3F mice, we were able to examine the status of peripheral T cells in this setting. The number and frequency of peripheral CD4 and CD8 T cells in SLP76-/-Cbl-/-Y3F mice were comparable with those in wild-type mice (data not shown). However, these peripheral T cells were markedly unresponsive to TCR stimulation by anti-CD3/anti-CD28. Under conditions equivalent to those that generated robust tyrosine phosphorylation responses in SLP76-/-Cbl-/-Y3F thymocytes, neither tyrosine phosphorylation nor proliferation was induced in peripheral T cells (data not shown). Although the explanation for this discordance has not been established, it may reflect the documented differences in expression of Cbl and Cbl-b in the thymus and periphery (33). These two related ubiquitin-protein isopeptide ligases have multiple and significantly overlapping molecular targets. In the thymus, Cbl is expressed at high levels and Cbl-b at relatively low levels. Thus, inactivation of Cbl results in a high degree of overall depletion of Cbl family ubiquitin ligase activity in the thymus, with consequent abrogation of normally Cbl-mediated regulatory effects. In contrast, Cbl-b is expressed at high levels in the periphery and Cbl at low levels. Inactivation of Cbl might therefore have limited impact in the periphery, where intact Cbl-b could exert substantial regulatory effects. Deficiency of both Cbl and Cbl-b results in embryonic lethality (33), and complex strategies involving conditional inactivation of Cbl family members would therefore be required to resolve this possibility. However, Naramura et al. (14) suggested that Cbl mutation, by altering thymic TCR signaling, resulted in altered selection of thymocytes, giving rise to peripheral T cells with an attenuated ability to respond to TCR stimulation. Therefore, the abnormality observed in SLP76-/-Cbl-/-Y3F peripheral T cells might be the result of altered selection in the thymus for T cells with reduced capacity for signal transduction.
The studies presented here demonstrate that Cbl normally enforces the SLP76 dependence of early T cell development by suppressing a signaling pathway that does not require SLP76 association with Vav, Nck, or Itk. This pathway has been shown to involve tyrosine phosphorylation of signaling intermediates commonly regarded as both upstream and downstream of SLP76 in pre-TCR/TCR signal transduction. The molecular complexes involved in signaling through the TCR are both complex and highly dynamic (34). Cbl, a ubiquitin-protein isopeptide ligase, interacts with numerous substrates, including multiple known mediators of T cell signaling (13). It is therefore likely that the potent effects demonstrated here for Cbl modulation of T cell development reflect the net effect of complex Cbl-mediated alterations in molecular signaling networks.
Supplementary Material
Acknowledgments
We are grateful to Genevieve Sanchez-Howard and staff at BIOQUAL, Inc., for expert animal care and breeding. We thank Drs. Alfred Singer and Larry Samelson for their critical review of this manuscript and helpful comments on this work.
This work was supported, in whole or in part, by a National Institutes of Health grant (NCI Intramural Research Program). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: TCR, T cell receptor; MHC, major histocompatibility complex; DN, double negative; DP, double positive; SH2, Src homology 2; SP, single positive; ERK, extracellular signal-regulated kinase; PLC-γ1, phospholipase C-γ1; Ab, antibody; WT, wild type.
References
- 1.Mazza, C., and Malissen, B. (2007) Semin. Immunol. 19 225-235 [DOI] [PubMed] [Google Scholar]
- 2.Yamasaki, S., and Saito, T. (2007) Trends Immunol. 28 39-43 [DOI] [PubMed] [Google Scholar]
- 3.Koretzky, G. A., and Myung, P. S. (2001) Nat. Rev. Immunol. 1 95-107 [DOI] [PubMed] [Google Scholar]
- 4.Laky, K., Fleischacker, C., and Fowlkes, B. J. (2006) Immunol. Rev. 209 274-283 [DOI] [PubMed] [Google Scholar]
- 5.Clements, J. L., Yang, B., Ross-Barta, S. E., Eliason, S. L., Hrstka, R. F., Williamson, R. A., and Koretzky, G. A. (1998) Science 281 416-419 [DOI] [PubMed] [Google Scholar]
- 6.Kadlecek, T. A., van Oers, N. S., Lefrancois, L., Olson, S., Finlay, D., Chu, D. H., Connolly, K., Killeen, N., and Weiss, A. (1998) J. Immunol. 161 4688-4694 [PubMed] [Google Scholar]
- 7.Zhang, W., Sommers, C. L., Burshtyn, D. N., Stebbins, C. C., DeJarnette, J. B., Trible, R. P., Grinberg, A., Tsay, H. C., Jacobs, H. M., Kessler, C. M., Long, E. O., Love, P. E., and Samelson, L. E. (1999) Immunity 10 323-332 [DOI] [PubMed] [Google Scholar]
- 8.Pivniouk, V., Tsitsikov, E., Swinton, P., Rathbun, G., Alt, F. W., and Geha, R. S. (1998) Cell 94 229-238 [DOI] [PubMed] [Google Scholar]
- 9.Koretzky, G. A., Abtahian, F., and Silverman, M. A. (2006) Nat. Rev. Immunol. 6 67-78 [DOI] [PubMed] [Google Scholar]
- 10.Griffiths, E. K., and Penninger, J. M. (2002) Sci. STKE 2002, RE3. [DOI] [PubMed]
- 11.Kumar, L., Pivniouk, V., de la Fuente, M. A., Laouini, D., and Geha, R. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 884-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myung, P. S., Derimanov, G. S., Jordan, M. S., Punt, J. A., Liu, Q. H., Judd, B. A., Meyers, E. E., Sigmund, C. D., Freedman, B. D., and Koretzky, G. A. (2001) Immunity 15 1011-1026 [DOI] [PubMed] [Google Scholar]
- 13.Thien, C. B., and Langdon, W. Y. (2005) Biochem. J. 391 153-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naramura, M., Kole, H. K., Hu, R. J., and Gu, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 15547-15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thien, C. B., Bowtell, D. D., and Langdon, W. Y. (1999) J. Immunol. 162 7133-7139 [PubMed] [Google Scholar]
- 16.Chiang, Y. J., Sommers, C. L., Jordan, M. S., Gu, H., Samelson, L. E., Koretzky, G. A., and Hodes, R. J. (2004) J. Exp. Med. 200 25-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers, M. D., Dragone, L. L., and Weiss, A. (2005) J. Cell Biol. 170 285-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan, M. S., Sadler, J., Austin, J. E., Finkelstein, L. D., Singer, A. L., Schwartzberg, P. L., and Koretzky, G. A. (2006) J. Immunol. 176 2430-2438 [DOI] [PubMed] [Google Scholar]
- 19.Kaye, J., Hsu, M. L., Sauron, M. E., Jameson, S. C., Gascoigne, N. R., and Hedrick, S. M. (1989) Nature 341 746-749 [DOI] [PubMed] [Google Scholar]
- 20.Shortman, K., Wilson, A., Pearse, M., Gallagher, P., and Scollay, R. (1988) Immunol. Cell Biol. 66 423-433 [DOI] [PubMed] [Google Scholar]
- 21.Nakayama, T., Singer, A., Hsi, E. D., and Samelson, L. E. (1989) Nature 341 651-654 [DOI] [PubMed] [Google Scholar]
- 22.Alonso, A., Bottini, N., Bruckner, S., Rahmouni, S., Williams, S., Schoenberger, S. P., and Mustelin, T. (2004) J. Biol. Chem. 279 4922-4928 [DOI] [PubMed] [Google Scholar]
- 23.Fagerholm, S., Hilden, T. J., and Gahmberg, C. G. (2002) Eur. J. Immunol. 32 1670-1678 [DOI] [PubMed] [Google Scholar]
- 24.Manetz, T. S., Gonzalez-Espinosa, C., Arudchandran, R., Xirasagar, S., Tybulewicz, V., and Rivera, J. (2001) Mol. Cell. Biol. 21 3763-3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunnell, S. C., Singer, A. L., Hong, D. I., Jacque, B. H., Jordan, M. S., Seminario, M. C., Barr, V. A., Koretzky, G. A., and Samelson, L. E. (2006) Mol. Cell. Biol. 26 7155-7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seet, B. T., Berry, D. M., Maltzman, J. S., Shabason, J., Raina, M., Koretzky, G. A., McGlade, C. J., and Pawson, T. (2007) EMBO J. 26 678-689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, M. S., Smith, J. E., Burns, J. C., Austin, J. E., Nichols, K. E., Aschenbrenner, A. C., and Koretzky, G. A. (2008) Immunity 28 359-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogin, Y., Ainey, C., Beach, D., and Yablonski, D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6638-6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas, J. A., Atherly, L. O., and Berg, L. J. (2002) J. Immunol. 168 6142-6151 [DOI] [PubMed] [Google Scholar]
- 30.Han, J., Das, B., Wei, W., Van Aelst, L., Mosteller, R. D., Khosravi-Far, R., Westwick, J. K., Der, C. J., and Broek, D. (1997) Mol. Cell. Biol. 17 1346-1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawash, I. Y., Kesavan, K. P., Magee, A. I., Geahlen, R. L., and Harrison, M. L. (2002) J. Biol. Chem. 277 5683-5691 [DOI] [PubMed] [Google Scholar]
- 32.Rao, N., Miyake, S., Reddi, A. L., Douillard, P., Ghosh, A. K., Dodge, I. L., Zhou, P., Fernandes, N. D., and Band, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3794-3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naramura, M., Jang, I. K., Kole, H., Huang, F., Haines, D., and Gu, H. (2002) Nat. Immunol. 3 1192-1199 [DOI] [PubMed] [Google Scholar]
- 34.Balagopalan, L., Barr, V. A., Sommers, C. L., Barda-Saad, M., Goyal, A., Isakowitz, M. S., and Samelson, L. E. (2007) Mol. Cell. Biol. 27 8622-8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.