Abstract
Despite the quantity of high-throughput data available nowadays, the precise role of many proteins has not been elucidated. Available methods for classifying proteins and reconstructing metabolic networks are efficient for finding global categories, but do not answer the biologist’s specific and targeted questions. Following Yamanishi [Yamanishi, Y, Vert, JP, Nakaya, A, and Kaneisha, M (2003). “Extraction of correlated clusters from multiple genomic data by generalized kernel canonical correlation analysis.” Bioinformatics 19, Suppl. 1, i323–i330] we used a kernel canonical correlation analysis (KCCA) to predict the role of the bacterial peptidase PepF. We integrated five existing data types: protein metabolic networks, microarray data, phylogenetic profiles, distances between proteins and incomplete two-dimensional-gel data (for which we propose a completion strategy), available for Lactococcus lactis to determine relationships between proteins. The predicted relationships were then used to guide our laboratory work which proved most of the predictions correct. PepF had previously been characterized as a zinc dependent endopeptidase [Nardi, M, Renault, P, and Monnet, V (1997). “Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis.” J. Bacteriol. 179, 4164–4171; Monnet, V, Nardi, M, Chopin, MC, and Gripon, JC (1994). “Biochemical and genetic characterization of PepF on oligoendopeptidase from Lactococcus lactis.” J. Bio. Chem. 269, 32070–32076]. Analyzing a PepF mutant, we confirmed its participation in protein secretion through a strong relationship between the signal peptidase I and PepF predicted by the KCCA. The global nature of our approach made it possible to discover pleiotropic roles of the protein which had remained unknown using classical approaches.
Although a lot of high-throughput data are accumulated in databases, a large proportion of known proteins remains uncharacterized until targeted experiments prove their role. In place of analyzing global data, the biologists usually run experiments based on their own knowledge. When the role of a protein is difficult to identify due to the absence of clue or due to inconclusive laboratory results, two different approaches can indeed be considered: run experiments to detect protein interactions or use existing data to predict relationships that guide experiments.
Besides classical experiments to determine the role of single proteins, methodologies based on two-hybrid approaches (Ito et al., 2001) and mass spectrometry of multiprotein complexes (Ho et al., 2002) have been developed to detect protein-protein interactions in yeast. The two-hybrid method requires multiple experimental steps (cloning of the genes into a prey and a bait vector, transformation of two-hybrid strains with both type of plasmids, mating reactions of all possible combinations and PCR of positive colonies to decode interactions) in order to obtain a satisfactory result (Ito et al., 2001). Knowledge in protein interactions in bacteria has not reached the same level as in yeast (Noirot and Noirot-Gros, 2004). One reason for this difference is certainly due to the fact that this method is time consuming and the results contain many false positives that have to be eliminated through further experiments or analysis. On the other hand, identification of multiprotein complexes needs to set up and run quite heavy experiments, i.e., immunoaffinity purification followed by SDS-PAGE electrophoresis and mass spectrometry (Ho et al., 2002).
Instead of using a technique like the two-hybrid or performing targeted experiments without a clear line of action, scientists can explore existing data, available in databases and unsupervised or supervised approaches to reconstruct protein networks. Several methods based on utilization of different data sources of high-throughput data have been proposed so far (Akesson et al., 2004; Qi et al., 2005; Covert et al., 2004; Aerts et al., 2006; Werhli and Husmeier, 2007).
The protein we are interested in is the zinc dependent oligoendopeptidase PepF. Its possible participation in protein turnover and sporulation had been evoked (Monnet et al., 1994; Nardi et al., 1997; Kanamaru et al., 2002), but no precise role had been determined. Nevertheless, the importance of the protein due to a double copy found in L. lactis NCDO 763 and L. lactis SK11 (Monnet et al., 1994; Siezen et al., 2005) is intriguing. PepF is found in nearly all low GC bacterial species: (Staphylococcus sp., Bacillus sp., Streptococcus sp., Lactobacillus sp., Mycoplasma sp., Clostridium sp., etc), Spyrochetes, Proteobacteria (Agrobacterium sp., Escherichia coli, Salmonella sp., Yersinia sp., etc.), Archaea (Halobacterium sp., Methanosarcina sp., etc), Protozoa (Plasmodium sp.), and others (Thermus sp., Deinococcus sp., Rhodopirellula sp., etc). The different studies done on PepF in several bacteria have shown it interfering in important cell functions and its inactivation having pleiotropic effects. It is important to specify the role of this widespread bacterial protein with an apparent global and pleiotropic function, in order to control and improve strains used as model organisms as well as in many industrial applications.
To determine the role of the oligoendopeptidase PepF we conducted a study in two parts: (1) inferring possible partners of the protein by a global statistical analysis of existing high throughput data and (2) validating the predicted possible relationships by experimental work. The inference of possible partners of PepF was obtained by constructing a network for all potential proteins coded in the Lactococcus lactis IL1403 genome, based on different types of data. The kernel canonical correlation analysis (KCCA) (Yamanishi et al., 2003) we used allowed us to obtain distances between all proteins of the bacterium and place PepF as well as other proteins of the organism in this network. To do this, we integrated four types of data available for all proteins fromL. lactis: microarray data, phylogenetic profiles, distances between genes (coding for all potential proteins of L. lactis) on the chromosome and two-dimensional (2D)-gel data, our standard data set being the protein metabolic network. For 2D-gel data, we designed a new kernel taking care of missing data in such a way that no proteins, available for the other types of data but not present in 2D gel data, have to be discarded from the learning set.
Using KCCA (Yamanishi et al., 2003) we defined 63 possible partners of PepF, belonging to numerous functional categories. Four of them were predominant: protein secretion, pyruvate metabolism, peptidoglycan synthesis and cell division. We experimentally validated PepF’s implication in most of these functions. The study of PepF negative mutants confirmed its participation in protein secretion as well as in other predicted functions.
MATERIALS AND METHODS
During the first phase of our study, we did a kernel canonical correlation analysis to predict possible relationships of the peptidase PepF with other proteins of L. lactis using the method of Yamanishi et al. (2003). In subsequent phases of the work, we compared pepF mutants with the wild type strain with special attention to the main functional categories the potential partners of PepF, inferred by the KCCA, belong to.
Inference of possible relationships by kernel canonical correlation analysis
The KCCA is based on classical canonical correlation analysis (CCA) used to measure linear relationships between two groups of variables y and z. The goal is to find linear combinations a1 and a2 of y and z that are maximally correlated: . The linear combinations are found by eigenvector decomposition and are ordered decreasingly. The KCCA is a regularized CCA based on kernels. This means that the two groups of variables y and z used in CCA are replaced by kernels, inner products between objects, the objects being in our case the proteins. More precisely, the data set S is represented by a square matrix of pair wise comparisons K=[k(x,x′)]x,x′∊S×S (Schölkopf et al., 2004). Then a classical CCA is done between the images of y and z. The advantage of using kernels is that many different data types can be represented as a comparison between objects and that once the kernel obtained different data types are represented in the same way and can be integrated into the same analysis.
We used five existing data types: protein metabolic networks, microarray data, phylogenetic profiles, distances between proteins and 2D-gel data available for L. lactis:
The protein metabolic network was constructed from networks representing several metabolic pathways obtained from the database KEGG (Kanehisa et al., 2002). We constructed a unique graph, composed of 333 proteins, our golden standard, with all available proteins in version KGMḺv0.6.
The available microarray data belong to studies on L. lactis IL1403 comparing mutants and wild type strains or two growth conditions. They were obtained in a database harbored at the server of the MIG department at INRA: http:∕∕genome.jouy.inra.fr∕efp∕base∕www and at the Gene Expression Omnibus at NCBI; we also used the data of Guedon et al. (2001). We treated 51 experiments possessing a different number of repetitions, making 115 hybridizations in total.
The genetic profiles (binary presence∕absence vectors for the genes of L. lactis) were constructed for all genes fromL. lactis IL1403. The presence of a gene was evaluated by similarity (BLAST) in 276 completely sequenced bacteria. The ARCT 0.9 program (http:∕∕genomics.senescence.info∕software∕) included in HAGR (de Magalhães et al., 2005) was used to construct the profiles. If the sequence similarity had an E-value lower than 10−5 the gene was declared present, otherwise the gene was declared absent from the organism. These data inform about the co-evolution of genes, which is possibly related to a common function.
The position of the genes on the chromosome has been used to calculate the distance between them. We calculated the number of base pairs between the end of one gene and the beginning of the next one, so as to use this measure as distance. This data type has been included because generally neighboring genes participate to the same function in bacteria.
We had 2D gel data from 13 experiences (1 or 2 repetitions) at our disposal, revealing the protein quantity (expressed in volume percentage) of proteins with an isoelectric point between 4 and 7, on two different strains: L. lactis IL1403, as all other data used, and L. lactis NCDO763. All gels were run in the bacterial biochemistry laboratory (Unité de Biochimie Bactérienne) at INRA (France). Data on ten of these gels had already been published (Guillot et al., 2003 and Gitton et al., 2005). We conducted a test of maximum mean discrepancy described by Borgwardt et al. (2006), in order to determine if the data of the strain NCDO763 could be used together with that of IL1403. The conclusion was that data from both strains belong to the same distribution and therefore can be used together. This type of data contains many missing pieces of data compared to the transcriptomic data, phylogenetic profiles and distance on the chromosome. We used a completion strategy in order to deal with the missing data.
To apply a kernel method such as the KCCA, the first step is to define a valid kernel for each type of data. Herein kernels are, as described before, gene similarity matrices. We have at our disposal information on the genes on one hand and the protein metabolic network on the other hand. In order to represent the undirected graph of the protein metabolic network we used a Laplacian exponential diffusion kernel (Kondor and Lafferty, 2002). For the other data we used Gaussian and polynomial kernels. The most appropriate kernels for each type of data are listed in Table 1. Before the KCCA was done, all kernels were normalized.
Table 1.
Kernels used for each type of data.
| Datatype | Kernel | |||
|---|---|---|---|---|
| Kernel type | Kernel function | Explanations | Parameters | |
| Metabolic network | Diffusion kernel | KI=eα1L | where i∼j means proteins i and j are joined in the graph and di is the number of proteinsjoined to gene i. | α10.01–0.1 |
| Phylogenetic profiles | Polynomial kernel | where Φ2 is a vector of features the size of which depends on the power of the polynomial inK2(x,y)=(⟨x,y)⟩+α2)d=Φ2(x)TΦ2(y)This kernel construction has been chosen to give a higher weight to the interactionsbetween two genes, than to the interactions of higher order. | α2=40 d=5 | |
| Distancebetween genes | Gaussian kernel | K3=e−α3∗dispos2 | where dispos is the distance in base pairs between the end and the beginning of two genes. | α30.0005 |
| Transcriptomic data | Gaussian kernel | K4=e−α4∗DD2 | where DD is the norm between the gene expression profiles. | α40.001–0.011 |
| 2D-gel data | Completion of a Gaussian kernel | where d is the norm betweenthe protein volume profiles.See the text for K5,a and K5,b. | α50.55 | |
Parameters α1, α3, α4, α5 as well as component number and regularization parameter (δ) in KCCA were determined with a grid search leave-one-out cross validation (see supplementary data). In each of the 333 iterations, the set of 333 proteins in the golden standard was split into a training set and a test set composed of one protein. The feature space was trained on the training set. The graph was built progressively and compared with the original protein metabolic network, the golden standard. The parameters retained were those that made it possible to find known relationships with the lowest error. The minimal error in regard of the test proteins was calculated using the false positives (f) and the true positives (h), . The most adequate values were chosen to minimize this error.
The 2D-gel data are the protein volumes of the observed protein spots. This quantity was normalized with the total protein volume on the gel to obtain a volume percentage for each protein. We transformed these quantities following the recommendations of Chich et al. (2007), transforming the volume percentage (%V) into T(%V) as follows: T(%V)=(%V)1∕3. If we denote n the number of proteins for which the information is available in the three datasets used to construct K2, K3, and K4, only n1<n proteins are present in the dataset used to construct K5.
Our strategy consists in completing kernel K5(n1×n1) to give kernel K5(n×n) of the same size of the other kernels. The most simple completion of K5 would be to replace the missing data by zero (K5zeros). This means that a neutral value (the mean similarity) replaces the missing similarity values after centering of the kernel: where n is the number of proteins present in all datasets and n1 the proteins detected on the 2D gels.
Nevertheless, we have some information about the missing data, i.e., the proteins not detected on the gel, helpful in completing this kernel in a more “informative” way. We propose to create a kernel where missing data will be completed with qualitative data taking into account the information we have about the missing proteins (K5quali). We know that some of the proteins are absent because it is not possible to detect them due to the experimental conditions, and that some others are absent but could have been detected. We supposed the proteins to belong to three object families: X=X1∪X2∪X3. X1 were the observed proteins, X2 were the observable proteins (with a pH between 4 and 7 in the case of our dataset) which were undetected on the gels and X3 were the proteins that were unobservable. We constructed a kernel for the objects belonging to X1, kernel . In order to complete the missing data we considered all possible interactions and replaced the missing value by a value reflecting the interactions between each type of pair (Table 2). The result is K5quali(n×n). The similarity between observed and observable proteins as well as between observable and non-observable (ε) was chosen to be 0.01. The similarity between two observable proteins (θ) was chosen to be 0.02. This means that K5quali(n×n) contains the values corresponding to the kernel for proteins detected on the gels and qualitative values or mean values for the proteins that were not detected (Table 2). Other possibilities for the data completion could be considered, for example, different values can be used instead of uniform values.
Table 2.
Construction of K5quali with values for the protein similarities based on the information about missing and available data. Each cell of the table contains the similarity between two proteins x and x′. The values to complete missing data were ε=0.01, θ=0.02, the mean similarity of the detected protein x∊X1 with other proteins inside X1 (mx) and the overall mean similarity for the comparison of two nonobservable proteins (X3). As the similarity between the protein and itself is maximal, the diagonal of K5quali is composed of ones.
| Observed X1 | ObservableX2 | Nonobservable X3 | |
|---|---|---|---|
| Observed X1 | ε | ||
| Observable X2 | ε | θ>ε | ε |
| Nonobservable X3 | ε | ||
At this point there was no guarantee that the K5quali was a positive definite kernel (PDK). This was achieved minimizing the Frobenius distance between K5quali and a PDK. The use of this distance has already been described in Yamanishi and Vert (2007). In the present case a PDK (K5) based on the original and the kernel K5qualiresults from minimizing the Frobenius distance K5=arg minK∊I‖K5quali−K‖, where I is the set of positive semidefinite matrix of size n×n. The resulting PDK is givenby:
The goal of KCCA is to find correlations between two datasets. One data set is the golden standard (KI) and the second data set is composed of microarray data, phylogenetic profiles, etc., aggregated as KII=K2+K3+K4. A representation of all proteins is constructed in a way that both data sources are as closely correlated as possible. The proteins making up part of each dataset, the protein metabolic network in one hand and of the integrated dataset on the other hand, are represented in a way that reflects the distance between them. To construct this representation we used the method proposed by Yamanishi et al. (2003). Given kernels KI and KII, to each x∊S corresponds a feature ΦI(x) [respectively ΦII(x)] belonging to a functional space H1 (respectively H2). We searched for a direction f1 in H1 (resp. f2 in H2) such that the generalized canonical correlation ρ(f1,f2) between uI(x)=⟨ΦI(x),f1⟩ and uII(x)=⟨ΦII(x),f2⟩ is maximized, where . It is possible to show that f1=∑x′∊Sαx′ΦI(x′), resp. f2=∑x′∊Sβx′ΦII(x′), and, resp. . So cov(uIuII)=αTKIKIIβ, , and ‖f1‖2=αTKIα, ‖f2‖2=βTKIIβ. Several orthogonal directions can be considered for summarizing the feature space. If we denote , resp. the directions, then gene x is represented by, resp. . Therefore the distance between gene x and gene x′ will be the Euclidian distance between uII(x) and uII(x′).
We verified the validity of different kernel combinations using the parameters obtained by the leave-one-out validation reconstructing known relations for more than one protein at the same time testing different combinations of kernels. The combination of all kernels, which gave the least false positives, was used to make predictions on our protein of interest, PepF.
To define the possible partners of PepF, the threshold was chosen as follows: the highest distance found between proteins known to be neighbors in the protein metabolic network was calculated and chosen to be the maximum distance to accept a relationship between two proteins.
Experimental validation of possible relationships
The bacterial strains and plasmids used in this study are listed in Tables 3, 4. L. lactis strains were grown at 30 °C in M17 (Difco) medium supplemented with 5% glucose. The chemical minimal medium contained only seven amino acids essential for all lactococci (Cocaign-Bousquet et al., 1995) as well as arginine (1.2 g∕l) and threonine (2.3 g∕l) essential for L. lactis IL1403.
Table 3.
Bacterial trains used in this work.
| Strain | Plasmid content | Resistance | Reference |
|---|---|---|---|
| E. coli TG1 repA+ | pGhost9-pepF deleted (pTIL 120) | Ery | Nardi et al. (1997) |
| L. lactis IL1403 | — | — | — |
| L. lactis IL1403 Δ pepF | — | — | This work |
| L. lactis IL1403 Δ pepF comp | pILN13-pepF low copy | Ery | This work |
| L. lactis NZ9000-pSEC1 | pSEC1 | Cm | de Ruyter et al. (1996),Chatel et al. (2001) |
| L. lactis NZ9000 Δ PepF | pSEC1 | Cm | This work |
| L. lactis NZ9000 Δ pSEC1 comp | pSEC1+pILN13-pepF low copy | Ery | This work |
| L. lactis NZ9000-pSEC1+pepF | pSEC1+pILN13-pepF high copy | Cm, Ery | This work |
| E. coli BL21 (DE3) Gold | — | — | Stratagen |
| E. coli BL21 (DE3) Gold-pET | pET28-pepF | Km | This work |
Table 4.
Plasmids used in this work.
| Plasmid | Characteristics | Resistance | Reference |
|---|---|---|---|
| pTIL 120 | pGhost 9-pepF deleted: 162 b.p. deletion including the active site | Ery | Nardi et al. (1997) |
| pSEC1 | expression under PnisA encodes SPUsp:NucB | Cm | Chatel et al. (2001) |
| pILN13 | allows switch to low copy number | Ery | Renault et al. (1996) |
| pET28 | conceived for heterologous protein production | Km | Novagen |
The following antibiotics were added as selective agents when appropriate: erythromycin (5 μg ml−1 for L. lactis, 150 μg ml−1 for E. coli), chloramphenicol (5 μg ml−1 forL. lactis; 20 μg ml−1 for E. coli) and ampicillin (100 μg ml−1 for E. coli), kanamycin (20 μg ml−1 for E. coli).
Molecular cloning techniques were performed using standard procedures (Sambrook and Russel, 2001). Plasmids were extracted by using a QIAprep Spin miniprep kit (Qiagen). Total L. lactis and E. coli DNA was isolated as described previously (Hoffman and Winston, 1987). Restriction enzymes (New England Biolabs), T4 DNA ligase from the Fast-link ligation kit, (Epicentre), Taq DNA polymerase MP (Qbiogene) and the TripleMaster PCR system (Eppendorf) were used according to the suppliers’ recommendations. PCRs were run using a Mastercycler gradient thermal cycler (Eppendorf). All constructions were verified by sequencing with an Applied Biosystems 310 automated DNA sequencer using the ABI PRISM Dye Terminator Cycle Sequencing Kit (Perkin Elmer). Primers for L. lactis were selected on IL1403 (Bolotin et al., 2001) and for E. coli on K12 (Blattner et al., 1997) genome sequences. The oligonucleotides were purchased from Invitrogen. Annealing was performed between 52 and 60 °C, depending on the primers used.
We constructed pepF mutants by replacement with a deleted pepF gene as explained in Nardi et al. (1997), electroporating pTIL120 into L. lactis IL1403 and NZ9000. Integration of pTIL120 into the chromosome and subsequent excision was achieved using the thermosensibility of the plasmid. Mutant strains were screened first on their resistance to erythromycin and second on the size of the fragment amplified by PCR with primers FpepF and RpepF (Table 5). The presence of a correct insertional event and absence of the vector was further verified by Southern blotting using and ECL detection system (Amersham).
Table 5.
Primers used in this work (the introduced restriction enzyme sites are underlined)
| Primer name | Sequence |
|---|---|
| FpepF | GCGGATATTAAGTTACCTATGGT |
| RpepF | TTTGGCAATTACTTCTAAAGGAT |
| PetPepF-For | CATGCCATGGTTGCTAAGAATAGAAATGAAAT |
| PetPepF-Rev | GGAAGATCTAAGATGGACTCCTTTTTCAA |
| PilPepF-For | CTGCAGGCAGAAGAAGGATATGAATGAATG |
| PilPepF-Rev | GCGGCCGCATTTTTAAAGATGGACTCCTTTTTCAAC |
A sequence corresponding to pepF was amplified by PCR from L. lactis IL1403 total DNA with the primers PetPepF-For and PetPepF-Rev (Table 5) containing NcoI and BglII sites. PCR fragments were digested and cloned in-frame upstream of the hexa-His pET28 vector (Novagen). E. coli BL21 (DE3) Gold competent cells were transformed with the resulting plasmids.
The whole pepF gene with its promoter region was introduced into the plasmid pILN13 (or pILNew) (Renault et al., 1996). PILNew is a high copy number plasmid. It is possible to transform it into a low copy number plasmid restoring the replication repressor by a KpnI restriction and further ligation. We used this construction for two purposes: (i) in high copy number to study the effects on protein secretion of the overproduction of PepF in the NZ9000-pSEC1 strain and (ii) in low copy number to complement our mutants. The primers used to amplify the gene with its promoter region and to introduce the needed restriction sites, PstI and NotI, were PilPepF-For and PilPepF-Rev (Table 5).
Cellular extracts were prepared to analyze PepF activity, presence of peptides, and metabolites of the pyruvate metabolism. Once the cultures had been harvested at OD600 nm=0.6 by centrifugation and washed with phosphate buffer 0.2 M, cell lysis was achieved with a cell disruptor (Constant System Ltd). The cytoplasmic fraction was obtained by ultracentrifugation at 4 °C and 50 000 rpm for 20 min (Centrikon T-1080, Kontron intruments).
PepF activity was measured by its hydrolytic activity with a fluorescent quenched substrate: Mc-Pro-Leu-Gly-Pro-Lys-(DNP)OH. The fluorescence is emitted when the peptide is cleaved (Tisljar et al., 1990) and was followed over 100 s on a spectrofluorometer 25 (Kontron instruments).
Pyruvate, acetate, lactate and formate in cell extracts were quantified by HPLC on an Aminex-HPX-87H column (BioRad) with an isocratic elution with 5 mM H2SO4 at a flow rate of 0.35 ml∕min at 35 °C. Proteins had previously been precipitated with H2SO4 (2% final concentration). The peak surfaces obtained were integrated and the quantity of acid was calculated by comparison with the calibration curve of each acid of interest.
Peptidoglycan from L. lactis IL1403 and its pepF mutant was prepared from an exponentially growing culture (OD600 nm=0.3) according to the protocol of Atrih et al. (1999). Briefly, cells were boiled in 4% (w/v) sodium dodecyl sulfate (SDS) for 30 min. The insoluble cell wall was washed six times with distilled water to wash out the SDS. To remove the proteins, the cell wall pellet was treated with Pronase (200 μg ml−1) for 16 h at 37 °C, then with trypsin (200 μg ml−1) for 16 h at 37 °C. The cell wall was then treated with fluorohydric acid to eliminate teichoic acids. Once a final digestion with muramidase had been completed, the muropeptides were reduced with borate and analyzed by HPLC on a C18 Hypersyl PEP100 column. Muropeptides were identified by comparison with a standard in which peptides had been identified by mass spectrometry. Once the number of the relative quantity (peak surface) of dimers (d), trimers (t), and tetramers (te) of muropeptides had been obtained, a cross-linking index reflecting the peptidoglycan reticulation level was calculated as follows (Glauner, 1988): Cross-linkage=1∕2∑d+2∕3∑t+3∕4∑te∕∑muropeptides.
Secretion was studied exclusively in the strain NZ9000-pSEC1 which allows the induction of a secreted nuclease (NucB) of Staphylococcus aureus possessing the signal peptide of the lactococcal protein Usp45 and under control of the nisin promoter. As NZ9000 possesses the nisRK genes on its chromosome it is possible to induce the production and further secretion of NucB by addition of nisin to the growth medium. We tested four different nisin doses: 1, 2.5, 5, and 7.5 ng∕ml. Protein cellular extracts were prepared from 200 ml culture after 3 h nisin induction, which we had started at OD600 nm=0.5. Cell lysis was achieved by disruption (Constant System Ltd.) in NaP04 20 mmol buffer containing protease inhibitor cocktail P8465 (Sigma-Aldrich). After ultracentrifugation, the supernatant contained the intracellular extract and the pellets containing the envelope fraction were resuspended in the same NaP04 buffer. In order to compare the profiles between wild type and mutant, with induction of 5 ng∕ml nisin, a constant number of cells was analyzed on 4–12% polyacrylamide gels (NuPAGE).
The secreted proteins were prepared from 5 ml supernatant (after centrifugation of 6 ml), precipitated with TCA (20% final concentration), incubated for 30 min at 4 °C and centrifuged for another 30 min at 4 °C. Once the supernatant had been eliminated, the pellet was washed with cold acetone, air dried and resuspended in 500 μl NaOH 50 mM before analysis on 4–12% polyacrylamide gels (NuPAGE).
This test is based on the measurement of mono- and dinucleotides released by the DNA hydrolysis activity of NucB. The incubation was carried out at 37 °C in 500 ml buffer (Tris 25 mM pH 8,8; CaCl2 10 mM; BSA 0,1 mg∕ml) containing 1 mg∕ml sonicated salmon sperm DNA (Sigma) with 10 μl supernatant. The reaction was stopped by the addition of perchloric acid, which precipitates non-hydrolyzed DNA. After incubation for 15 min and centrifugation for 7 min, the optical density corresponding to the liberated nucleotides was measured at 260 nm.
The first step in determining the accumulation of the signal peptide was a separation of intracellular and envelope proteins of wild type and mutant by one-dimensional sodium dodecyl sulfate-polyacrylamide (4–12%) gel electrophoresis (SDS-PAGE) using the NuPage system (Invitrogen). We then cut the gel in the molecular weight range between 0 and 6 kDa. Peptides were obtained from these fragments through three subsequent washes with ACN 50% followed by TFA 0.1% without digestion. The obtained peptides were pooled and dried in a SpeedVac concentrator for 1 h, and then resolubilized in 25 μl of HPLC loading buffer (0.08% TFA and 2% ACN) and then analyzed by LC-MS-MS. The peptide mixtures (4 μl) were injected onto the precolumn PepMap C18 (300 μm ID×5 mm, 100 Å) with a flow rate of 20 μl∕min to remove salts. The peptides were analyzed in a 50 min gradient of 2–80% of acetonitrile in water containing 0.1% formic acid. A flow rate of 300 nl∕min was used to elute peptides from the C-18 PepMap100 reversed-phase nanocolumn (75 μm ID×15 cm, 3 μm, 100 Å) (LC Packings, Amsterdam, The Netherlands) to a PicoTipTMEMITER nanospray needle (360 OD×20 μm, 10 μm ID) (New Objective, USA) for ionization and peptide fragmentation on an ion trap mass spectrometer. MS/MS spectra were acquired for the 200–2000 m∕z range and batch processed by using Bioworks 3.2 software packages and searched against the L. lactis MG1363 (NZ9000 being a derivate of this strain) protein database using SEQUEST software.
A culture of E. coli containing the plasmid to induce heterologous production of PepF-6histidines under the T7 promoter was done in LB medium. The production was induced by adding IPTG at a final concentration of 1 mM to the culture at an OD650 nm of 0.5. Bacteria were grown at 37 °C until IPTG addition and were then transferred at 30 °C during the expression time (4 h) to avoid the formation of inclusion bodies. The cells were harvested by centrifugation and broken by one passage at a pressure of 1600 bar with a Constant Cell Disruption System. The soluble fraction containing the recombinant protein was collected by centrifugation at 15 000 g for 15 min at 4 °C. The hexa-His-tagged proteins were purified by affinity chromatography on Ni2+-nitrilotriacetic acid spin columns (Qiagen) according to the manufacturer’s instructions.
For the localization of PepF in the cell, polyclonal antibodies raised against PepF were produced by PARIS (Production d’Anticorps, Réactifs Immunologiques & Services, Compiegne, France). Once the antibodies had been tested by a Western Blot, cell cultures were fixed in 4% paraformaldehyde, then dehydrated for 1 h in ethanol 30% (at 4 °C), 50%, 70%, (at −20 °C), 90% and 100% (each at −35 °C) and immersed at −35 °C for 3 h in three baths of Lowicryl K4M (Delta microscopies-Labège-France)/ethanol 100% (1v∕2v, 1v∕1v, and 2v∕1v, respectively), followed by two baths in Lowicryl K4M (16 h and 2 h). Polymerization was done at 320 nm for 48 h at −35 °C, and increased temperature, for three days, up to 20 °C following Leica AFS procedure (Leica-Microsystems Rueil-Malmaison—France). Thin sections (90 nm) were mounted on nickel grids. After blocking reaction in buffer containing polybutene sulfone (PBS)-1% BSA-0.1% cold water fish skin gelatin, thin sections were incubated for 2 h at room temperature with the PARIS antibody raised against PepF, diluted 1:100 from solution at 4.45 mg∕ml, in buffer containing 0.1% PBS and 0.1% BSAc (Aurion-BioValley-France). The grids were rinsed in the same buffer and incubated for 30 mn in protein A (1:20) conjugated with gold particles of 10 nm (Aurion-BioValley-France). Once PepF molecules were observable, 50 independent cells of each culture (L. lactis NZ9000 nisin induced and not induced) were observed and the quantity of marked cells and of molecules per cell was counted.
To study cell division, cultures of IL1403, NZ9000-pSEC1 and their pepF mutants were harvested and fixed chemically with a solution of 2% glutaraldehyde and 0.1M sodium cacodylate, included in resin Epon, and then cut at room temperature. The cultures of NZ9000-pSEC1 were induced for 2 h with 5 ng∕ml nisin, which we started at OD600=0.3. All sections were examined with a Zeiss EM902 electron microscope operated at 80 kV and images were acquired with a charge-coupled device camera (Megaview III) and analyzed with ITEM Software (Eloïse, France; MIMA2 Platform, INRA-CRJ).
RESULTS
Our results include (1) the inference: validation of the KCCA on known relationships and (2) application of this method to infer new relationships and the experimental validation of the predicted relationships.
Inference
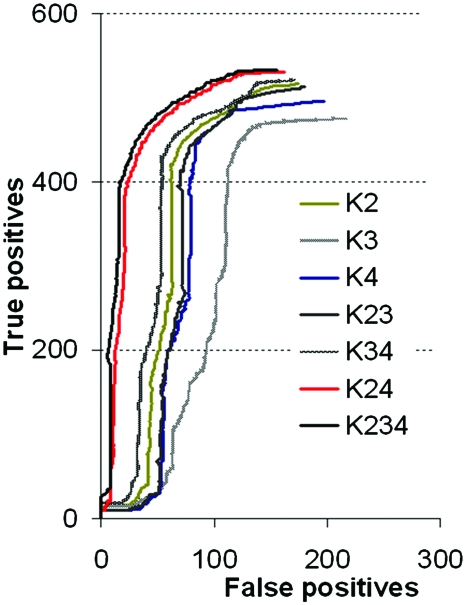
The protein metabolic network constructed from the metabolic pathways existing in KEGG for L. lactis contains 333 proteins. In order to test the performance of each kernel we evaluated their ability to reconstruct the known protein metabolic. In Fig. 1 we plotted the mean false positives against the mean true positives until the expected number of arrows (present in the known network) was placed. When kernels were used alone, their performances were not good, for example, when K3 was used alone the highest number of false positives was found. The combination of the kernels K2, K3, and K4 turned out to be the best one (Fig. 1) and was used later on to make the predictions for PepF.
Figure 1. Kernel performance represented by their capacity to find known relationships between proteins (true positives) in comparison with the wrongly detected relationships (false positives).
K2: polynomial kernel constructed on the phylogenetic profiles; K3: Gaussian kernel constructed on the distance between genes on the chromosome; K4: Gaussian kernel constructed on the gene expression profiles from microarray data. The combination of kernels (i.e., K23) was done by the addition of each kernel: K2+K3.
In order to evaluate the performance of K5 (proteomic kernel), we worked with a smaller group of 104 proteins for which 2D-gel data were available. We constructed the proteomic kernel K5ori for this group of proteins and tested its performance to reconstruct the network obtaining an error (percentage of false positives) of 0.385. We then split this dataset into two parts and used only 54 proteins to construct the kernel. Using the strategy of kernel completion, we constructed K5Q and obtained an error of 0.394. The completed kernel K5 has the highest error in comparison with the kernels constructed for the other data: K2, K3, and K4 (error=0.294 for K2; 0.356 for K3; and 0.256 for K4, Fig. 1). It should be noticed that all kernels, if used alone, have a low performance. The best results were obtained when data were fused by summation. Using all kernels together, we decreased the error from 0.18 (obtained for K234, Fig. 1) to 0.17 (obtained for K2345).
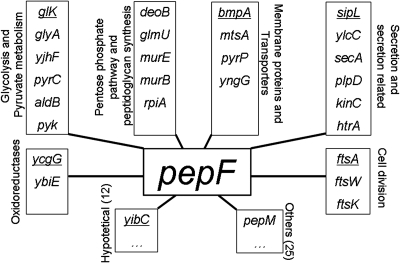
Using three kernels (K2, K3, and K4) (Table 1) 63 proteins were found to be potentially related to PepF (Fig. 2); see supplementary data for complete list of predicted relationships and exact distances. This means that the distance to these 63 proteins was below the chosen threshold (maximal obtained distance between two proteins known to be related on the protein metabolic network). Using the four kernels (K2, K3, K4, and K5) we found very similar results: 65 proteins potentially related to PepF; 61 proteins belong to the 63 found using K2, K3, and K4, and four were proteins which had not been identified before: MalE, DfP, ArsC, and FtsQ (see supplementary data).
Figure 2. Graphical representation of the predicted relationships found for PepF by KCCA organized by functional categories.
Experimental validation was done for the most represented functional groups. Proteins belonging to the first ten relationships of PepF and represented here are underlined. For the exact distances please refer to the supplementary data.
The proteins which were found to be related to PepF can be divided into two main categories: (i) proteins belonging to known metabolic pathways (represented on the network) and (ii) proteins not belonging to known metabolic pathways (not represented on the network). The relationships of PepF to proteins of the first category belong principally to two metabolic pathways: pyruvate metabolism and peptidoglycan synthesis. In the second category, we found enzymes responsible for cell division and protein secretion that are strongly represented. Moreover, the strongest relationship (the shortest distance) was found to the signal peptidase SipL. This relationship was reinforced by several common relations of these two enzymes with proteins belonging to both categories mentioned above (Fig. 2).
Experimental validation of predicted relationships for PepF
In order to determine possible relationships of PepF with the predicted enzymes, we constructed two deletion mutants of pepF in the L. lactis IL1403 and NZ9000 strains and compared the phenotypes of wild type and mutant. The L. lactis IL1403 strain is the strain for which the data for the predictions was used; L. lactis NZ9000 contains the pSEC1plasmid (Chatel et al., 2001) that carries the gene coding for an exported nuclease. We used this strain because current lactococci export only few proteins and we wanted to test our hypothesis in a strain where secretion could be boosted and regulated. This construction allowed us to overproduce a secreted nuclease (NucB) which had the signal peptide of Usp45 (sp45), the naturally most secreted protein in lactococci (Chatel et al., 2001). As nucB expression depended on a nisin inducible promoter, we were able to determine the effect of the absence of PepF in high secretion conditions.
The approach of Tisljar et al. (1990) using a quenched fluorescent substrate was used to assess the absence of PepF activity in the pepF mutant and pepF overproducing strain, as well as to check that activity was restored in complemented mutants (see supplementary data).
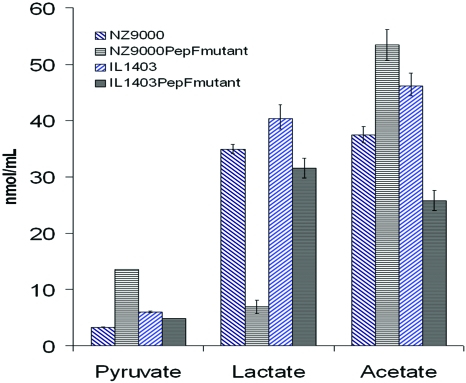
We analyzed the acids present in the supernatant by HPLC (Fig. 3). We observed that the PepF mutant strains produce less lactate. In L. lactis NZ9000-PSEC1 the pyruvate quantity of the mutant increases in comparison with the wild type. Modifications of acetate quantities were also observed but the two strains behave in different ways. These results confirmed an alteration of the pyruvate metabolism in the absence of PepF.
Figure 3. Quantity (in nm∕ml) of acids of the pyruvate metabolism in the culture supernantant determined by HPLC comparing wild type (blue) and mutant strains (black) of L. lactis IL1403 and NZ9000-PSEC1.
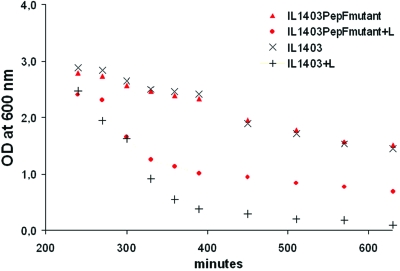
For L. lactis IL1403 we found that, in rich media, the mutant showed a more reticulated peptidoglycan. The relative measurement of dimers, trimers, and tetramers of muropeptides of peptidoglycan makes it possible to calculate the cross-linking index which was higher in the pepF mutant (45.26) than in the wild type (35.36), indicating a higher reticulation level of the cell wall in the mutant. In regards to the differences in peptidoglycan composition, we studied the resistance of mutant and wild type to 1 mg∕ml lysozyme. As expected, the mutant, with a more reticulated peptidoglycan, was more resistant than the wild type to lysozyme (Fig. 4).
Figure 4. Response of L. lactis IL1403 wild type and pepF mutant cultures at stationary phase to 1 mg∕ml lysozyme (+L).
The cell densities were measured at an optical density of 600 nm.
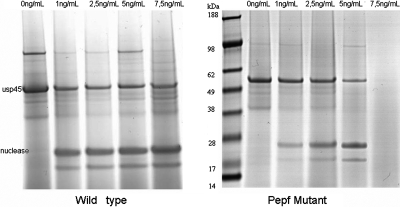
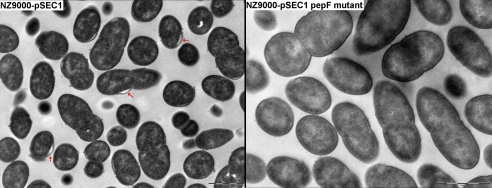
We measured the activity of the nisin induced secreted nuclease NucB in the supernatant and we did SDS-page gels of secreted proteins. We observed that the export of proteins was negatively affected in the pepF mutant when a high quantity of nuclease was produced (induction with 5 and 7.5 ng∕ml of nisin) (Fig. 5) and, correlated with this result, we observed a rundown in the nuclease activity in the supernatant (data not shown). We confirmed that this phenomenon was only provoked by the absence of PepF, since in the complemented mutant, secretion was restored. We also tested the effect of an overproduction of pepF by transforming L. lactis NZ9000-pSEC1 with a high copy number plasmid carrying the pepF gene. No obvious difference was observed in the overproducing strain, suggesting that the quantity of PepF present in the wild type was not limiting. Additionally, the observation of strain NZ9000-PSEC1 by electronic microscopy allowed us to observe a detachment of the cell wall from the cytoplasm (Fig. 6), which we attributed to the strongly induced secretion of the nuclease. It was possible to observe that this detachment did not occur in the mutant strain, in which, as we know, protein secretion was diminished or even absent.
Figure 5. SDS-PAGE electrophoresis of secreted proteins at different nisin concentrations comparing the wild type NZ9000-pSEC1 nuclease overproducing strain with its pepF mutant.
Figure 6. Electronic microscopy observations of L. lactis NZ9000-PSEC1 showing the detachment of the cell wall (gray arrows) in the nuclease overproducing strain at 5 mg∕ml nisin induction compared to its pepF mutant.
The analysis of wild type and mutant strains allowed us to detect a peptide in the low molecular weight fraction of the cell cytoplasm of the pepF mutant corresponding to the first ten amino acids of the signal peptide of Usp45. This signal peptide is the most abundant signal peptide present in the cell because of the induction of the expression of the nuclease having the signal peptide of Usp45. The sequence of this peptide is: MKKIISAILMSTVVLSAAAPLSGVYA. The detected peptide was: MKKIISAILM with two variants corresponding to different oxidation states of methionin: MKKIISAILMox (646.6837 kDa) and MoxKKIISAILMox (654.2787 kDa). These peptides were undetected in the wild type’s same fraction.
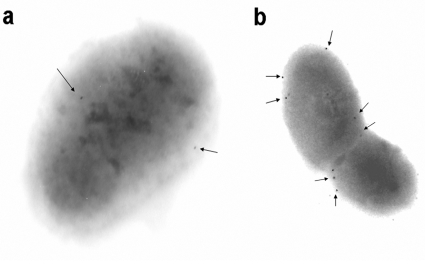
We were able to observe PepF in the periphery of the cell, in L. lactis NZ9000-pSEC1 (Fig. 7). Furthermore, in the nisin induced culture of NZ9000-pSEC1 we observed a higher percentage of bacterial sections with at least one PepF labeled molecule: approximately 70% in comparison to 30% of the non-induced ones. The number of PepF molecules detected in the not induced culture is also lower (Fig. 7).
Figure 7. Electronic microscopy observations of the immunogold-labeled peptidase PepF in L. lactis NZ9000-PSEC1.
When secretion is not induced PepF (a) then after induction with 5 ng∕mL nisin (b). The arrows indicate gold-labeled PepF molecules.
As far as cell division is concerned (i.e., morphology of the septum) no differences were observed in the electronic microscopy observations.
DISCUSSION
This research sought to assess the role of the bacterial peptidase PepF using a global statistical approach to guide experimental studies. Our approach took advantage of existing knowledge since available heterogeneous data had been analyzed before experimental work was started. The approach allowed us (i) to discover a global role of PepF and decipher consequences of the principal function of this protein and (ii) guide laboratory work in order to avoid useless and time consuming experiments.
The results obtained during the validation of the KCCA with known proteins making up part of the metabolic network proved the power of the method. The error rates on regaining the known protein metabolic networks are similar to the ones obtained by Yamanishi et al. (2003) and Yamanishi and Kanehisa (2004). The good quality of predictions is certainly due to both the possibility of integrating more than one data type and the training with a known network by correlation to the metabolic network. The use of different data types and the fact that we were able to introduce even data with missing values, allowed us to improve predictions and added even more flexibility to the KCCA than before. The strategy we introduce can be a starting point for any type of missing data. The kernel completion we propose for 2D gel data can be improved by the use of different similarity values instead of uniform values, for example.
The KCCA predictions allowed us to find a strong relationship with secretion proteins that had not been evoked in previous studies on PepF. SipL, the protein that was predicted to be the closest to PepF (see supplementary data) is the signal peptidase I that cleaves the signal peptide from secreted proteins other than lipoproteins. In the model for B. subtilis proposed by Tjalsma et al., (2000) the signal peptidase SipL cleaves the signal peptide of secreted proteins at the moment of translocation. SipL is thought to cut this peptide into two parts again separating both parts and allowing the hydrophilic fragment to reach the cytoplasm (Tjalsma et al., 2000), where the degradation activity by signal peptide peptidases (sppases) takes place (Novak et al., 1982). Using a pepF mutant we determined that PepF is needed to achieve the secretion of proteins. The blockage of protein secretion in the mutant and the presence of a hydrophilic fragment of the signal peptide in the pepF mutant support the hypothesis that PepF is a signal peptide peptidase (sppase). Its principal function would consist in hydrolyzing and recycling the liberated signal peptide. As PepF is an endopeptidase that hydrolyses peptides between 7 and 17 amino acids (Nardi et al., 1997) the hydrophilic fragment of the signal peptide is in its range of action. Furthermore, an inhibition of secretion by signal peptides has been observed in E. coli (Chen et al., 1987; Wicker et al., 1987). We think that the blockage of protein secretion observed in our pepF mutant is due to an accumulation of fragments of the signal peptide. As growth is not affected in the absence of PepF and the effects on protein secretion are only observed during strong induction, it seems that PepF is not the sole sppase in L. lactis. Similarly, two sppases exist in B. subtilis: the membrane bound SppA (yteI) and the cytoplasmic TepA (Tjalsma et al., 2000). The fact that we observed PepF localized in the periphery of the cell reinforces its participation in protein secretion, which occurs in the cell membrane. The fact that PepF is more abundant when secretion is induced also confirms its role in this process. In light of our results, the presence of a second copy of pepF on the lactose plasmids of L. lactis NCDO763 strains (Monnet et al., 1994; Siezen et al., 2005) together with proteins needed for growth in milk, as, for example, the cell-envelope- protease that has to be secreted and the peptide transport system OppCBFD (Siezen et al., 2005) becomes clearer. In a general manner, it is not surprising that an additional copy of pepF is required for the proper localization of membrane and surface proteins implicated in casein processing and peptide transport. Among the membrane proteins with possible relationships to PepF we found, in one of the first positions, BmpA, a basic membrane lipoprotein of unknown function that is in fact an outer membrane in Borrelia burgdorferi (Shin et al., 2004) and thus has to undergo secretion in this organism. In L. lactis it possesses a signal peptide of 27–30 amino acids as predicted by SignalP 3.0 (Bendtsen, 2004). It is therefore not surprising that this possibly secreted protein has a strong relationship with PepF.
OpdA from E. coli is a protein similar to PepF showing 53% similarity around the active site (positions in the amino acid sequence 378–439 and 457–525 of PepF and OpdA, respectively). OpdA has been described as being a possible sppase (Dev and Ray, 1990; Novak et al., 1982; Ichihara et al., 1984). OpdA mutants affect the secretion of several proteins (Emr and Bassford, 1982; Emr and Silhavy, 1980; Conlin et al. 1992). We tried to complement our pepF mutants with opdA from E. coli to prove that both proteins have the same function but opdA seems to be toxic in L. lactis, because we were not able to obtain cells containing the pILN13 plasmid containing the opdA gene (data not shown).
When studied in several bacteria, both PepF and OpdA were implicated in several functions. We have shown that peptidoglycan structure was modified in the pepF mutant in rich medium. We attribute this change to a collateral response to the absence of PepF that causes a stress. A change in the peptidoglycan structure has been documented in response to osmotic and nutritional stresses in Lactobacillus (Piuri et al., 2005) or in E. coli (Gyaneshwar et al., 2005). In regards to the relationships of PepF with enzymes of the pyruvate cycle, heterofermentation seems to be preferred in the deletion mutants. In the NZ9000-PSEC1 strain, acetate is produced at the expense of lactate. In the IL1403 strain it seems to be the production of acetolactate that is preferred, whether any more acetate nor formate is detected. The alteration of the pyruvate metabolism can explain the presence of oxidoreductases in the possible partners of PepF that could be responsible for rebalancing the redox potential due to a fermentation modification. We did not observe differences in cell division between wild type and pepF mutant observed by electronic microscopy. It is possible that the morphology of the mutant is not affected and that a technique other than the observation of dividing cells would reveal differences between wild type and pepF mutant. It is not surprising to find some of the proteins participating in cell division among our possible relationships of PepF. The cell division proteins (FtsW, FtsK, FtsA) of lactococci are homologous toB. subtilis sporulation proteins and in B. subtilis the overproduction of PepF inhibits sporulation apparently due to its possible participation in the maturation of a signaling peptide (Kanamaru et al., 2002). Recently Kavanaugh et al. (2007) demonstrated the involvement of an analogue of SipL in Staphylococcus aureus in the maturation of an autoinducing peptide implicated in quorum sensing the precursor of which is a signal peptide. Taking into account the strong relationship predicted between PepF and SipL, the involvement of SipL in quorum sensing in S. aureus and the fact that sporulation is affected by PepF (Kanamaru et al., 2002), we cannot exclude that the peptides maturated by PepF serve as extracellular signals (quorum sensing) or as intracellular signals (gene regulation) which could explain its indirect participation in different cellular functions.
We have shown that a global statistical analysis, with the flexibility of the KCCA, can be used to predict the role of a protein by inferring possible relationships with other proteins. This approach allows one to pose a hypothesis about the role of a single protein using existing data in order to guide the laboratory work. At the same time it enables one to find global relationships and pleiotropic roles that would not have been detected with other approaches. The experimental validations allowed us to confirm predicted roles and give biological sense to the predicted relationships. Nevertheless, even if a global approach was used, the complexity of biological systems impedes the complete elucidation of the role of a single protein. We increased our knowledge of PepF and confirmed its participation in protein secretion, but the precise mechanisms by which it interferes and other cellular functions we studied remains unclear. This situation encourages the development of complex and at the same time precise biological models that take into account pleiotropy and connectedness of all cellular functions.
Supporting information is available in an EPAPS document.
SUPPORTING INFORMATION
Supplemental Content S1.doc
Supplemental Content S2.doc
ACKNOWLEDGMENTS
This work is the result of collaboration between the Microbiology and the Applied Mathematics divisions of INRA and was financed by this institution. It was also financially supported by the Ile de France regional council, especially for the LC-MS-MS experiment. We thank Jean-Philippe Vert for providing the programs which made possible the combination of all our KEGG data into a sole graph. At INRA in Jouy en Josas we would like to thank: Gaëlle Bergot at the Unité de Biochimie Bactérienne for the measurements of metabolites of the pyruvate cycle and the group of Marie Pierre Chapot-Chartier for sharing with us their protocol of peptidoglycan analysis; at the Unité de Génetique Microbienne, we thank Marion Velten, Sophie Cheruel, and Patrice Polard for their assistance in the genetic construction for the heterologous production of PepF and the material they provided us with and Eric Guedon for sharing with us his microarray data; at the Unité d’Ecologie et de Physiologie du Système Digestif, we thank Philippe Langella for his advice and for providing us with the nuclease overproducing strain. We would also like to thank Alain Guillot at the Proteomic Platform (PAPSS) and Sophie Chat at the Microscopic Platform (MIMA2). Finally, we would like to thank Mireille Yvon, Kiên Kiêu, and Donald White for the revision of the manuscript and their helpful comments.
References
- Aerts, S, Lambrechts, D, Maity, S, Van, Loo P, Coessens, B, De Smet, F, Tranchevent, L C, De Moor, B, Marynen, P, Hassan, B, Carmeliet, P, and Moreau, Y (2006). “Gene prioritization through genomic data fusion.” Nat. Biotechnol. 34, 537–544. [DOI] [PubMed] [Google Scholar]
- Akesson, M, Förster, J, and Nielsen, J (2004). “Integration of gene expression data into genome-scale metabolic models.” Metab. Eng. 6, 285–293. [DOI] [PubMed] [Google Scholar]
- Atrih, A, Bacher, G, Allmaier, G, Williamson, M P, and Foster, S J (1999). “Analysis of peptidoglycan structure from vegetative cells of Bacillis subtilis 168 and Role of PBR 5 in Peptidoglycan Maturation.” J. Bacteriol. 181, 3956–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J D, Nielsen, H, von Heijne, G, and Brunak, S (2004) “Improved prediction of signal peptides: SignalP 3.0.” J. Mol. Biol. 10.1016/j.jmb.2004.05.028 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Blattner, F R, Plunkett, G, Bloch, C A, Perna, N T, Burland, V., Riley, M, Collado-Vides, J, Glasner, J D, Rode, C K, Mayhew, G F, Gregor, J, Davis, N W, Kirkpatrick, H A, Goeden, M A, Rose, D J, Mau, B, and Shao, Y (1997) “The complete genome sequence of Escherichia coli K-12.” Science 10.1126/science.277.5331.1453 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Bolotin, A, Wincker, P, Mauger, S, Jaillon, O, Malarme, K, Weissenbach, J, Ehrlich, S D, and Sorokin, A (2001). “The complete genome sequence of the lactic acid bacterium Lactococcus lactis.” Genome Res. 10.1101/gr.GR-1697R 11, 731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgwardt, K M, Gretton, A, Rasch, M J, Kriegel, H P, Schölkopf, B, and Smola, A J. (2006). “Integrating structured biological data by Kernel Maximum Mean Discrepancy.” Bioinformatics 10.1093/bioinformatics/btl242 22, 49–57. [DOI] [PubMed] [Google Scholar]
- Chatel, J M, Langella, P, Adel-Patient, K, Commissaire, J, Wal, J M, and Corthier, G (2001). “Induction of mucosal immune response after intranasal or oral inoculation of mice with Lactococcus lactis producing bovine beta-lactoglobulin.” Clin. Diagn. Lab Immunol. 8, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L, Tai, P C, Briggs, M S, and Gierasch, L M (1987). “Protein translocation into Escherichia coli membrane vesicles is inhibited by functional synthetic signal peptides.” Biol. Chem. 262, 1427–1429. [PubMed] [Google Scholar]
- Chich, J F, David, O, Villers, F, Schaeffer, B, Lutomsli, D, and Huet, S (2007). “Statistics for proteomics: experimental design and 2-DE differential analysis.” J. Chromatogr., B: Biomed. Appl. 849, 261–272. [DOI] [PubMed] [Google Scholar]
- Cocaign-Bousquet, M, Garrigues, C, Novak, L, Lindley, N D, and Loubiere, P (1995). “Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis.” J. Appl. Bacteriol. 79, 108–116. [Google Scholar]
- Conlin, C A, Trun, N J, Silhavy, T J, and Miller, C G (1992). “Escherichia coli prlC Encodes an Endopeptidase and is homologous to the Salmonella typhimurium opdA gene.” J. Bacteriol. 174, 5881–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert, M W, Knight, E M, Reed, J L, Herrgard, M J, and Palsson, B O (2004). “Integrating high-throughput and computational data elucidates bacterial networks.” Nature (London) 10.1038/nature02456 429, 92–96. [DOI] [PubMed] [Google Scholar]
- Dev, I K, and Ray, P H (1990). “Signal peptidases and signal peptide hydrolases.” J. Bioenerg. Biomembr. 22, 271–290. [DOI] [PubMed] [Google Scholar]
- Emr, S D, and Bassford, P J (1982). “Localization and processing of outer membrane and periplasmic proteins in Escherichia coli strains harboring export-specific suppressor mutations.” Biol. Chem. 257, 5852–5860. [PubMed] [Google Scholar]
- Emr, S D, and Silhavy, T J (1980). “Mutations affecting localization of Escherichia coli outer membrane protein, the bacteriophage λ receptor.” J. Mol. Biol. 10.1016/S0022-2836(80)80029-5 141, 63–90. [DOI] [PubMed] [Google Scholar]
- EPAPS Document No. E-HJFOA5-2-002801 for supplemental material. This document can be reached through a direct link in the online article’s HTML reference section via the EPAPS homepage (http://www.aip.org/pubservs/epaps.html).
- Gitton, C, Meyrand, M, Wang, J, Caron, C, Trubuil, A, Guillot, A, and Mistou, M Y (2005). “Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk.” Appl. Environ. Microbiol. 10.1128/AEM.71.11.7152-7163.2005 71, 7152–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner, B (1988). “Separation and quantification of muropeptides with high-performance liquid chromatography.” Anal. Biochem. 10.1016/0003-2697(88)90468-X 172, 451–464. [DOI] [PubMed] [Google Scholar]
- Guedon, E, Serror, P, Ehrlich, S D, Renault, P, and Delorme, C (2001). “Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis.” Med. Mundi 40, 1227–1239. [DOI] [PubMed] [Google Scholar]
- Guillot, A, Gitton, C, Anglade, P, and Mistou, M Y (2003). “Proteomic analysis of Lactococcus lactis, a lactic acid bacterium.” Proteomics 10.1002/pmic.200390047 3, 337–354. [DOI] [PubMed] [Google Scholar]
- Gyaneshwar, P, Paliy, O, McAuliffe, J, Popham, D L, Jordan, M I, and Kustu, S (2005). “Sulphur and nitrogen limitation in Escherichia coli K12, specific homeostatic responses.” J. Bacteriol. 10.1128/JB.187.3.1074-1090.2005 187, 1074–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y, Gruhler, A, Heilbut, A, Bader, G D, Moore, L, Adams, S L, Millar, A, Taylor, P, Bennett, K, and Boutilier, K (2002). “Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry.” Nature (London) 10.1038/415180a 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Hoffman, C S, and Winston, F (1987). “A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli.” Gene 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Ichihara, S, Beppu, N, and Mizushima, S (1984). “Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity.” Biol. Chem. 259, 9853–9857. [PubMed] [Google Scholar]
- Ito, T, Chiba, T, Ozawa, R, Yoshida, M, Hattori, M, and Sakaki, Y (2001). “A comprehensive two-hybrid analysis to explore the yeast protein interactome.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.061034498 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru, K, Stephenson, S, and Perego, M (2002). “Overexpression of the PepF oligopeptidase inhibits sporulation initiation in Bacillus subtilis.” J. Bacteriol. 10.1128/JB.184.1.43-50.2002 184, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M, Goto, S, Kawashima, S, and Nakaya, A (2002). “The KEGG databases at GenomeNet.” Nucleic Acids Res. 10.1093/nar/30.1.42 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondor, R I, and Lafferty, J (2002). “Diffusion kernels on graphs and other discrete input spaces.” International Conference for Machine Learning pp. 315–322.
- Kavanaugh, J S, Thoendel, M, and Horswill, A R (2007). “A role for type I signal peptidase in Staphylococcus aureus quorum sensing.” Mol. Microbiol. 10.1111/j.1365-2958.2007.05830.x 65, 780–798. [DOI] [PubMed] [Google Scholar]
- de Magalhães, J P, Costa, J, and Toussaint, O (2005). “HAGR: the Human Aging Genomic Resources.” Nucleic Acids Res. 33, Database issue D537-D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnet, V, Nardi, M, Chopin, A, Chopin, M C, and Gripon, J C (1994). “Biochemical and genetic characterization of PepF an oligoendopeptidase from Lactococcus lactis.” J. Biol. Chem. 269, 32070–32076. [PubMed] [Google Scholar]
- Nardi, M, Renault, P, and Monnet, V (1997). “Duplication of the PepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis.” J. Bacteriol. 179, 4164–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot, P, and Noirot-Gros, M F (2004). “Protein interaction networks in bacteria.” Curr. Opin. Microbiol. 10.1016/j.mib.2004.08.005 7, 505–512. [DOI] [PubMed] [Google Scholar]
- Novak, P, Ray, P H, and Dev, I K (1982). “Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide.” Biol. Chem. 261, 420–427. [PubMed] [Google Scholar]
- Piuri, M, Sanchez-Rivas, C, and Ruzal, S M (2005). “Cell wall modifications during osmotic stress in Lactobacillus casei.” J. Appl. Microbiol. 98, 84–95. [DOI] [PubMed] [Google Scholar]
- Qi, Y, Klein-Seetharam, J, and Bar-Joseph, Z (2005). “Random forest similarity for protein-protein interaction prediction from multiple sources.” Biocomputing: Proc. Pacific Symposium 10, Hawaii. [PubMed]
- Renault, P, Corthier, G, Goupil, N, Delorme, C, and Ehrlich, S D (1996). “Plasmid vectors for Gram-positive bacteria switching from high to low copy number.” Gene 183, 175–182. [DOI] [PubMed] [Google Scholar]
- de Ruyter, P G. G. A, Kuipers, O P, Beerthuyzen, M M., van Alen-Boerrigter, I, and de Vos, W M (1996). “Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis.” J. Bacteriol. 178, 3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J, and Russel, D W (2001). Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schölkopf, B, Tsuda, K, and Vert, J P (2004). Kernel Methods in Computational Biology, MIT Press, Cambridge, MA. [Google Scholar]
- Shin, J J, Bryksin, A V, Godfrey, H P, and Cabello, F C (2004). “Localization of BmpA on the exposed outer membrane of Borrelia burgdorferi by monospecific anti-recombinant BmpA rabbit antibodies.” Infect. Immun. 72, 2280–2287. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Siezen, R J, Renckens, B, van Swam, I, Peters, S., van Kranenburg, R, and de Vos, W M (2005). “Complete sequence of four plasmids of Lactococcus lactis subsp cremoris SK11 reveal extensive adaptation to the dairy environment.” Appl. Environ. Microbiol. 10.1128/AEM.71.12.8371-8382.2005 71, 8371–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisljar, U, Knight, C G, and Barrett, A J (1990). “An alternative quenched flourescence substrate for Pz-peptidase.” Anal. Biochem. 10.1016/0003-2697(90)90582-T 186, 112–115. [DOI] [PubMed] [Google Scholar]
- Tjalsma, H, Bolhius, A, Jongbloed, J DH, Bron, S, and van Dijl, J M (2000). “Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome.” Microbiol. Mol. Biol. Rev. 10.1128/MMBR.64.3.515-547.2000 64, 515–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werhli, A, and Husmeier, D (2007). “Reconstructing gene regulatory networks with Bayesian networks by combining expression data with multiple sources of prior knowledge.” Stat. Appl. Genetics Mol. Biol. 6, Issue 1, Article 15. [DOI] [PubMed]
- Wicker, W, Moore, K, Dibb, N, Geissert, D, and Rice, M (1987). “Inhibition of purified Escherichia coli leader peptidase by the leader (signal) peptide of bacteriophage M13 procoat.” J. Bacteriol. 169, 3821–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi, Y, Vert, J P, Nakaya, A, and Kaneisha, M (2003). “Extraction of correlated clusters from multiple genomic data by generalized kernel canonical correlation analysis.” Bioinformatics 19 Suppl. 1, i323–i330. [DOI] [PubMed] [Google Scholar]
- Yamanishi, Y, Vert, J P, and Kanehisa, M (2004). “Protein network inference from multiple genomic data: a supervised approach.” Bioinformatics 20, Suppl. 1, i363–i370. [DOI] [PubMed] [Google Scholar]
- Yamanishi, Y, and Vert, J P (2007). “Kernel Matrix Regression, Cornell University Library.” (submitted: http://arxivorg/abs/q-bio/0702054v1).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Content S1.doc
Supplemental Content S2.doc