Abstract
Targeted disruption of murine Cdk2ap1, an inhibitor of CDK2 function and hence G1/S transition, results in the embryonic lethality with a high penetration rate. Detailed timed pregnancy analysis of embryos showed that the lethality occurred between embryonic day 3.5 pc and 5.5 pc, a period of implantation and early development of implanted embryos. Two homozygous knockout mice that survived to term showed identical craniofacial defect, including a short snout and a round forehead. Examination of craniofacial morphology by measuring Snout Length (SL) vs. Face Width (FW) showed that the Cdk2ap1+/− mice were born with a reduced SL/FW ratio compared to the Cdk2ap1+/+ and the reduction was more pronounced in Cdk2ap1−/− mice. A transgenic rescue of the lethality was attempted by crossing Cdk2ap1+/− animals with K14-Cdk2ap1 transgenic mice. Resulting Cdk2ap1+/−:K14-Cdk2ap1 transgenic mice showed an improved incidence of full term animals (16.7% from 0.5%) on a Cdk2ap1−/− background. Transgenic expression of Cdk2ap1 in Cdk2ap1−/−:K14-Cdk2ap1 animals restored SL/FW ratio to the level of Cdk2ap1+/−:K14-Cdk2ap1 mice, but not to that of the Cdk2ap1+/+:K14-Cdk2ap1 mice. Teratoma formation analysis using mESCs showed an abrogated in vivo pluripotency of Cdk2ap1−/− mESCs towards a restricted mesoderm lineage specification. This study demonstrates that Cdk2ap1 plays an essential role in the early stage of embryogenesis and has a potential role during craniofacial morphogenesis.
Introduction
CDK2AP1 was initially identified as a cancer-related gene by using hamster oral cancer model [1]. CDK2AP1 is a highly conserved and ubiquitously expressed gene located on human chromosome 12q24 and is a 115-aa nuclear polypeptide that is downregulated in ∼70% of oral cancers [2], [3]. Murine Cdk2ap1 with only three amino acid deviations from the human CDK2AP1 is located at chromosome 5 [4], [5]. We have obtained significant amount of data demonstrating potential cellular function of CDK2AP1 in vitro and in vivo [5], [6], [7], [8], [9], [10], [11]. In addition to its role as a cell cycle regulatory molecule through two important cellular partners: CDK2 and DNA polymerase-alpha/primase, we have shown that CDK2AP1 has a role in TGF-β induced growth arrest, cisplatin induced genotoxicity, and cellular apoptosis [6], [7], [8], [10], [11]. Recently, we have shown the in vivo tumor regression effect of CDK2AP1 with reducing proliferation and increasing apoptotic indices in a xenograft mouse model of head and neck cancer [10]. Furthermore, we have demonstrated that overexpression of Cdk2ap1 in a transgenic mouse model resulted in gonadal atrophy, seminiferous tubule degeneration, and folliculogenesis abnormalities in vivo [12]. Despite the exact in vivo physiological role of Cdk2ap1 remains to be determined, there are several indirect evidences that implicate its role in development. Cdk2ap1 has been identified as one of stem cell specific genes that are enriched in both embryonic and adult stem cells [13]. More interestingly, it has been reported through microarray analysis that Cdk2ap1 has been categorized as one of genes that are expressed in an early stage preimplantation embryos and its expression gradually decreases as the embryo further develops [14]. Through a global expression map of cell cycle regulators in mitosis and meiosis, Cdk2ap1 was found as one of genes with postnatal testis maturation-associated decrease [15]. In addition, Cdk2ap1 mRNA has been found to be elevated upon estrogen treatment during early implantation process, suggesting its role in uterine decidualization where the cells stop proliferation and start differentiation [16].
Proper development of organisms including vertebrates requires spatial and temporal orchestration between many different molecules. Uncontrolled or deleted expression of certain gene can cause abnormal embryo development leading to lethality [17], [18], [19], [20], [21]. These are classes of molecules that are known to play essential roles during development. However though their associated molecular mechanisms have been extensively studied, our understanding towards developmental process especially during the early embryogenesis remains largely elusive. Early stage of the embryo development is defined as a period of rapid cellular proliferation to suffice the needed number of cells in a short period of time. This is most likely related to either silenced or reduced activity of cell cycle regulatory pathways observed in embryonic stem cells [22], [23], [24], [25]. Several well-known cell cycle regulatory molecules including pRb are known to remain inactive during early gestation period until embryonic stem cells start to differentiate into specified cell lineages [26], [27], [28]. The physiological function of a given molecule or its significance in development in vivo can be studied by taking a knockout approach in mouse. In this paper, we examined the developmental role of Cdk2ap1 through a knockout approach via a specific deletion of Cdk2ap1 in mouse to understand physiological significance of Cdk2ap1 during development. Our data clearly demonstrate that Cdk2ap1 is an essential gene during early embryo development since the targeted homozygous disruption of Cdk2ap1 resulted in the lethality of the early embryos. Interestingly, significant craniofacial abnormality was observed in the Cdk2ap1 knockout model, implicating its potential role of Cdk2ap1 in skeletal morphogenesis.
Materials and Methods
Targeted disruption of p12Cdk2ap1 and generation of knockout mice
Murine embryonic stem cells (ES-LW1) were grown on a gelatin coated plate and maintained in DMEM supplemented with 15% FBS, 0.1 µM β-mercaptoethanol, 1% L-Glutamine, 0.2% (v/v) LIF, 1% (v/v) PSF, and 0.1% (v/v) gentamycin sulfate. Generation of Cdk2ap1 knockout mESCs was described by Kim et al. [5]. Established two Cdk2ap1+/− cells were independently injected into blastocysts from C57/BL6. Chimeric animals were identified by genotyping and Southern analysis and populated to establish Cdk2ap1+/− mouse line. All the animals were maintained and taken care of under the husbandry guidelines supervised by the UCLA Chancellor's Animal Research Committee.
Genotyping and Southern analysis of Cdk2ap1 knockout mice
Tail biopsies from weaned animals were collected according to the guidelines from UCLA Animal Research Committee. Genomic DNA was extracted by incubating in 17 mM Tris-HCl (pH 7.5), 17 mM EDTA, 170 mM NaCl, 0.85% SDS, and 0.2 mg/ml proteinase K overnight at 55°C. Final DNA pellet was dissolved in TE and subjected to a multiplex PCR analysis by using Advantage 2 PCR kit (Clontech, Palo Alto, CA). Primer set 1 [CDU2 (5′GCCTTCTTGACGAGTTCTTCTGAG3′) and CDD2 (5′TGGATGTGGAATGTGTGCGAG3′)] amplifies a 460 bp fragment within the cytosine deaminase gene in the targeting vector and primer set 2 [I2U (5′ATGGGGATGGATGTGTGAGAGG3′) and I2D (5′TGGGTTCAAGGAAGTGGACTAATG3′)] generates a 239 bp fragment from intron 2 which is absent in a successful homologous recombination). For Southern analysis, 5 µg of DNA was digested with Bsu36I resulting in 8.4 and 5.6 kb fragments.
Timed pregnancy analysis and genotyping
For timed pregnancy analysis, heterozygous females were mated with heterozygous males with or without hormonal stimulation. At day 2.5 or 3.5 dpc, females were sacrificed and the embryos were flushed out of the uterus. Genomic DNA was isolated by using blastocyst lysis buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl, 2 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, 60 µg/ml proteinase K) and subjected to multiplex PCR analysis with or without genomic DNA amplification by using GenomiPhi DNA Amplification kit (GE Healthcare, Piscataway, NJ) according to the manufacturer's instruction. For later gestation time points, the embryos were fixed in 4% paraformaldehyde, rinsed under tap water, stored in 70% EtOH and paraffin-embedded for sectioning onto metal LCM slides. LCM slides were deparaffinized 5 min in xylene and rehydrated in 100% EtOH for 2 min, 95% EtOH for 2 min, 70% EtOH for 2 min, 50% EtOH for 2 min and dH2O for 2 min. Slides were stained with methyl green for 30 sec and washed in dH2O for 1–5 min. Finally slides were air dried and a portion of embryo was microdissected into PCR caps containing 20 µl of digestion buffer (Arcturus DNA Extraction Kit). Tissue was digested overnight as suggested by Arcturus protocol. A 5 µl of digest was used per PCR reaction by using Advantage 2 PCR kit (Clontech, Palo Alto, CA). Two different sets of primers were used for genotyping analysis: For homologous recombinant DNA, CD3F (TGTCGTATCCCAACGGTGAAGC) and CD1R (CCAGGGCGAAGGTTTTATGC) or CD3F and CD3R (ACATCATCGTGACCAAAGCAGACG) were used. For the wild type DNA, Int4F (TTGCTTTGCTTGTTTCCTGGGC) and IntR (GGCTGGGGTTTGGCTCATAGAATC) were used.
Generation of Cdk2ap1−/−:K14-Cdk2ap1 hybrid animals
Transgenic K14-Cdk2ap1 mice were generated as described previously [12]. Heterozygous Cdk2ap1+/− mice were mated with K14-Cdk2ap1 mice and pups were analyzed by PCR based genotyping on tail biopsies. Confirmed heterozygous Cdk2ap1+/−:K14-Cdk2ap1 mice were intercrossed to examine the viability of homozygous Cdk2ap1−/−:K14-Cdk2ap1. In addition, the expression of transgenic Cdk2ap1 was confirmed by RT-PCR analysis with total RNA from ear-snips. Briefly, frozen earsnips were powdered and mixed with lysis buffer. Total RNA was purified by using RNeasy mini kit from Qiagen (Valencia, CA). For RT-PCR analysis, equal amount of total RNA was subjected to reverse transcription by using Superscript III RT and oligo(dT) primer. The synthesized cDNA was amplified by using a pair of primers specific to mouse Cdk2ap1. In addition, GAPDH primer was used for normalization.
Blastocyst outgrowth analysis
Embryos were harvested at 3.5 dpc as described for timed pregnancy analysis and cultured in gelatin coated 96 well plate in blastocyst culture medium ((DMEM, 20% FBS, 2 mM glutamine, 4 mg/ml BSA, 0.1 mM β-ME, PSF). Culture was maintained for 5 days and the hatching and outgrowth of blastocyst was photomicrographed for comparison. At the end of culture, cells were lysed in blastocyst lysis buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl, 2 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, 60 µg/ml proteinase K) for isolation of total genomic DNA. Genotyping analysis was performed as described above.
Mesenchymal cell culture
Primary bone marrow mesenchymal cell were isolated and established by flushing bone marrows from the tibiae, femora, and humeri of 4 week-old animals with cold DMEM using a 27-gauge needle. Cells were then washed with cold PBS and cultured to confluence in a 100 mm plate in α-MEM with 10% FBS. At confluence cells were collected for further analysis.
Results
Targeted disruption of p12CDK2-AP1 in mice leads to early embryonic lethality
Significant amount of data has been gathered demonstrating the importance of Cdk2ap1 in cellular regulation in vitro and also in vivo [5], [8], [9], [10]. However, the normal physiological role of Cdk2ap1 has not been fully elucidated. As a way of evaluating the importance of Cdk2ap1 in the animal development and physiology, we generated a conventional knockout mouse model. Heterozygous and homozygous Cdk2ap1 knockout murine embryonic stem cells were generated as reported previously [5]. Targeting vector was designed to recombine with the introns 1 and 4, resulting in the substitution of the exons 2 and 3 with neor and cytosine deaminase genes (Fig. 1). Electroporated ES-LW1 cells (129sv background) were screening for gaining of resistance to G418. Selected heterozygous Cdk2ap1 knockout cells were further screened for homozygous knockout through mitotic crossing over event under high dose of G418 selection. The expression of Cdk2ap1 was examined by RT-PCR, northern and western analysis. Homozygous knockout cells showed a complete absence of Cdk2ap1 expression [5]. To generate Cdk2ap1 knockout mice, heterozygous Cdk2ap1 knockout cell line 272 or 284 was independently injected into the blastocysts from C57/B6 mice. The injected blastocysts were transferred into the uterus of the pseudo-pregnant mouse. Chimeras were verified by PCR analysis on tail DNA and southern blot analysis. Chimeric animals were mated to generate heterozygous Cdk2ap1 knockout mouse lines and heterozygosity was verified by multiplex PCR and southern analysis on tail biopsies (Fig. 1). We then attempted to generate homozygous line by mating proven heterozygous pairs. From extensive breeding and genotyping analysis, we have found the knockout of Cdk2ap1 resulted in very high penetrating rate of embryonic lethality. As shown in Table 1, we had 2/399 (from 84 litters) homozygous knockout mice. The genotyping result of the two survived homozygous Cdk2ap1 knockout mice is shown in Fig. 2. To define a window of lethality and gain insight into the role of Cdk2ap1 during embryogenesis, we have performed a detailed timed pregnancy analysis by using heterozygous animals. The result showed that there were 2.5 and 3.5 dpc embryos genotyped as homozygous knockout (7/24 and 11/66, respectively), but we have not observed any viable Cdk2ap1−/− embryos after 3.5 dpc. These data suggest that the lethality occurs after 3.5 dpc blastocyst stage and a knockout of Cdk2ap1 may have an effect on the proper hatching of blastocyst, the implantation of embryo, or very early differentiation of embryos.
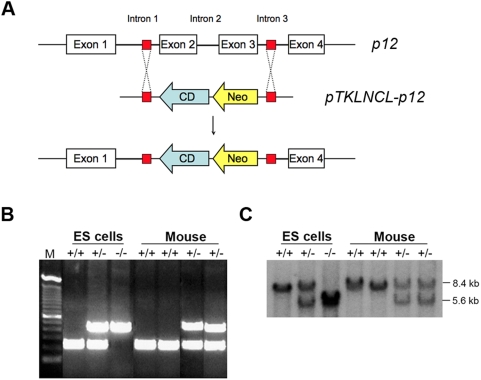
Figure 1. Generation of Cdk2ap1 knockout mice.
A. Targeting vector construct pTKLNCL-Cdk2ap1 was designed to knockout Cdk2ap1 exon2 and 3 by homologous recombination. Cytosine deaminase (CD) and neomycin resistance (Neo) genes were placed in to disrupt Cdk2ap1 gene in mouse chromosome 5. B. Multiplex genotyping analysis was used to screen recombinant ES cells as described in Materials and Methods. The top band of 460 bp is amplified from cytosine deaminase in the targeting vector and the bottom band of 239 bp is amplified from the introns of wild type Cdk2ap1. Mouse tail DNA was also analyzed for screening of Cdk2ap1 knockout mice by multiplex PCR. C. The multiplex PCR genotyping data was confirmed by Southern blot analysis. The wild type Cdk2ap1 showed 8.4 kb fragment, while the recombinant showed 5.6 kb fragment.
Table 1. Timed pregnancy analysis by Het x Het matings.
| Age | Litter | Total Number | Genotype | Resorbed | ||
| +/+ | +/− | −/− | ||||
| 4 weeks | 56 | 399 | 137 | 260 | 2 | N/A |
| E10.5 | 5 | 48 | 21 | 27 | 0 | 6 |
| E7.5 | 5 | 29 | 12 | 17 | 0 | 4 |
| E5.5 | 5 | 24 | 8 | 16 | 0 | N/A |
| E3.5 | 9 | 66 | 21 | 34 | 11 | N/A |
| E2.5 | 4 | 24 | 5 | 12 | 7 | N/A |
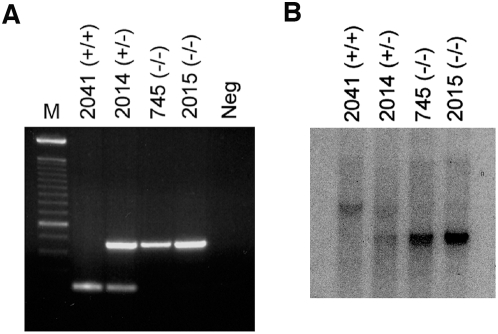
Figure 2. Genotyping of survived homozygous Cdk2ap1 knockout mice.
Two mice (M745 and F2015) were born live out of 399 pups analyzed from crossing of heterozygous Cdk2ap1 knockout mice. A. The genotype of both animals was analyzed by multiplex PCR on tail DNA with no DNA negative control. B. The PCR genotyping result was also confirmed by Southern analysis. Both M745 and F2015 mice showed homozygous knockout genotype compared to WT type (F2041) and heterozygous (F2014) animals.
To determine if the deletion of Cdk2ap1 has any effect on hatching process of blastocysts, we have performed blastocyst outgrowth analysis. Embryos at E3.5 were collected from timed pregnancy analysis and cultured in vitro to observe hatching and outgrowth of embryonic stem cells. At the end of observation, cells were genotyped to match with the phenotypes. As shown in Fig. 3, blastocysts properly hatched and outgrew regardless of the genotype. There were no significant changes observed among Cdk2ap1+/+, Cdk2ap1+/−, and Cdk2ap1−/− blastocysts. This result suggests that the lethality from the deletion of Cdk2ap1 is not due to the abrogation in the hatching process.
Figure 3. Blastocyst outgrowth analysis of Cdk2ap1 knockout embryos.
To examine the hatching and outgrowth of Cdk2ap1−/− embryos, blastocyst outgrowth analysis was performed. Heterozygous females and males were mated for timed pregnancy analysis and E3.5 blastocysts were collected. A. Individual blastocyst was cultured in a 96 well plate for 4 days. Photomicrograph was taken every day to monitor the hatching and outgrowth of blastocysts. B. After final recording of morphology, outgrown cells were lysed and genomic DNA was isolated for multiplex genotyping analysis as described in Materials and Methods. The PCR products were analyzed on a 1.5% agarose gel and the genotype was matched to the phenotype from A.
Rescue of lethality by crossing Cdk2ap1+/− with transgenic K14-Cdk2ap1
The process of generating knockout mice involves an extensive in vitro screening of recombinant embryonic stem cells, including an extended period of culturing and a selection with high dose of antibiotics. This could potentially impose the surmount level of pressure to the cells and lead to an unwanted bias in the analysis. The most appropriate approach to avoid this kind of bias would be using a conditional knockout model. This is currently in progress and we have already generated heterozygous floxed Cdk2ap1 knockout mice by crossing chimeras. Meanwhile, to ensure that the lethality we observed with homozygous Cdk2ap1 knockout mice was due to the specific inactivation of the gene, we tried to rescue the lethal phenotype of Cdk2ap1−/− mice by taking a transgenic approach. To complement the loss of Cdk2ap1 expression in Cdk2ap1 knockout mice, we crossed the heterozygous Cdk2ap1 knockout (Cdk2ap1+/−) mice with the K14-Cdk2ap1 transgenic (Cdk2ap1+/−:K14-Cdk2ap1) mice that overexpress Cdk2ap1 by using human keratin 14 promoter [12]. We first generated and identified F1 hybrid mice with Cdk2ap1+/− genotype and K14-Cdk2ap1 transgene integrated into the genome (Cdk2ap1+/−:K14-Cdk2ap1). As shown in Fig. 4A, we were able to generate hybrid mice of Cdk2ap1−/− genotype with K14-Cdk2ap1 transgene integrated into the genome. The resulting heterozygous Cdk2ap1+/−:K14-Cdk2ap1 hybrids were then intercrossed to generate homozygous Cdk2ap1 knockout with K14-Cdk2ap1 transgene (Cdk2ap1−/−:K14-Cdk2ap1). We investigated the effect of transgenic rescue on the development of Cdk2ap1−/−:K14-Cdk2ap1 mice in terms of survival and proper development. The resulting weaned pups of 4 weeks of age were analyzed by genotyping on tail biopsies. We have analyzed 36 pups from 11 litters and the transgenic rescue approach resulted in an improved birth of homozygous Cdk2ap1 knockout mice (Table 2). The rate of the birth of Cdk2ap1−/− was increased to 16.7% from 0.5%. The expected incidence of homozygous pups should be 25% based on Mendelian genetics. Therefore, it is about 67% of the expected number, which means while the rescue was not achieved with 100% efficiency. It still represents significantly improved rescue of lethality. During this hybrid mating process, we have not obtained any homozygous Cdk2ap1 knockout mice without the integrated Cdk2ap1 transgene. This appears especially important because we can rule out the strain dependent genetic modifier effect with this finding. To see if the integrated transgene through the hybrid approach is expressed in the mice, we examined Cdk2ap1 expression in Cdk2ap1−/−:K14-Cdk2ap1 hybrid animals by RT-PCR analysis on total RNA isolated from ear-snips. As shown in Fig. 4B, we detected the expression of Cdk2ap1 mRNA in Cdk2ap1+/+ and Cdk2ap1+/− mice from mating of heterozygous Cdk2ap1+/− mice. The Cdk2ap1+/−:K14-Cdk2ap1 mouse from mating of Cdk2ap1+/−:K14-Cdk2ap1 hybrids also expressed Cdk2ap1 mRNA. We observed that there was Cdk2ap1 mRNA expressed in Cdk2ap1−/−:K14-Cdk2ap1 hybrids, probably due to the expression of transgenic Cdk2ap1. As anticipated the Cdk2ap1−/−:K14-Cdk2ap1 mice also expressed considerable level of Cdk2ap1 mRNA detected by RT-PCR, while Cdk2ap1−/− mESCs used as a control did not show any expression of Cdk2ap1 mRNA. These evidence confirm that the complementary expression of Cdk2ap1 has been achieved in Cdk2ap1−/−:K14-Cdk2ap1 hybrids.
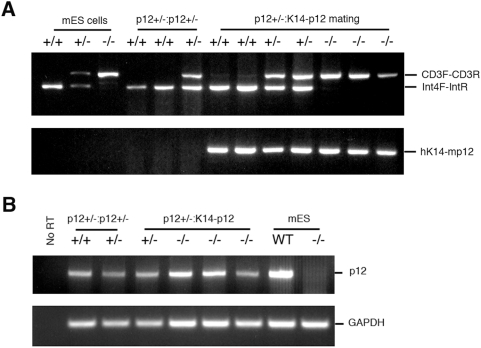
Figure 4. Genotyping and expression analysis of Cdk2ap1−/−:K14-Cdk2ap1 hybrids.
Hybrid mice were generated by crossing heterozygous Cdk2ap1 knockout mice and K14-Cdk2ap1 transgenic mice. A. The resulting pups were genotyped to identify hybrids by using PCR. Wild type Cdk2ap1 gene was amplified by using Int4F and IntR primers against the Cdk2ap1 intron sequence. The recombinant Cdk2ap1 was detected by using CD3F and CD3R primers against cytosine deaminase gene. In addition, the presence of transgenic Cdk2ap1 was detected by using hK14 and mCdk2ap1 primers against human K14 promoter and mouse Cdk2ap1 transgene. By intercrossing heterozygous Cdk2ap1+/−:K14-Cdk2ap1 mice, we were able to generate homozygous Cdk2ap1−/−:K14-Cdk2ap1 mice. Mouse ESCs (Cdk2ap1+/+, Cdk2ap1+/−, and Cdk2ap1−/−) and pups (WT and heterozygous) from mating of heterozygous Cdk2ap1 knockout mice were used as controls. B. The expression of Cdk2ap1 was examined by RT-PCR analysis on total RNA from earsnips as described in Materials and Methods. Compared to no RT or Cdk2ap1−/− mESC control, comparable amount of Cdk2ap1 mRNA was detected in Cdk2ap1−/−:K14-Cdk2ap1 mouse tissues, which showed that the expression of Cdk2ap1 was achieved from the Cdk2ap1 transgene in the mice with Cdk2ap1−/− genotype.
Table 2. Transgenic rescue result from genotyping of Cdk2ap1+/−:K14-Cdk2ap1 mating.
| Age | Litter | Total Number | Genotype | ||
| +/+ | +/− | −/− | |||
| 4 weeks | 11 | 36 | 10 | 20 | 6 |
Craniofacial abnormalities in survived Cdk2ap1−/− mice
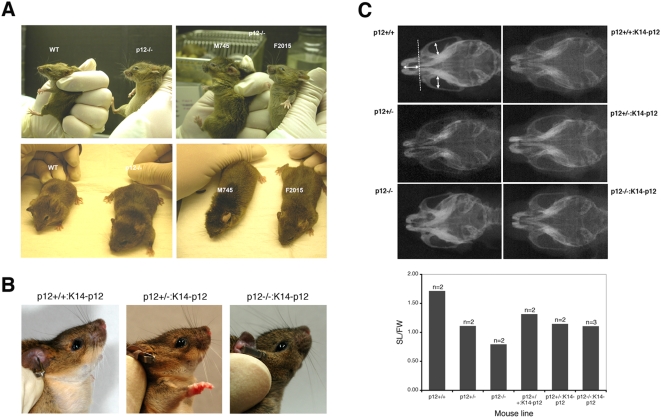
One of the interesting findings was that the two homozygous Cdk2ap1 knockout mice (M745 and F2015) born and lived exhibited similar craniofacial defects with a noticeable short snout and a round forehead compared to wild type (Fig. 5A). We further reasoned that if Cdk2ap1 plays a specific role in craniofacial development, the hybrid mice with transgenic Cdk2ap1 expression should overcome the craniofacial abnormality observed in Cdk2ap1−/− mice. As shown in Fig. 5B, the hybrid Cdk2ap1−/−:K14-Cdk2ap1 mice did not show much phenotypic difference from Cdk2ap1+/+:K14-Cdk2ap1 or Cdk2ap1+/−:K14-Cdk2ap1 mice. To examine this phenotype in more detail, we have performed X-ray cephalometry of the animals. We analyzed the morphometric difference of the facial skeletons of the animals by comparing the Snout Length vs. Face Width (SL/FW) (Fig. 5C). As shown in the X-ray result, Cdk2ap1−/− animal showed a noticeable reduction of SL/FW ratio compared to the wild type (Table 3). The wild type Cdk2ap1+/+ mice showed an average SL/FW ratio of 1.72, while Cdk2ap1−/− mice showed an average SL/FW ratio of 0.80, which is only 47.0% of the wild type. This clearly shows that the survived Cdk2ap1−/− mice were born with a reduced snout length compared to wild type mice. Furthermore, we also observed the reduction of SL/FW ratio in heterozygous Cdk2ap1 knockout mice compared to WT. The SL/FW ratio in heterozygous Cdk2ap1 knockout mice was 1.11, which is 65.0% of the wild type. To see if the abnormal craniofacial morphology is specifically due to the loss of Cdk2ap1 expression, we also examined the SL/FW ratio of Cdk2ap1−/−:K14-Cdk2ap1 mice rescued by the transgenic complementation of Cdk2ap1 expression with K14-Cdk2ap1 mice. As summarized in Table 3, we observed that the average SL/FW ratio of Cdk2ap1−/− hybrid mice (SL/FW = 1.10) were returned to that of Cdk2ap1+/− hybrid mice (SL/FW = 1.18) after complementation of Cdk2ap1 expression by transgenic Cdk2ap1. In both Cdk2ap1+/−:K14-Cdk2ap1 and Cdk2ap1−/−:K14-Cdk2ap1 mice, the SL/FW ratio was still smaller than Cdk2ap1+/+:K14-Cdk2ap1 mice (89.0% and 83.0% of the Cdk2ap1+/+:K14-Cdk2ap1 mice, respectively). Comparison of the SL/FW ratios from heterozygous mice and homozygous mice without or with transgenic complementation showed that the ratio was improved in the hybrid animals (increased from 73% without complementation to 93% with complementation), which implies that the complementary expression of Cdk2ap1 in Cdk2ap1−/−:K14-Cdk2ap1 mice rescued the craniofacial deformity to the state of the heterozygous Cdk2ap1+/−:K14-Cdk2ap1 mice.
Figure 5. Craniofacial abnormality of survived Cdk2ap1−/− mice.
A. The craniofacial morphology was noticeably different between wild type (WT) and Cdk2ap1−/− mice. Two survived Cdk2ap1−/− mice (M745 and F2015) were also compared side-by-side and both mice showed similar craniofacial abnormality compared to WT. B. The complement expression of Cdk2ap1 in Cdk2ap1−/− mice by transgenic approach resulted in an increased incidence of homozygous Cdk2ap1 knockout mice. The rescued mice (Cdk2ap1−/−:K14-Cdk2ap1) also showed the recovery of the craniofacial deformity to the state of the heterozygous (Cdk2ap1+/−:K14-Cdk2ap1) mice. C. The craniofacial differences between animals were analyzed by measuring the Snout Length (SL) and Facial Width (FW) ratio measured on the X-ray films obtained in a standardized head position as indicated by the arrows. The average ratio was depicted and compared between Cdk2ap1+/+ and Cdk2ap1−/− mice, and also the hybrid animals with Cdk2ap1+/+:K14-Cdk2ap1, Cdk2ap1+/−:K14-Cdk2ap1, and Cdk2ap1−/−:K14-Cdk2ap1 genotype.
Table 3. Snout Length (SL) and Facial Width (FW) ratio.
| Mouse line | Cdk2ap1+/+ | Cdk2ap1+/− | Cdk2ap1−/− | Cdk2ap1+/+:K14-Cdk2ap1 | Cdk2ap1+/−:K14-Cdk2ap1 | Cdk2ap1−/−:K14-Cdk2ap1 |
| SL/FW | 1.82 | 1.07 | 0.67 | 1.42 | 1.18 | 1.22 |
| 1.61 | 1.15 | 0.92 | 1.21 | 1.18 | 0.98 | |
| 1.12 | ||||||
| Average | 1.72 | 1.11 | 0.80 | 1.32 | 1.18 | 1.10 |
| Vs. +/+ | 1 | 0.65 | 0.47 | 1 | 0.89 | 0.83 |
Expression of Cdk2ap1 in mesenchymal stem cells in rescued hybrid animals
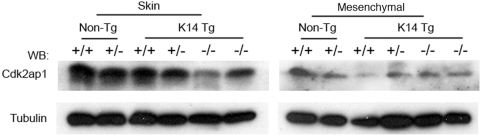
Since K14 promoter is known to be active in epithelial lineage cells, it is necessary to demonstrate if the rescue of craniofacial defect by transgene is through its expression in mesenchymal cells. To address this, we have isolated mesenchymal stem and progenitor cells from bone marrow in rescued hybrid animals. As shown in Fig. 6, the expression of Cdk2ap1 was detected in skin tissues and mesenchymal cells from Cdk2ap1+/+ and Cdk2ap1+/− mice. It was also found that the expression of transgenic Cdk2ap1 was detected in both skin tissues and mesenchymal cells from Cdk2ap1−/−:K14-Cdk2ap1 mice. This demonstrates that the expression of K14-Cdk2ap1 transgene is not limited to epithelial cell lineages, but also it is actively expressed in mesenchymal cells from bone marrow of Cdk2ap1−/−:K14-Cdk2ap1.
Figure 6. Expression of K14-Cdk2ap1 in mesenchymal cells in the rescued mice.
A. Expression of K14-Cdk2ap1 transgene was detected in skin tissues from Cdk2ap1−/−:K14-Cdk2ap1 mice. Earsnips from non-transgenic or transgenic mice (Cdk2ap1+/+:K14-Cdk2ap1, Cdk2ap1+/−:K14-Cdk2ap1, Cdk2ap1−/−:K14-Cdk2ap1) were harvested and subjected to Western analysis with anti-Cdk2ap1 antibody. The loading was confirmed by staining with anti-tubulin antibody. B. To examine if the K14-Cdk2ap1 transgene is expressed in mesenchymal lineage cells, bone marrow was isolated from Cdk2ap1−/−:K14-Cdk2ap1 mice. Cultured mesenchymal stem and progenitor cells were subjected to Western analysis to examine the expression of K14-Cdk2ap1 in Cdk2ap1−/−:K14-Cdk2ap1 mice.
Altered in vivo pluripotency in Cdk2ap1−/− mESCs
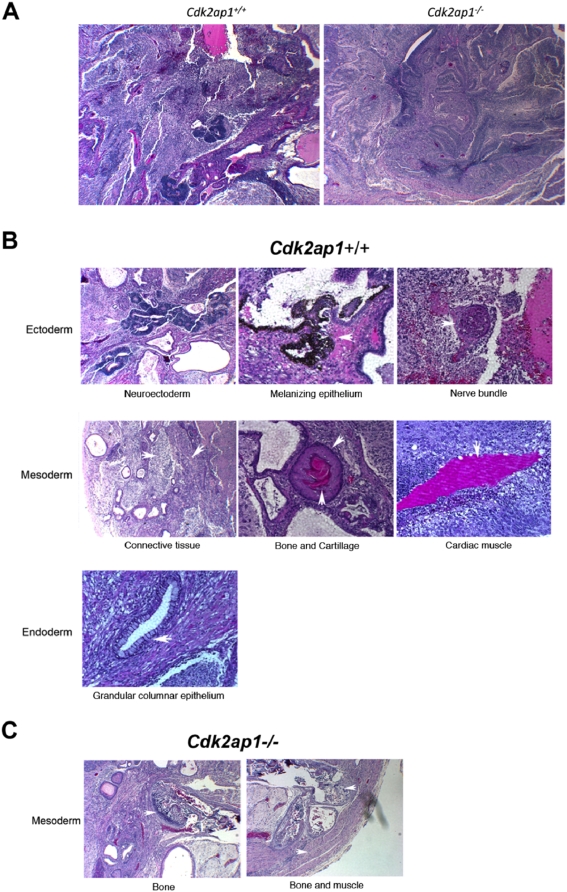
Our extensive in vitro studies with Cdk2ap1−/− mESCs showed that Cdk2ap1 plays a role in mESC differentiation by modulating the expression of stem cell marker genes and also differentiation-related genes, especially involved in trophoblast and mesoderm differentiation (Kim et al., manuscript in review). The in vivo pluripotency of Cdk2ap1−/− mESCs was examined by teratoma formation analysis. Either Cdk2ap1+/+ or Cdk2ap1−/− mESCs (0.5–1.0×106 cells) were transplanted into the testis of SCID mice. After 4–6 weeks, mice were euthanized and tumors were extracted for histological analysis [29]. As shown in Fig. 7, both Cdk2ap1+/+ and Cdk2ap1−/− mESCs formed teratomas. However, there was noticeable difference between tumors from Cdk2ap1+/+ and Cdk2ap1−/− mESCs at low magnification. Tumors from Cdk2ap1−/− mESCs did not show definable differentiation compared to tumors from Cdk2ap1+/+ mESCs (Fig. 7A). Cdk2ap1+/+ mESCs showed proper differentiation and commitment into three different lineages as shown in the Fig. 7B. Interestingly, only types of tissues we observed in tumors from Cdk2ap1−/− mESCs were bones and muscles, which are mesoderm lineage tissues (Fig. 7C). This result demonstrates that the deletion of Cdk2ap1 led to a compromised in vivo pluripotency in mESCs. This is very intriguing finding that suggests a potential role of Cdk2ap1 in mouse embryo development and also it provides us an insight on a potential mechanism of the early embryonic lethality in Cdk2ap1−/− mice. Taken together, this suggests that the deletion of Cdk2ap1 could lead to an abrogated differentiation of mESCs during early embryo development and further result in the early embryonic lethality potentially through defect in cellular differentiation and development.
Figure 7. Cdk2ap1−/− mESCs showed an abrogated in vivo pluripotency.
In vivo pluripotential competence of Cdk2ap1−/− mESCs was evaluated by teratoma formation analysis. Cdk2ap1+/+ or Cdk2ap1−/− mESCs were transplanted into the testis of SCID mice in duplicate as described by Conway et al. (29). After 4 weeks, tumors were extracted and subjected to fixation and sectioning. The slides were stained with H&E and examined under bright field microscope. A. Gross examination of teratoma sections from Cdk2ap1+/+ and Cdk2ap1−/− mESCs (×4 magnification). B. Specified three lineages committed from Cdk2ap1+/+ mESCs. C. A restricted commitment of Cdk2ap1−/− mESCs to a certain mesoderm lineage.
Discussion
Data presented in this paper clearly demonstrate that Cdk2ap1 is an essential gene in the proper development of mouse embryos. It remains to be seen what kind of role Cdk2ap1 precisely play during embryogenesis. We have obtained significant amount of data supporting that the lethality is due to an abrogated differentiation of mESCs, which is involved in placenta development (Kim et al., manuscript in review). We also demonstrated that a specific deletion of Cdk2ap1 leads to the aberrant craniofacial development. This finding may be noteworthy even though we only have two survived animals showing this phenotype due to the high penetrance of lethality in Cdk2ap1−/− animal. It is well known that the transforming growth factor-beta (TGF-β) and Smad pathway is involved in craniofacial morphogenesis [30], [31], [32], [33]. We have extensive lines of evidence demonstrating the function of Cdk2ap1 in cell cycle regulation, apoptosis, and also growth arrest and tumor regression [5], [8], [9], [10], [34]. Interestingly, we have also demonstrated that the functional involvement of Cdk2ap1 in TGF-β induced growth arrest. It has been shown that Cdk2ap1 is activated by TGF-β-Smad2 and mediates TGF-β induced growth arrest in normal diploid cells and the loss of Cdk2ap1 expression in squamous cell carcinoma is correlated with disrupted TGF-β-Smad signaling pathway [9], [11]. The role of the TGF-β family in normal embryonic development, specifically in development of the craniofacial region, has been well documented [30], [33]. The function of these molecules is vital to development of the secondary palate. They regulate maxillary and palate mesenchymal cell proliferation and extracellular matrix synthesis. The function of this growth factor family is particularly critical in that perturbation of either process results in a cleft of the palate. The cellular and phenotypic effects of TGF-β on embryonic craniofacial tissue have been extensively examined, but the specific genes that function as downstream mediators of TGF-β in maxillary/palatal development are poorly defined. We have used a transgenic approach to rescue the lethality by crossing with the K14-Cdk2ap1 transgenic mice. It is well documented that K14 promoter is active in epithelial tissues, including skin, palate, tongue, and ovary [35], [36], [37], [38], [39], [40]. In the previous paper demonstrating the phenotype of K14-Cdk2ap1 mice, we have observed the overexpression of Cdk2ap1 in epithelial tissues [12]. Now the question we have is how the overexpression of Cdk2ap1 directed in epithelial tissues in K14-Cdk2ap1 mice rescued craniofacial defects in mesenchymal origin. It is known that epithelial cells can undergo transition to mesenchymal cells through the process called epithelial-to-mesenchymal transition (EMT) [41], [42]. The roles of EMT have been described in embryonic development and morphogenesis. In embryonic development, it is known that EMT affects tissues on a global scale, such as gastrulation and also formation of a three-layered embryo. It has an effect on organogenesis of heart, musculoskeletal system, and craniofacial structures such as palate [42]. It is also known that TGF-β signaling is significantly associated with the induction and maintenance of EMT [43], [44], [45]. Based on these, we speculate that the overexpression of Cdk2ap1 in epithelial cells in Cdk2ap1 knockout and K14-Cdk2ap1 hybrid embryo could have an effect on the embryo development and on the craniofacial development possibly through the EMT process.
From in vitro study using Cdk2ap1 knockout mouse embryonic stem (mES) cells, we are beginning to unveil potential roles of Cdk2ap1 during differentiation process (Kim et al., manuscript in review). We have observed significantly compromised differentiation potential in Cdk2ap1−/− mESCs and we are in the process of detailing a mechanism that underlies the cellular and molecular significance of Cdk2ap1 during the early embryogenesis. Even though this is the first report demonstrating the importance of Cdk2ap1 in the embryogenesis, other lines of evidences also suggest the potential involvement of Cdk2ap1 in stem cell biology and also implantation process [13], [14], [16]. Once our ongoing attempt to generate an inducible conditional Cdk2ap1 knockout mouse model is complete, we will be able to address standing questions regarding the essential role of Cdk2ap1 in the animal development and its underlying mechanisms.
Acknowledgments
The authors thank the UCLA DLAM staff, Dr. Marcelo Cuoto, Michelle Steel and Joanna Gallino for the maintenance of the animals and help with timed pregnancy test described in this paper. We also thank Drs. Greg Lawson and Noh-Jin Park for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by PHS Grants R01 DE 14857 [D.T.W.] and T32 DE 007296-08 [Y.K.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Todd R, McBride J, Tsuji T, Donoff RB, Nagai M, et al. Deleted in oral cancer-1 (doc-1), a novel oral tumor suppressor gene. Faseb J. 1995;9:1362–1370. doi: 10.1096/fasebj.9.13.7557027. [DOI] [PubMed] [Google Scholar]
- 2.Tsuji T, Duh FM, Latif F, Popescu NC, Zimonjic DB, et al. Cloning, mapping, expression, function, and mutation analyses of the human ortholog of the hamster putative tumor suppressor gene Doc-1. J Biol Chem. 1998;273:6704–6709. doi: 10.1074/jbc.273.12.6704. [DOI] [PubMed] [Google Scholar]
- 3.Shintani S, Mihara M, Terakado N, Nakahara Y, Matsumura T, et al. Reduction of p12DOC-1 expression is a negative prognostic indicator in patients with surgically resected oral squamous cell carcinoma. Clin Cancer Res. 2001;7:2776–2782. [PubMed] [Google Scholar]
- 4.Kim Y, Tsuji T, Elovic A, Shintani S, Mihara M, Salih E, Kohno Y, Chin BR, Patel V, Wong DTW, Todd R. Murine doc-1 cDNA cloning, sequencing and expression in normal adult tissues. Int J Oral Biol December. 2001;2001:87–91. [Google Scholar]
- 5.Kim Y, McBride J, Zhang R, Zhou X, Wong DT. p12(CDK2-AP1) mediates DNA damage responses induced by cisplatin. Oncogene. 2005;24:407–418. doi: 10.1038/sj.onc.1208222. [DOI] [PubMed] [Google Scholar]
- 6.Matsuo K, Shintani S, Tsuji T, Nagata E, Lerman M, et al. p12(DOC-1), a growth suppressor, associates with DNA polymerase alpha/primase. Faseb J. 2000;14:1318–1324. doi: 10.1096/fj.14.10.1318. [DOI] [PubMed] [Google Scholar]
- 7.Kohno Y, Patel V, Kim Y, Tsuji T, Chin BR, et al. Apoptosis, proliferation and p12(doc-1) profiles in normal, dysplastic and malignant squamous epithelium of the Syrian hamster cheek pouch model. Oral Oncol. 2002;38:274–280. doi: 10.1016/s1368-8375(01)00055-0. [DOI] [PubMed] [Google Scholar]
- 8.Shintani S, Ohyama H, Zhang X, McBride J, Matsuo K, et al. p12(DOC-1) is a novel cyclin-dependent kinase 2-associated protein. Mol Cell Biol. 2000;20:6300–6307. doi: 10.1128/mcb.20.17.6300-6307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu MG, Hu GF, Kim Y, Tsuji T, McBride J, et al. Role of p12(CDK2-AP1) in transforming growth factor-beta1-mediated growth suppression. Cancer Res. 2004;64:490–499. doi: 10.1158/0008-5472.can-03-2284. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo ML, Kim Y, St John MA, Wong DT. p12CDK2-AP1 gene therapy strategy inhibits tumor growth in an in vivo mouse model of head and neck cancer. Clin Cancer Res. 2005;11:3939–3948. doi: 10.1158/1078-0432.CCR-04-2085. [DOI] [PubMed] [Google Scholar]
- 11.Peng H, Shintani S, Kim Y, Wong DT. Loss of p12CDK2-AP1 expression in human oral squamous cell carcinoma with disrupted transforming growth factor-beta-Smad signaling pathway. Neoplasia. 2006;8:1028–1036. doi: 10.1593/neo.06580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueiredo ML, Dayan S, Kim Y, McBride J, Kupper TS, et al. Expression of cell-cycle regulator CDK2-associating protein 1 (p12CDK2AP1) in transgenic mice induces testicular and ovarian atrophy in vivo. Mol Reprod Dev. 2006;73:987–997. doi: 10.1002/mrd.20458. [DOI] [PubMed] [Google Scholar]
- 13.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 14.Sharov AA, Piao Y, Matoba R, Dudekula DB, Qian Y, et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diederichs S, Baumer N, Schultz N, Hamra FK, Schrader MG, et al. Expression patterns of mitotic and meiotic cell cycle regulators in testicular cancer and development. Int J Cancer. 2005;116:207–217. doi: 10.1002/ijc.21034. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Lee SA, Shim C, Khang I, Lee KA, et al. Identification of estrogen-regulated genes in the mouse uterus using a delayed-implantation model. Mol Reprod Dev. 2003;64:405–413. doi: 10.1002/mrd.10232. [DOI] [PubMed] [Google Scholar]
- 17.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 18.LeCouter JE, Kablar B, Whyte PF, Ying C, Rudnicki MA. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development. 1998;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- 19.Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 20.Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, et al. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bair SR, Mellon SH. Deletion of the mouse P450c17 gene causes early embryonic lethality. Mol Cell Biol. 2004;24:5383–5390. doi: 10.1128/MCB.24.12.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, et al. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- 23.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 24.Adham IM, Sallam MA, Steding G, Korabiowska M, Brinck U, et al. Disruption of the pelota gene causes early embryonic lethality and defects in cell cycle progression. Mol Cell Biol. 2003;23:1470–1476. doi: 10.1128/MCB.23.4.1470-1476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PL, Liu F, Cai S, Lin X, Li A, et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- 27.Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- 28.White J, Stead E, Faast R, Conn S, Cartwright P, et al. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conway AE, Lindgren A, Galic Z, Pyle AD, Wu H, et al. A Pluripotency and Self-Renewal Program Controls the Expansion of Genetically Unstable Cancer Stem Cells in Pluripotent Stem Cell-Derived Tumors. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunker N, Krieglstein K. Targeted mutations of transforming growth factor-beta genes reveal important roles in mouse development and adult homeostasis. Eur J Biochem. 2000;267:6982–6988. doi: 10.1046/j.1432-1327.2000.01825.x. [DOI] [PubMed] [Google Scholar]
- 31.Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 32.Greene RM, Pisano MM. Recent advances in understanding transforming growth factor beta regulation of orofacial development. Hum Exp Toxicol. 2005;24:1–12. doi: 10.1191/0960327105ht492oa. [DOI] [PubMed] [Google Scholar]
- 33.Dudas M, Kaartinen V. Tgf-beta superfamily and mouse craniofacial development: interplay of morphogenetic proteins and receptor signaling controls normal formation of the face. Curr Top Dev Biol. 2005;66:65–133. doi: 10.1016/S0070-2153(05)66003-6. [DOI] [PubMed] [Google Scholar]
- 34.Cwikla SJ, Tsuji T, McBride J, Wong DT, Todd R. doc-1–mediated apoptosis in malignant hamster oral keratinocytes. J Oral Maxillofac Surg. 2000;58:406–414. doi: 10.1016/s0278-2391(00)90924-8. [DOI] [PubMed] [Google Scholar]
- 35.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 36.Gallicano GI. Composition, regulation, and function of the cytoskeleton in mammalian eggs and embryos. Front Biosci. 2001;6:D1089–1108. doi: 10.2741/gallican. [DOI] [PubMed] [Google Scholar]
- 37.Hafner M, Wenk J, Nenci A, Pasparakis M, Scharffetter-Kochanek K, et al. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–181. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- 38.Strothmann K, Simoni M, Mathur P, Siakhamary S, Nieschlag E, et al. Gene expression profiling of mouse Sertoli cell lines. Cell Tissue Res. 2004;315:249–257. doi: 10.1007/s00441-003-0834-x. [DOI] [PubMed] [Google Scholar]
- 39.Plikus M, Wang WP, Liu J, Wang X, Jiang TX, et al. Morpho-regulation of ectodermal organs: integument pathology and phenotypic variations in K14-Noggin engineered mice through modulation of bone morphogenic protein pathway. Am J Pathol. 2004;164:1099–1114. doi: 10.1016/S0002-9440(10)63197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 41.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 42.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 43.Mizuno S, Kurosawa T, Matsumoto K, Mizuno-Horikawa Y, Okamoto M, et al. Hepatocyte growth factor prevents renal fibrosis and dysfunction in a mouse model of chronic renal disease. J Clin Invest. 1998;101:1827–1834. doi: 10.1172/JCI1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 45.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, et al. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]