Abstract
A century ago, Ramón y Cajal proposed that the brain consisted of huge numbers of neurons that communicated with each other through junctions called synapses. Today we routinely monitor single neuron and single synapse responses, and we have elaborate maps of connections between different regions of the brain. What we lack is a way to bridge these two scales of representing neuronal circuits. The challenges in doing so are formidable: even a small mammalian neuronal circuit has many thousands of neurons and millions of synapses. Can we keep track of individual cells and synapses in this crowd? Here we examine how two recent techniques may complement each other to do so. The recent “Brainbow” method is a way to color-code cells and their projections, so we can see which cells come near each other, but cannot be sure they connect. Functional circuit mapping tells us about connections between cells, but we cannot identify more than a handful at a time. Together these methods may fill in each other’s blanks and give us brain wiring diagrams that combine scale and precision.
A brain wiring diagram may mean many things: what regions are connected together, what cells are connected together, and the exact weights of these connections as they evolve over time. Even if we knew everything about single-cell physiology, it would take the last and most detailed wiring diagram to do what one routinely does in electronic circuits: begin to understand how function emerges from the network. Real brains pose extra problems compared to electronic circuits: not only are all brains different, but even the same brain changes over time. Many of the most interesting properties of the brain—how it develops, memory, attention—are dynamic ones. It is, therefore, particularly interesting to explore methods for working out circuitry in live brains. In this commentary we discuss new optical methods for connection tracing and recording that begin to achieve this goal.
ANATOMICAL MAPPING
Golgi staining
The earliest methods used to map out neural circuitry were anatomical techniques utilizing the Golgi stain. Cajal raised this technique to an art, and on the basis of observations of Golgi stained preparations, he hypothesized that sites of contact between nerve cells, later termed synapses, were fundamental in the processing of information by the brain. Despite the resolution limitations of early 1900s microscopy, this method was instrumental in determining projection patterns between neuronal layers (Cajal, 1904, 2000; Fairen, 2005).
EM circuit reconstruction
With the advent of electron microscopy, synapses could finally be visualized (Blackstad, 1965) and attempts at reconstructing circuitry followed soon after (reviewed in Briggman and Denk, 2006).
A new technique, serial block face scanning electron microscopy (Denk and Horstmann, 2004), promises to make EM reconstruction a much faster process. Instead of cutting and preserving thin serial sections, the block of tissue itself is imaged on one face with a scanning electron microscope (SEM), following which that section is shaved off and the next layer can then be imaged. This approach is much faster and easier to automate, as it eliminates the alignment and ordering stages of serial transmission electron microscopy. Even though the lateral resolution is poorer than transmission electron microscopy (TEM)—30 nm versus 10 nm—it is sufficient to resolve synaptic sites and thin processes such as spine necks (Denk and Horstmann, 2004). The primary drawback of EM methods is that they take a long time. For example, serial TEM reconstruction of the entire nervous system of a relatively simple animal like C. elegans took 15 years (White et al., 1986; Briggman and Denk, 2006) and, even with serial block-face SEM, the reconstruction of a single cortical column would take approximately a year.
Multicolor labeling and Brainbow
A major problem with light microscopic reconstruction of neuronal circuits was that it is virtually impossible to reconstruct a circuit of densely stained neurons owing to the lack of resolution of light microscopy, even with optical sectioning techniques such as confocal imaging. An elegant solution to this problem was to label neurons in multicolor so that the processes of individual neurons could be distinguished from each other. This was first achieved by delivering beads coated with random combinations of three spectrally distinct lipophilic dyes—DiO, DiI, and DiD—into neurons in slice cultures by ballistic loading with a gene gun (Gan et al., 2000). As a result of this, neurons with multiple colors (up to 7) could be distinguished. The limitation on the number of colors was because differential diffusion of the three dyes in the cell membrane resulted in a spread of colors rather than a single color for any given neuron.
Exploiting spectral variants of the green fluorescent protein (GFP), the same principle was recently extended to multi-color labelling of genetically targeted neurons in mice by the Lichtman–Sanes group (Livet et al., 2007) using a set of constructs termed Brainbow. They made use of the Cre-lox system of inducible Cre recombinase expression to stochastically recombine these Brainbow constructs, resulting in the expression of any one out of three or four fluorescent proteins (XFPs). Combinatorial expression of these XFPs increased the color range, resulting in some 100 discernable colors. In addition, Brainbow expression was restricted to pyramidal neurons since it was under the control of the Thy1 promoter.
-
1.
This technique has the great advantage that it can potentially be used to trace out anatomical wiring diagrams in living tissue, and with far greater speed than serial electron microscopy, because light microscopy can be used to image larger fields of view.
-
2.
Optical microscopy has lower resolution, which prevents conclusive identification of all synaptic contacts between neurons. This could be circumvented somewhat by the use of synaptic markers to identify synaptic contacts.
FUNCTIONAL INPUT-OUTPUT MAPPING
Functional circuit mapping techniques share the same basic premise: synapses can be measured by stimulating presynaptic neurons or fibers and monitoring postsynaptic responses in output neurons. There are a number of stimulation and recording methods available. Intracellular methods for electrical recording and stimulation are precise and can activate individual axonal projections and monitor individual synapses. Extracellular methods can electrically activate small groups of axons and monitor spiking activity of tens of individual neurons, and work in vivo. Optical methods can stimulate individual neurons or even synaptic spines and can monitor hundreds of neurons. Several permutations of input and output methods have been attempted. For example, paired simultaneous intracellular recordings enable precise stimulation and recording from identified pre- and post-synaptic neurons. If the cells are connected, then synaptic response amplitudes provide a measure of synaptic weights. However, the technique does not scale very well: up to seven cells have been simultaneously patched (Silberberg and Markram, 2007), which are a very small proportion of the neurons in any given circuit. A more scalable method is to use optical stimulation to systematically scan the input field of a single intracellularly recorded neuron to generate a projection diagram. Both glutamate uncaging (Brivanlou et al., 2004; Briggs and Callaway, 2005) and Channelrhodopsin-2 stimulation (Petreanu et al., 2007) have been used to do so.
Optical methods and scaling
Optical methods have the potential to combine the best of both worlds for large-scale input-output mapping. These methods can stimulate and record from volumes of the order of a single dendritic spine and can also scale to hundreds or more neurons. The number of potential synapses monitored should scale as the product of individual inputs and outputs. We now consider some of the technical capabilities of these optical methods.
-
1.
Neurotransmitter uncaging—optical uncaging of neurotransmitters, in particular, glutamate, is an effective tool to stimulate individual synapses, or groups of synapses, on identified neurons. In this technique, UV light is used to flash-photolyse a “caged” (complexed and inactive) form of a neurotransmitter; for example, MNI-glutamate. This results in the release of free neurotransmitter, which will activate synapses in the vicinity of the uncaging location (Petitt et al., 1997). The glutamate uncaging method has already been shown to be useful for single neuron input activation (Nikolenko et al., 2007).
-
2.
Channelrhodopsin-2—neurons can be made light-sensitive by genetic incorporation of a light-sensitive cation channel, channelrhodopsin-2 (ChR2), which is derived from an algal species (Boyden et al., 2005). When ChR2-expressing neurons are irradiated with blue light, the channel opens, resulting in depolarization of the membrane and firing of action potentials. As labeling is under genetic control, one can achieve targeted stimulation of a subset of cells (Adamantidis et al., 2007). ChR2 opto-simulation has already been used to map connectivity both within (Wang et al., 2007) and between different neuronal layers (Petreanu et al., 2007) by coupling it with intracellular recordings. All optical stimulation and recording, with ChR2 and calcium sensitive dyes, has also been demonstrated (Zhang et al., 2007). One drawback of ChR2-based stimulation is that the channel seems to have a very small two-photon cross-section (Zhang and Oertner, 2007). This is a problem, since precise stimulation of cells deeper inside tissue would not be feasible.
-
3.
Voltage sensitive dye (VSD) imaging—the membrane potential is a highly desirable index of neuronal activity, because it would give the ability to monitor spikes, spike times, and graded potentials. The most commonly used VSDs are charged amphipathic molecules which get incorporated into the cell membrane. A change in membrane potential results in a concomitant change in fluorescence or absorbance properties of the dye. However, signals from existing VSDs have poor signal-to-noise ratios (Fisher et al., 2005) and are prone to phototoxicity (Spors and Grinvald, 2002).
-
4.
Calcium sensitive dye (CSD) imaging—nearly all neurons possess voltage gated calcium channels that open in response to membrane depolarizations such as spikes. In addition, Ca2+ permeable NMDA receptor channels are present at most glutamatergic synapses. When these channels open, the intracellular calcium concentration rises from ∼100 nM to ∼10 μM (Sabatini et al., 2002). This change can be detected by intracellular calcium sensitive dye reporters. However, calcium imaging has low temporal resolution due to the slow time-scales of calcium responses, especially efflux. Thus, this technique cannot be used to resolve spikes at high firing frequencies, although estimates of firing rate changes are possible (Yaksi and Friedrich, 2006; Ramdya et al., 2006). Several genetically encoded calcium sensors exist, which are essentially calcium binding proteins such as calmodulin (Miyawaki et al., 1997) or muscle troponin C (Heim and Griesbeck, 2004) tagged to a FRET pair (e.g., CFP-YFP). Binding of calcium induces a conformational change, which in turn alters FRET efficiency. While recent genetic sensors are quite sensitive, their temporal resolution is still poor (∼1 s) (Heim et al., 2007).
COMBINING ANATOMICAL AND PHYSIOLOGICAL DATA
To recapitulate, anatomical mapping techniques provide a great deal of information about neuronal morphology and the dendritic localization of synapses, while functional mapping techniques provide information about physiological parameters such as synaptic weights and probabilities of release. These two approaches can complement each other remarkably well (Table 1) and, thus, it would be very useful to combine them in circuit mapping studies.
Table 1.
Comparison between an anatomical, and a functional mapping method. Parameters where the two techniques are likely to complement each other are bold.
| Mapping parameters | Brainbow multicolor imaging | Functional mapping using optical methods |
|---|---|---|
| Mapping in living tissue | √ | √ |
| Distinguishing between neurons | 100 colorsa | √ |
| Distinguishing between axons and dendrites | √ | √ |
| Distinguishing between branches of processes | Cannot resolve very thin (<200 nm) fibres | × |
| Confirmation of the presence of synapses | ×, Contact based; but perhaps by synaptic labelling | √ |
| Tracking long range projections | √ | √, by stimulating cut axons |
| Determination of synaptic weights | × | √ |
| Determining positions of multi-synaptic connections | √, but possible false positives | By single spine imaging, doesn’t scale well |
| Determining weights of multi-synaptic connections between pairs of cells | × | By single spine imaging, doesn’t scale well |
| Scaling—how many synapses? | Limited by slicing technique | Electrical readout—sparse Optical—dense |
| Speed of mapping | Fast—minutes to hours | Slow—hours |
According to the birthday problem, the probability of at least two neurons sharing the same color out of a sample of n multicolored neurons is given by the 1−(the probability of all neurons having different colors). For 100 colors, this works out to a probability close to 1 for just n=30 neurons.
Here we consider two of the most interesting aspects of this complementarity, as highlighted in Table 1 and Fig. 1.
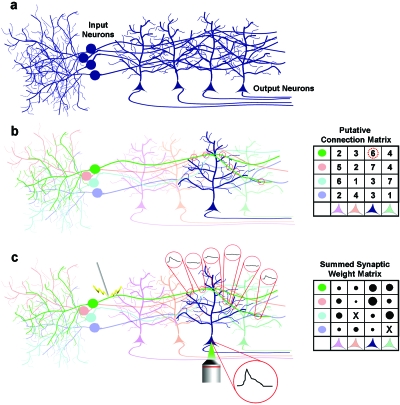
Figure 1. Combined anatomical and functional mapping of synapses in a hypothetical feed-forward neuronal circuit.
Axons from the input layer of cells (positioned on the left) synapse onto the apical dendrites of the output cell layer (somas positioned at the bottom of each figure). (a) Using conventional labelling methods, it is virtually impossible to trace out the processes of each neuron. (b) Following combinatorial Brainbow labelling, each neuron and its processes assume a different color from that of neighboring neurons, and a pairwise putative synaptic connection matrix can be drawn up, on the basis of proximity between axons and dendritic spines. (c) Using functional mapping techniques, each axon can be electrically or optically stimulated using ChR2 and the resultant response measured at the soma of each cell. The somatic response represents the sum of all synaptic responses, which can also be measured independently by calcium imaging at single spines.
Functional synapses
While Brainbow-based anatomical mapping can predict the positions of synapses, it cannot confirm whether the predicted sites contain functional synapses and, if so, their synaptic weight. Functional mapping, by ChR2-based stimulation and calcium imaging in single spines, can be used to confirm both synaptic presence and synaptic weight. Since Brainbow mapping can also be used to trace out the dendritic tree structure of many neurons, the position of these synapses with respect to this underlying structure will be known. This sort of data is useful for modelling realistic neurons in networks and also to understand how synaptic inputs will summate with each other.
Scaling and speed of mapping
A problem with functional mapping studies is the time taken for systematic stimulation and recording, to interrogate every possible synapse. Such data are usually noisy due to both instrumentation noise and inherent synaptic noise because of probabilistic release. Therefore, several repetitions are required to accurately measure synapses (Oertner et al., 2002). Brainbow-based anatomical mapping can help enormously by first identifying putative synaptic sites, which can then be systematically tested with functional mapping. Thus, live anatomical mapping can help narrow down the connection space one needs to test with stimulate-record type techniques.
SUMMARY
New techniques that combine genetics, microscopy, and optical stimulation are poised to provide neuroscientists with a new level of data about neuronal connectivity and function. Between them, these methods combine high throughput and high resolution. The resulting abundance of data may lead to profound changes in how we study the brain.
ACKNOWLEDGMENTS
AD and USB wrote the article. USB is the recipient of funds from the DBT and DAE∕SRC. We thank M Modi and P Jain for their input.
References
- Adamantidis, A R, Zhang, F, Aravanis, A M, Deisseroth, K, and de Lecea, L (2007). “Neural substrates of awakening probed with optogenetic control of hypocretin neurons.” Nature 450(7168), 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstad, T W (1965). “Mapping of experimental axon degeneration by electron microscopy of Golgi preparations.” Cell Tissue Res. 67(6), 819–834. [DOI] [PubMed] [Google Scholar]
- Boyden, E S, Zhang, F, Bamberg, E, Nagel, G, and Deisseroth, K (2005). “Millisecond-timescale, genetically targeted optical control of neural activity.” Nat. Neurosci. 8(9), 1263–1268. [DOI] [PubMed] [Google Scholar]
- Briggman, K L, and Denk, W (2006). “Towards neural circuit reconstruction with volume electron microscopy techniques.” Curr. Opin. Neurobiol. 16(5), 562–570. [DOI] [PubMed] [Google Scholar]
- Briggs, F, and Callaway, E M (2005). “Laminar patterns of local excitatory input to layer 5 neurons in macaque primary visual cortex.” Cereb. Cortex 15(5), 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou, I H, Dantzker, J L M, Stevens, C F, and Callaway, E M (2004). “Topographic specificity of functional connections from hippocampal CA3 to CA1.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0308577100 101(8), 2560–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal, S R (2000). Texture of the Nervous System of Man and the Vertebrates, Volume 2. Springer Verlag, Wien. [Google Scholar]
- Denk, W, and Horstmann, H (2004). “Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure.” PLoS Biol. 2(11), online publication 19 October 2004, doi: 10.1371/journal.pbio.0020329 10.1371/journal.pbio.0020329, e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairen, A (2005). “Pioneering a golden age of cerebral microcircuits: the births of the combined Golgi-electron microscope methods.” Neuroscience 136(3), 607–614. [DOI] [PubMed] [Google Scholar]
- Fisher, J A, Salzberg, B M, and Yodh, A G (2005). “Near infrared two-photon excitation cross-sections of voltage-sensitive dyes.” J. Neurosci. Methods 10.1016/j.jneumeth.2005.06.027 148(1), 94–102. [DOI] [PubMed] [Google Scholar]
- Gan, W, Grutzendler, J, Wong, W T, Wong, R O. L, and Lichtman, J W (2000). “Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations.” Neuron 10.1016/S0896-6273(00)00031-3 27(2), 219–225. [DOI] [PubMed] [Google Scholar]
- Heim, N, and Griesbeck, O (2004). “Genetically encoded indicators of cellular calcium dynamics based on Troponin C and green fluorescent protein.” J. Biol. Chem. 10.1074/jbc.M312751200 279(14), 14280–14286. [DOI] [PubMed] [Google Scholar]
- Heim, N, Garaschuk, O, Friedrich, M W, Mank, M, Milos, R I, Kovalchuk, Y, Konnerth, A, and Griesbeck, O (2007). “Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor.” Nat. Methods 4(2), 127–129. [DOI] [PubMed] [Google Scholar]
- Livet, J, Weissman, T A, Kang, H, Draft, R W, Lu, J, Bennis, R A, Sanes, J R, and Lichtman, J W (2007). “Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system.” Nature 450(7166), 56–62. [DOI] [PubMed] [Google Scholar]
- Miyawaki, A, Llopis, J, Heim, R, McCaffery, J M, Adams, J A, Ikura, M, and Tsien, R Y (1997). “Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin.” Nature 10.1038/42264 388(6645), 882–887. [DOI] [PubMed] [Google Scholar]
- Nikolenko, V, Poskanzer, K E, and Yuste, R (2007). “Two-photon photostimulation and imaging of neural circuits.” Nat. Methods 4(11), 943–950. [DOI] [PubMed] [Google Scholar]
- Oertner, T G, Sabatini, B L, Nimchinsky, E A, and Svoboda, K (2002). “Facilitation at single synapses probed with optical quantal analysis.” Nat. Neurosci. 5(7), 657–664. [DOI] [PubMed] [Google Scholar]
- Petreanu, L, Huber, D, Sobczyk, A, and Svoboda, K (2007). “Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections.” Nat. Neurosci. 10(5), 663–668. [DOI] [PubMed] [Google Scholar]
- Pettit, D L, Wang, S S, Gee, K R, and Augustine, G J (1997). “Chemical two-photon uncaging: a novel approach to mapping glutamate receptors.” Neuron 10.1016/S0896-6273(00)80361-X 19(3), 465–471. [DOI] [PubMed] [Google Scholar]
- Ramdya, P, Reiter, B, and Engert, F (2006). “Reverse correlation of rapid calcium signals in the zebrafish optic tectum in vivo.” J. Neurosci. Methods 157(2), 230–237. [DOI] [PubMed] [Google Scholar]
- Sabatini, B L, Oertner, T G, and Svoboda, K (2002). “The life cycle of Ca2+ ions in dendritic spines.” Neuron 10.1016/S0896-6273(02)00573-1 33(3), 439–452. [DOI] [PubMed] [Google Scholar]
- Silberberg, G, and Markram, H (2007). “Disynaptic inhibition between neocortical pyramidal cells mediated by Martinotti cells.” Neuron 53(5), 735–746. [DOI] [PubMed] [Google Scholar]
- Spors, H, and Grinvald, A (2002). “Spatio-temporal dynamics of odor representations in the mammalian olfactory bulb.” Neuron 10.1016/S0896-6273(02)00644-X 34(2), 301–315. [DOI] [PubMed] [Google Scholar]
- Wang, H, et al. (2007). “High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice.” Proc. Natl. Acad. Sci. U.S.A. 104(19), 8143–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J G, Southgate, E, Thomson, J N, and Brenner, S (1986). “The structure of the nervous system of the nematode caenorhabditis elegans.” Philos. Trans. R. Soc. London, Ser. B 10.1098/rstb.1986.0056 314(1165), 1–340. [DOI] [PubMed] [Google Scholar]
- Yaksi, E, and Friedrich, R W (2006). “Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging.” Nat. Methods 3(5), 377–383. [DOI] [PubMed] [Google Scholar]
- Zhang, F, et al. (2007). “Multimodal fast optical interrogation of neural circuitry.” Nature 10.1038/nature05744 446(7136), 633–639. [DOI] [PubMed] [Google Scholar]
- Zhang, Y, and Oertner, T G (2007). “Optical induction of synaptic plasticity using a light-sensitive channel.” Nat. Methods 4(2), 139–141. [DOI] [PubMed] [Google Scholar]