Abstract
As a naturally occurring nanocapsule abundantly expressed in nearly all-eukaryotic cells, the barrel-shaped vault particle is perhaps an ideal structure to engineer for targeting to specific cell types. Recombinant vault particles self-assemble from 96 copies of the major vault protein (MVP), have dimensions of 72.5 × 41 nm, and have a hollow interior large enough to encapsulate hundreds of proteins. In this study, three different tags were engineered onto the C-terminus of MVP: an 11 amino acid epitope tag, a 33 amino acid IgG-binding peptide, and the 55 amino acid epidermal growth factor (EGF). These modified vaults were produced using a baculovirus expression system. Our studies demonstrate that recombinant vaults assembled from MVPs containing C-terminal peptide extensions, display these tags at the top and bottom of the vault on the outside of the particle, and can be used to specifically bind the modified vaults to epithelial cancer cells (A431) via the epidermal growth factor receptor (EGFR), either directly (EGF modified vaults) or as mediated by a monoclonal antibody (anti-EGFR) bound to recombinant vaults containing the IgG-binding peptide. The ability to target vaults to specific cells represents an essential advance towards using recombinant vaults as delivery vehicles.
Keywords: vaults, nanoparticles, delivery, targeting, Z domain, cryo-electron microscopy, EGF receptor
Major challenges for treating a variety of diseases are the lack of suitable methods to deliver therapeutic agents (e.g. drugs, proteins, or nucleic acids) to specific host tissues. Encapsulation of these agents into nanoparticles is a promising approach that could increase delivery to specific tissues and reduce systemic side effects by altering the pharmacokinetics of the encapsulated compound.1-4 Nanoparticle targeting can be passive, where the drug circulates longer because it is in a protective environment and these particles accumulate preferentially in tumor tissues through an enhanced permeability and retention effect.5,6 This effect is due to increased vascularization and decreased drainage of the tumor to the lymphatic system.7 Active targeting of nanocapsules can be accomplished by coupling them to antibodies, peptides, or small molecules that bind to cell surface receptors. Antibody-mediated targeting to cell surface receptors has been used to target dendrimers to the EGFR receptor,8 immunoliposomes to HER2-overexpressing cells,9 and to broaden the host specifity of adenoviruses.10 Peptide ligands that bind to cell surface receptors have been used to target L particles (EGF),11 adenovirus and nanoparticles (RGD),12-14 and prostate-specific peptides identified by phage display.15,16 Small molecules have also been attached to nanoparticles for cell-specific targeting.17,18 Finally, direct conjugation of cytotoxic drugs to a ligand that recognizes a well-characterized cell surface marker has also been used to target immunotoxins to specific cancer cells.19,20
Vaults are highly conserved, barrel-shaped, 13 MDa ribonucleoprotein particles, found in phylogeny as diverse as mammals, avians, amphibians, echinaderms, kinetoplasts, and amoebas.21 There are between 104-106 vault particles per mammalian cell and they do not trigger autoimmunity. Their large structure and size, coupled with the potential to encompass hundreds of proteins, have led to the proposal that they could be utilized as natural nanocapsules for drug, nucleic acid, or protein delivery.22 Native vaults are formed from 96 copies of an ∼100 kDa protein, termed the major vault protein (MVP),23 and two vault-associated proteins; vault poly-ADP-ribose polymerase (VPARP, 193 kDa)24 and telomerase associated protein one (TEP1, 290 kDa),25 and a small untranslated RNA, vRNA.26-28 Although vaults have been implicated in many cellular pathways including cell signaling, nuclear-cytoplasmic transport, multi-drug resistance, and innate immunity, their precise function remains unclear.29
All of the information for vault particle assembly is inherent in the MVP protein sequence.30 When insect cells are infected with a baculovirus containing an MVP cDNA, large quantities of recombinant vaults are produced; demonstrating that multimerization of this single protein is sufficient to form the exterior shell of the particle. Cryo-electron microscopy (cryoEM) and image reconstruction of recombinant vaults, formed from modified MVPs, revealed that they form empty capsules with an overall structure virtually identical to native vaults.31 Difference mapping indicated that all of the MVP N-termini come together around the particle waist and extend toward the interior of the particle. A crystal structure of the vault shell at ∼ 9 Å resolution has been used to model an MVP monomer as it extends from the particle waist to the cap.32 A vault-targeting peptide sequence has been identified at the C-terminus of the vault interacting protein VPARP (aa 1563-1724).24,33 This sequence has been designated, mINT, for the minimal interaction domain. When heterologous proteins are fused to mINT, they are directed to the inside of recombinant vaults, and these packaged proteins retain their native properties.22 Difference mapping has shown that the mINT domain binds to the inside of recombinant vaults at two locations, above and below the waist of the particle. The exterior shell of the vault acts as a protective barrier, as either fluorescent or enzymatic mINT containing fusion proteins are quenched with slower kinetics when packaged inside of vaults. However, recombinant vaults are not rigid structures, as they can package the vault-associated proteins, VPARP and/or TEP1, in cell lysates even after the MVP shell of vaults has formed.34 In addition, recombinant vaults will dissociate into halves at pH<4.35,36 Vaults have been shown to encapsulate a semiconducting polymer and pH lability of cross-linked vaults maybe useful for controlled release of encapsulated materials.37,38 The dynamic nature of recombinant vaults suggests a mechanism whereby vaults can interact with their environment. These properties have led us to explore their use as biocompatible nanocapsule carriers. Previously we have shown that vaults are non-specifically taken into HeLa cells probably by endocytosis suggesting that vaults maybe useful as a general delivery vehicle. However, one of our goals is to use vaults to target cancer cells for destruction without affecting normal cells. Thus their utility as delivery vehicles requires that a means of targeting vaults be developed. In this study, we show that when C-terminal peptide extensions are added to MVP, recombinant vaults are formed with the peptide accessible on the exterior surface of the particles at the caps. These peptides were used to target vaults to the cell surface receptor for epidermal growth factor, EGFR, either directly or when complexed with an antibody that binds to the external portion of the receptor. We chose EGFR as it is up-regulated in numerous cancer cell types. Thus both specific (peptide-directed) and general (antibody-mediated) methods can be used to target recombinant vault particles to cell surface receptors, representing a necessary step towards developing vaults as nanodelivery vehicles.
RESULTS AND DISCUSSION
Structural characterization of C-terminally tagged vaults
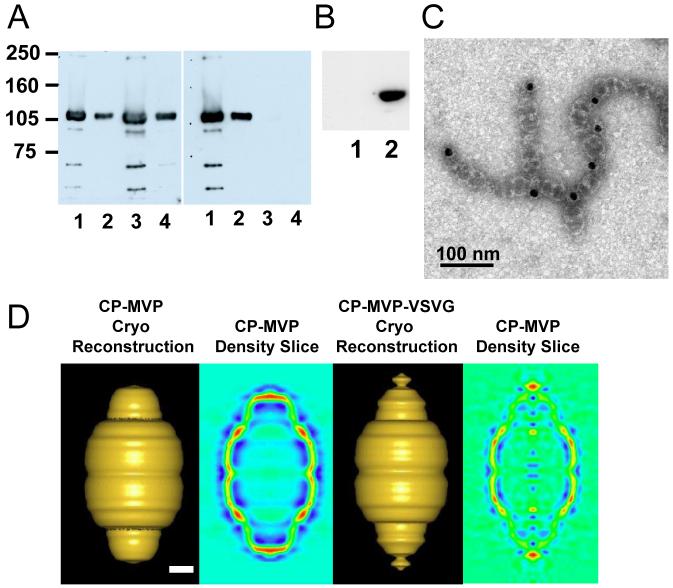
The N-terminus of MVP is located at the waist of the vault particle, extending into the interior of the barrel wall, as determined by cryoEM difference mapping31 and as modeled in a 9 Å resolution crystal structure of recombinant vaults.32 By deduction then the C-terminus is likely localized in the caps of the barrel. An 11 amino acid epitope tag derived from the vesticular stomatitis virus glycoprotein (VSVG) was fused to the C-terminus of MVP that also contained an N-terminal cysteine-rich (CP) peptide to form CP-MVP-VSVG. It was previously demonstrated that addition of the CP peptide to the MVP N-terminus results in vaults with greater particle regularity and stability.31 When the CP-MVP-VSVG fusion protein was expressed in insect cells using the baculovirus system, recombinant vault particles formed, and these particles were purified as previously described.30 The penultimate step during vault purification is fractionation on a discontinuous sucrose gradient; generally vaults fractionate in the 40 and 45% layer depending on whether or not the particles are empty or filled with mINT fusion proteins. Empty particles are found in higher concentrations in the 40% layer and filled particles fractionate in the 45% layer. Recombinant CP-MVP-VSVG (Fig. 1A, lanes 1 and 2) and CP-MVP (Fig. 1A, lanes 3 and 4) vaults, purified from the 40 and 45% sucrose gradient fractions, were analyzed by immunoblotting, with either an anti-MVP antibody (left panel) or an anti-VSVG antibody (right panel), and the ∼100 kDa CP-MVP-VSVG tagged protein was readily detected confirming that the C-terminal tag was fused in frame (right panel). To demonstrate the specificity and accessibility of the C-terminal epitope tag, pull-down experiments with anti-VSVG or anti-T7 (a control to evaluate non-specific binding) antibody bound beads were carried out. A crude extract from CP-MVP-VSVG infected Sf9 cells was incubated with either anti-T7 (Fig. 1B, lane 1) or anti-VSVG antibody (Fig. 1B, lane 2) plus protein G beads, and after washing, vault proteins bound to the beads were detected by immunoblotting with an anti-MVP antibody (Fig. 1B). MVP was only detected in precipitates containing the anti-VSVG antibody (lane 2) and not from precipitates containing the anti-T7 antibody (lane 1) suggesting that the C-terminal tag is exposed on the surface of the particles. Purified CP-MVP-VSVG vaults were incubated with anti-VSVG antibody and 15-nm gold-labeled secondary antibody, applied to carbon grids, stained with uranyl acetate, and visualized by transmission electron microscopy (Fig. 1C). Antibody bound vaults were observed to form chains that sometimes branch (Fig. 1C) indicating that the C-termini are exposed and are being linked by antibody. This was further supported by the high incidence of gold at the vault-vault interfaces. As each anti-VSVG antibody has two antigen binding sites, each antibody can bind to two VSVG tags, this results in the formation of chains of vaults even in the absence of secondary antibody (data not shown). No chaining is seen in the absence of antibody (data not shown). A cryoEM single-particle reconstruction of the CP-MVP-VSVG vault provides visual evidence that the ends of the vault are perturbed in the presence of a C-terminal tag on MVP. A set of 273 particle images of CP-MVP-VSVG vaults were selected from cryomicrographs and processed to yield a 3D reconstruction at 38 Å resolution (Fig. 1D). The major difference between cryoEM structures of CP-MVP and CP-MVP-VSVG vaults is the presence of additional density at the ends of the caps. The shape of the cap in the CP-MVP-VSVG vault also appears slightly distorted compared to the cap of CP-MVP vaults. A vertical density slice through the CP-MVP-VSVG vault reconstruction confirms the presence of additional density at the external ends of the caps. The cryoEM reconstruction indicates a location for the C-terminal tag in the vicinity of the vault cap, while the pull-down assay with anti-VSVG antibodies and the chaining of vault particles in the presence of anti-VSVG antibodies indicates that at least a portion of the added C-terminal VSVG peptides are located externally. Together these results suggest that the addition of peptide tags to the C-terminus of MVP could be a means of targeting vaults to cell surface receptors.
Figure 1. The C-terminal peptide tag is located on the exterior of the caps of recombinant vaults.
A. Western blot analysis comparing purified CP-MVP-VSVG (lanes 1 and 2) and CP-MVP (lanes 3 and 4) recombinant vaults to confirm the presence of the VSVG peptide tag. Lanes 1 and 3 represent vaults purified from the 40% sucrose gradient layers and lanes 2 and 4 are from the 45% sucrose gradient layer. The left panel was probed with an anti-MVP polyclonal antibody and the right panel was probed with an anti-VSVG monoclonal antibody. Protein standards are indicated. B. S20 extracts from CP-MVP-VSVG infected Sf9 cells were immunoprecipitated with either anti-T7 (lane 1) or anti-VSVG (lane 2) monoclonal antibodies complexed with protein G sepharose, proteins bound to the washed beads were analyzed by Western blotting with an anti-MVP polyclonal antibody. C. Electron micrograph of uranyl acetate-stained CP-MVP-VSVG vaults incubated with anti-VSVG monoclonal antibody followed by 15 nm gold labeled anti-mouse IgG. D. Comparison of cryoEM single-particle reconstructions of CP-MVP 31 and CP-MVP-VSVG recombinant vaults. Surface views and vertical density slices through the reconstructions are shown with both reconstructions filtered to 38 Å resolution. In the slices the strongest density is in red and the weakest in blue. Scale bar, 100 Å.
Z domain binding specificity
To confer IgG-binding activity on the vault particle, a small synthetic version of the staphylococcal protein A-derived Z domain, Z33, optimized to bind the Fc portion of human IgG1, was fused to the C-terminus of CP-MVP to form CP-MVP-Z.10,39,40 The recombinant MVP containing the Z domain was expressed in insect cells using the baculovirus expression system. To confirm that the IgG-binding activity of the Z domain was retained in assembled particles, pull-down experiments with IgG Sepharose were carried out (Fig. 2A). Crude extracts from either CP-MVP (lane 1) or CP-MVP-Z (lane 2) infected Sf9 cells or purified recombinant CP-MVP (lane 3) or CP-MVP-Z (lane 4) vaults were incubated with IgG-Sepharose beads and after washing, vault bound proteins were detected by immunoblotting with anti-MVP antibody. The ∼100 kDa CP-MVP-Z protein was readily detected in IgG-Sepharose pellets, but not in pellets from CP-MVP vaults. An ELISA was used to show that only vaults containing the Z domain (CP-MVP-Z) bind either rabbit or mouse IgG, whereas vaults lacking the Z domain (CP-MVP-VSVG) do not bind either of these IgG’s (Fig. 2B). The binding affinity of various species of IgG (human, rabbit and mouse) for CP-MVP-Z recombinant vaults was compared using an ELISA (Fig. 2C). As expected the human IgG1 bound CP-MVP-Z recombinant vaults with the highest affinity, with an apparent half-maximal binding affinity of 0.035 nM, whereas rabbit IgG’s bound CP-MVP-Z recombinant vaults with an apparent half-maximal binding affinity of 0.25 nM. Mouse IgGs bound with a much lower affinity in our more stringent ELISA binding conditions. However, all species and isotypes of IgG will bind to the Z domain (data not shown) and this ELISA can be used to determine the binding conditions by varying the antibody concentration, temperature, and length of the binding reaction. The variability in antibody isotype binding to the Z domain may reflect the variable affinity of protein A itself for different antibody isotypes. These data indicate that the Z domain fused to the C-terminus of MVP is accessible in recombinant vaults and binds IgG with broad specificity.
Figure 2. The Z domain is functional.
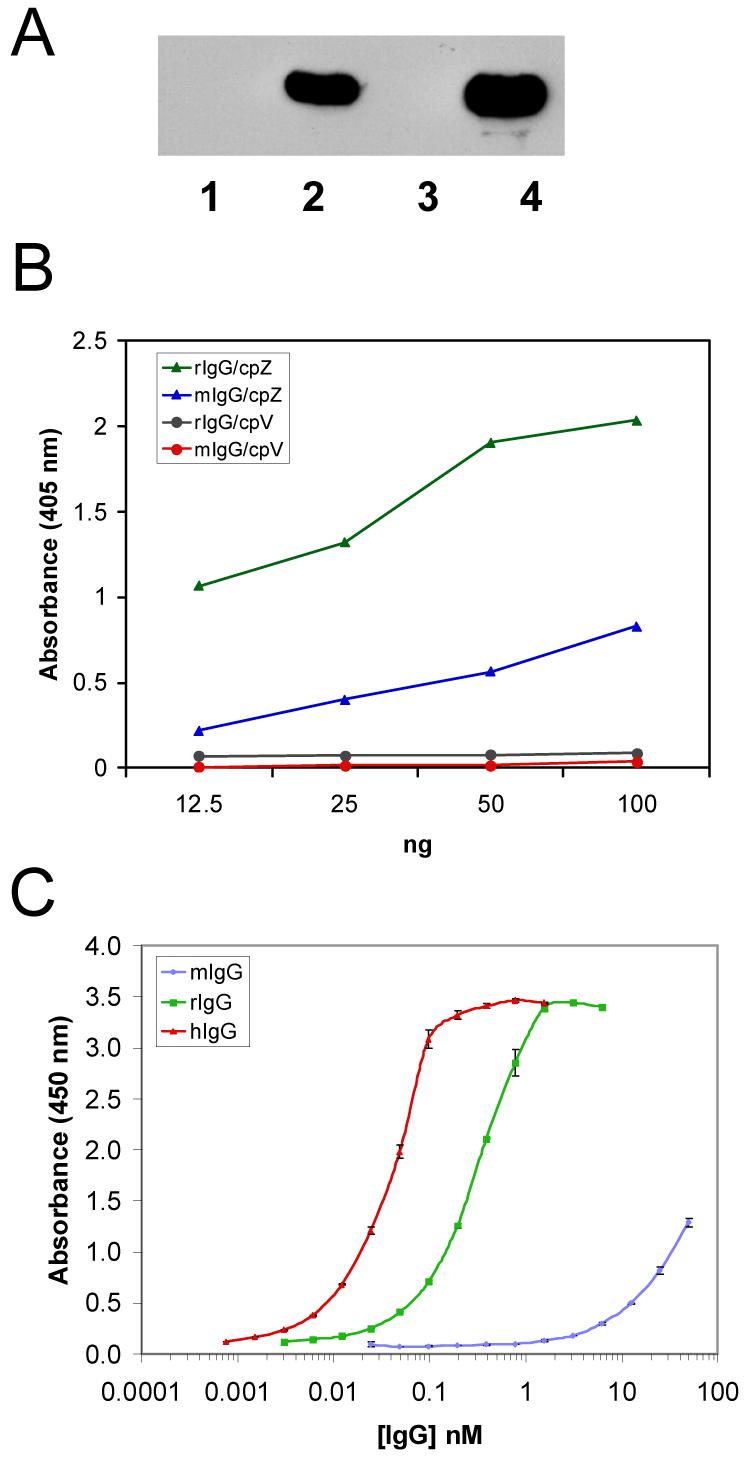
A. Western blot analysis of IgG sepharose affinity purified proteins from either S20 extracts (lanes 1 and 2) or purified recombinant vaults (lanes 3 and 4). Lane 1 and 3 are from CP-MVP and lanes 2 and 4 are from CP-MVP-Z infected Sf9 cells. B. ELISA comparing antibody binding to recombinant vaults with and without the Z domain. Microtiter plates were coated with serial dilutions (100 to 12.5 ng) of recombinant vaults, without the Z domain, CP-MVP-VSVG (cpV, closed circles) and with the Z domain, CP-MVP-Z (cpZ, closed triangles). Z binding assays were performed with 0.5 μg/ml of rabbit IgG (green or black) and 1.67 μg/ml of mouse IgG (blue or red). Antibody binding reactions were carried out at ambient temperature for 1 h, washed, and incubated with the appropriate peroxidase-conjugated secondary antibody for 1 h at ambient temperature. Following washing, antibody binding was detected using a colorimetric reagent, and the absorbance was determined using a plate reader (see Materials and Methods). C. Comparative analysis of CP-MVP-Z binding affinities by human, rabbit, and mouse IgG’s by ELISA. Microtiter plates were coated with 50 ng of CP-MVP-Z recombinant vaults. Z binding assays were performed with 1 μg/ml of human IgG1 (red), 4 μg/ml of rabbit IgG (green), and 32 μg/ml of mouse IgG (blue) in a two-fold dilution series. Antibody binding reactions were carried out as described in B.
Antibody-mediated vault targeting
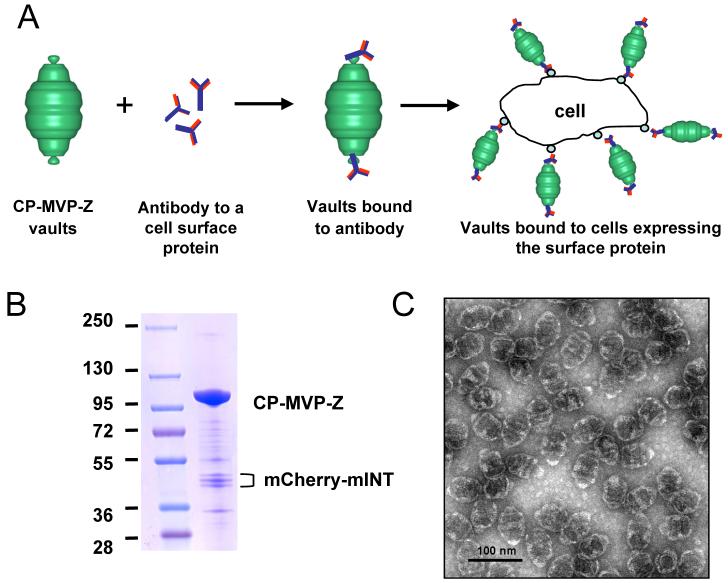
Our next goal was to use antibodies to target vaults to specific cell surface receptors, a model of antibody-mediated vault targeting is depicted (Fig. 3A). Previously the Z domain was inserted into the adenovirus fiber protein and was used to broaden the variety of cells that adenovirus could be targeted to using specific antibodies.10 For these experiments CP-MVP-Z vaults were made that contained a fluorochrome, either mCherry41 or an enhanced green fluorescence protein,22 attached to the previously defined vault-targeting domain, mINT (see above). Purified recombinant vaults (CP-MVP-Z/mCherry-mINT) were analyzed by Coomassie staining (Fig. 3B) and the particles were examined by negative-stain TEM, demonstrating the abundant, barrel-shaped recombinant particles (Fig. 3C). The presence of the stripes on either side of the particle waist is indicative of mINT packaged internal vault contents, here mCherry-mINT.22 When CP-MVP-Z vaults are incubated with antibody and visualize by negative-stain TEM, no chaining of vault particles is observed (data not shown). This was anticipated as the Z domain binds to the Fc portion of IgG.
Figure 3. Model for antibody-mediated targeting of vaults.
A. Model. B. Coomassie stain of purified CP-MVP-Z recombinant vaults containing mCherry-mINT fractionated on 4-15% SDS-PAGE. The major protein bands are the ∼100 kDa CP-MVP-Z and the ∼45 kDa mCherry-mINT (note, mCherry-mINT is a triplet that is likely caused by leaky translation due to the presence of 3 inframe methionines in the first 17 amino acids of mCherry). C. An electron micrograph of the negative-stained recombinant vault particles.
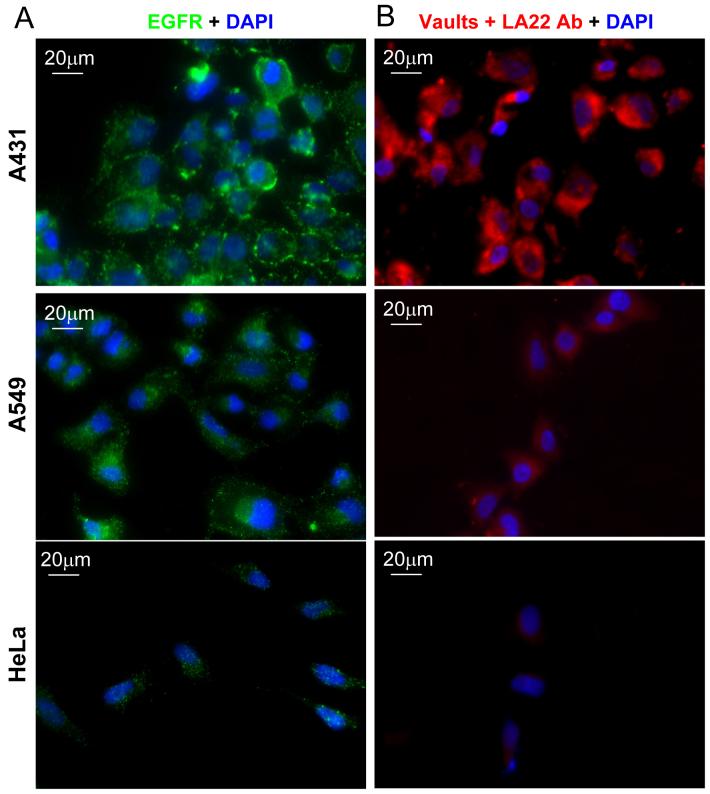
The epidermal growth factor receptor (EGFR) is overexpressed on most cancer cells,42 the epidermal carcinoma cell line (A431) contains upwards of 106 receptors per cell on its surface, versus A549 or HeLa cells that have ∼104 surface EGFRs per cell, this difference was confirmed by indirect immunofluorescence using an anti-EGFR monoclonal antibody (LA22) (Fig. 4A). To target vaults to EGFR, this antibody, an IgG2a isotype, was complexed with purified vaults containing mCherry (CP-MVP-Z/mCherry-mINT) by incubating overnight at 4°C to saturate binding. The antibody bound vaults were then added to A431, HeLa, or A549 cells at 4°C for 1 h. We observed significant vault binding to the A431 cells, detected by the mCherry-mINT red fluorescence compared with binding to either HeLa or A549 cells (Fig. 4B). In the absence of the anti-EGFR antibody very little binding to A431 cells was observed (Supporting Information Fig. S1). These experiments demonstrate that anti-EGFR antibodies bound to Z domain-containing vaults can mediate particle binding to EGFRs.
Figure 4. Antibody-mediated binding of vaults to EGFR.
A. Comparison of EGFR receptors abundance on the A431, A549, and HeLa cells. Cells were plated on glass coverslips overnight, following serum starvation for 1 h the cells were immunostained with the anti-EGFR monoclonal antibody (LA22) (green) and the nuclei were stained with DAPI (blue). B. Visualization of antibody-mediated binding of CP-MVP-Z/mCherry-mINT vaults to EGFR in the indicated cell lines. Cells were plated on glass coverslips overnight. Following serum starvation for 1 h, the cells were incubated with antibody-bound vaults (red, LA22+CP-MVP-Z/mCherry-mINT) for 1 h at 4°C, fixed and the nuclei were stained with DAPI (blue). The observed red fluorescence is the intrinsic fluorescence from the mCherry protein packaged inside of the vaults.
Dendritic cells (DC) are professional antigen presenting cells with a high capacity for phagocytosis of external proteins. To determine whether we could increase the uptake of vaults by DC, we cultured bone marrow derived DC with vaults containing a green fluorescent protein (GL-INT).22 These vaults (CP-MVP-Z/GL-INT, henceforth referred to as GL-vaults) were bound to either rabbit IgG (rIgG) or mouse IgG (mIgG). Vault uptake was quantified by analysis of green fluorescence using flow cytometry. DC exposed to GL-vaults pre-incubated with rIgG (Fig. 5A) displayed an increased uptake (92%) compared with GL-vaults pre-incubated with mIgG (Fig. 5B, 52% total). As expected, no fluorescence was observed in the cells incubated with media only (Fig. 5C). Further incubation of vaults with mouse IgG did not significantly increase the incorporation of GL-vaults (data not shown) revealing that only an antibody that binds to the Z domain with high affinity is capable of modifying vault uptake by DC. In addition, GL-vault uptake in the absence of antibody requires greater numbers of vaults and occurs at a slower rate (data not shown). Taken together, this quantitative assay confirms our vault ELISA results where we observed that Z containing vaults had a higher binding affinity for rIgG compared with mIgG (Fig. 2C). In conclusion, these data indicate that vaults bound to IgG, led to an increased uptake of the particles by DC likely through binding to the Fc receptor. The Z domain binds to the Fc portion of IgG at a separate location from the Fc receptor binding site.
Figure 5. Dendritic cells efficiently incorporate antibody bound vaults.
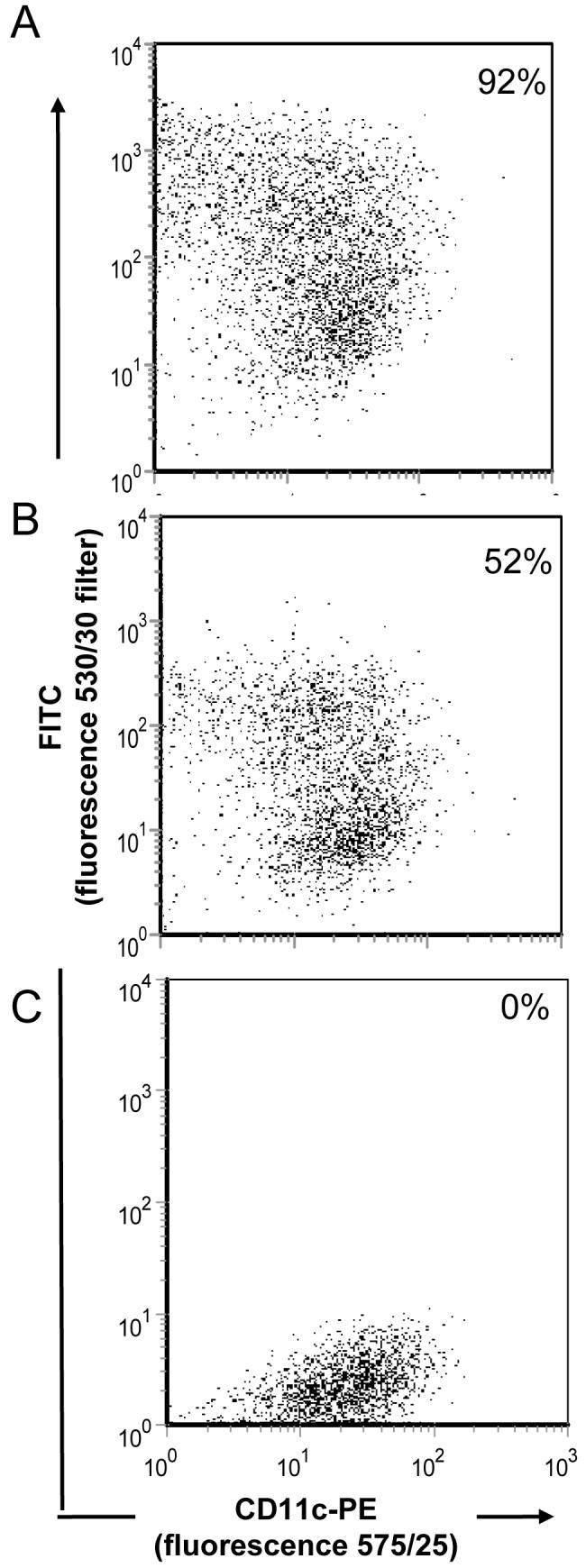
CP-MVP-Z/GL-INT vaults (500 μg) were pre-incubated with either (A) rabbit IgG (250 μg) or (B) mouse IgG (250 μg) for 30 minutes at 37°C. Bone marrow dendritic cells (1×106) were then mixed with the antibody coated vaults (A & B) or media alone (C). Cells were stained for a marker of all dendritic cells, CD11c-PE. Cells were gated on CD11c+DCs and displayed as dotplots.
EGF mediated vault targeting
Direct targeting of vaults to EGFR was evaluated by adding the 55 amino acids encoding mature EGF to the C-terminus of CP-MVP to form CP-MVP-EGF. However, analysis of recombinant vaults produced in Sf9 cells infected with the CP-MVP-EGF baculovirus revealed that the vaults were insoluble. As EGF is highly structured we reasoned that the 48 copies of EGF on the C-terminus of the recombinant vaults could interfere with vault particle formation and might be responsible for the insoluble nature of these particles. To alleviate the potential steric hinderance at the C-terminus we turned to the pFastBac Dual expression system, where we could make a baculovirus that expressed both an N-terminal VSVG-MVP (vMVP) from the p10 promoter (Pp10) and CP-MVP-EGF from the polyhedrin promoter (PPH) to form a composite vault (vMVP/CP-MVP-EGF) containing fewer copies of EGF per vault (Supporting Information Fig. S2). The recombinant MVPs containing these various tags were produced and the number of copies of CP-MVP-EGF in the dual vaults vMVP/CP-MVP-EGF (henceforth referred to as EGF vaults) was estimated using an ELISA with anti-EGF antibody where we plated serial dilutions of recombinant human EGF (rhEGF) and EGF vaults for comparison (data not shown). We determined that EGF vaults contain 6-8 copies of the CP-MVP-EGF incorporated into the particles.
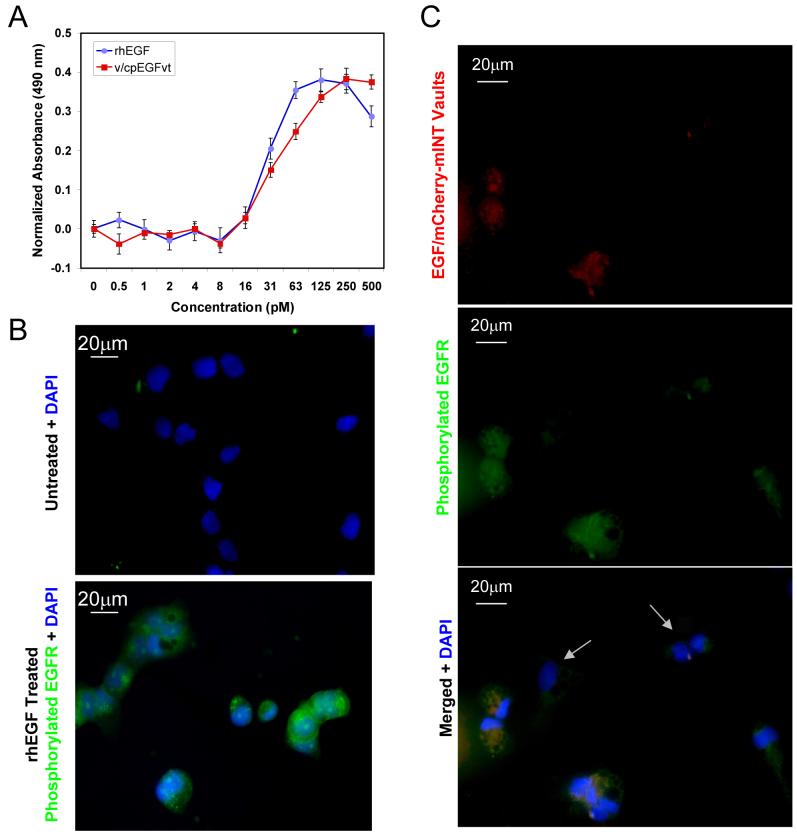
To determine whether or not EGF remains functional in the context of the assembled vault particle a cell proliferation assay was carried out. HeLa cells were treated with equimolar concentrations of either EGF vaults or rhEGF. As shown in Fig. 6A, stimulation of HeLa cell growth by the EGF vaults parallels the activity seen when cells were treated with rhEGF, this data indicates that EGF fused to the C-terminus of MVP retains its native conformation and is functionally accessible on the outside of the assembled vault particle. EGF binding to the cell surface receptor EGFR activates its intrinsic tyrosine kinase activity leading to tyrosyl-phosphorylation of itself and numerous other effector molecules.42 The major autophosphorylation site of human EGFR is tyrosine 1173. Antibodies that specifically recognize phoshorylated Tyr1173 on EGFR were used to show that EGF vaults also activate tyrosine kinase activity leading to autophoshorylation. Serum starved A431 cells were either not treated or incubated with rhEGF (Fig. 6B). As expected only cells treated with rhEGF contain phosphorylated EGFR as shown by immunofluorscence with an anti-phosho-EGFR (Tyr1173) Alexa Fluor 488 conjugated antibody (green). Likewise serum starved A431 cells treated with EGF/mCherry-mINT vaults (red, visualized by the intrinsic fluorescence from the packaged mCherry-mINT) also stimulate EGFR phosphorylation, as shown by immunofluorescence with the antibody described above (green, Fig. 6C). Merged immunofluorescence images revealed that only those cells that bind the EGF/mCherry-mINT vaults phosphorylate EGFR (Fig. 6C) where coincident staining appears as yellow, suggesting that receptor bound to vaults is being phosphorylated. Importantly there are several cells in the field that are only revealed by DAPI staining (Fig. 6C, white arrows) and these cells do not contain phoshorylated EGFR. These data indicate that EGF vaults have a functional activity that directs specific binding of the particles to cell surface receptors and triggers receptor activation in a manner similar to rhEGF, initiating a cascade of signaling pathways as demonstrated here by increased cell proliferation and autophoshorylation of EGFR.
Figure 6. EGF vaults are bioactive.
A. HeLa cell proliferation assay comparing equimolar amounts of recombinant human EGF (rhEGF) with V-MVP/CP-MVP-EGF (v/cp-EGFvt) recombinant vaults indicates that EGF attached to vaults is as bioactive as rhEGF. B. Serum-starved A431 cells were either not treated or treated with 500 ng/ml of rhEGF as indicated. Cells were examined by immunofluorescence with an anti-phosho-EGFR (Tyr1173) antibody conjugated to Alexa Fluor 488 (green) and the nuclei were stained with DAPI (blue). Only cells treated with rhEGF are green. C. Similarly serum-starved A431 cells were treated with 200 μg/ml of purified EGF vaults containing mCherry-mINT (red). Following vault binding, the cells were immunostained with the anti-phosph-EGFR (Tyr1173) antibody conjugated to Alexa Fluor 488 (green). Merged images show coincident staining as yellow indicating that only those cells that bound to the EGF/mCherry-mINT vaults contain phosphorylated EGFR. The white arrows indicate cells in the field that the vaults did not bind to, therefore, they do not contain phoshorylated EGFR.
CONCLUSION
In this work, we show that recombinant vaults containing C-terminal modifications can be specifically targeted to a cell surface receptor. Both specific (EGF mediated) and general (antibody directed) targeting strategies were employed. The latter approach, which utilizes antibodies to target vaults to cell surface molecules, could have wide applications, especially with the growing availability of a wide range of cell-surface binding-human Fc chimera’s.
MATERIALS AND METHODS
Construction of recombinant plasmids
The 11 amino acid sequence of the VSVG tag is YTDIEMNRLGK. A reverse primer encoding this peptide V-Kpn-CCCGGTACCCTACTTTCCCAGTCGGTTCATCTCGATGTCGGTGTACTTCTGTGCTGGCGGCTGAC was designed. All primers used in this study were purchased from Invitrogen. The 3′ end of MVP was PCR amplified using a forward primer MVP1812-GCCTCTGTCACCTTTGATGAC, V-Kpn, and CP-MVP pFastBac plasmid as the template. The PCR product (containing the 3′ end of the MVP cDNA fused in frame to the VSVG DNA) was purified on a Qiagen column, digested with XhoI and KpnI, and ligated to XhoI/KpnI digested CP-MVP pFastBac DNA to form CP-MVP-V pFastBac. All constructs were confirmed by DNA sequence analysis carried out by Laragen. The amino acid sequence used for addition of the Z33 peptide to the C terminus of CP-MVP was FNMQQQ RRFYEALHDPNLNEEQRNAKIKSIRDD40. The nucleotide sequence encoding this peptide was derived from primers used by Volpers and colleagues 10. Three overlapping reverse primers (together resulting in the complete Z33 DNA sequence) were designed as follows: R1-GCAGGGCCTCGTAAAAGCGGCGCTGCTGCTGCATGTTAAACTTCTGTGCTGGCGGCTGACC; R2-ATCTTGGCGTTGCGCTGCTCCTCGTTCAGGTTGGGGTCGTGCAGGGCCTCGTAAAAGC; R3-CTGAGGTACCCTAGTCGTCGCGAATGCTCTTAATCTTGGCGTTGCGCTGCTCC. The first round of PCR amplifications was carried out using a forward primer MVP1812 (see above), R1, and CP-MVP pFastBac plasmid as the template. The resulting PCR product (containing C terminal MVP cDNA fused in frame to part of Z33 DNA) was gel purified and used as a template for a second set of PCR amplifications performed with primers MVP1812 and R2. The product of this PCR amplification was gel purified and used as a template for a final set of PCR that employed MVP1812 and R3 as primers. The product of this reaction was purified, cut with XhoI and KpnI, and ligated to XhoI/KpnI cleaved CP-MVP pFastBac, to generate CP-MVP-Z pFastBac. A synthetic EGF DNA template was constructed by annealing the following primers followed by filling in with Klenow. EGF1-AACTCTGACTCCGAATGCCCGCTGTCTCACGACGGTTATTGCCTGC; EGF2-GTCCAGAGCTTCGATATACATACAAAACACCATCATGCAGGCAATAACCGTCG; EGF3-TATCGAAGCTCTGGACAAATATGCTTGCAACTGTGTTGTTGGTTACATCGGTGAGCG; EGF4-ACGCAGTTCCCACCATTTCAGGTCGCGATACTGGCAACGCTCACCGATGTAACC. EGF was fused onto the C-terminus of MVP by PCR ligation using the following primers: MVP1812 (see above); MVP/EGF forward GTCAGCCGCCAGCACAGAAGAACTCTGACTCCGAATGCCC; MVP/EGF reverse GGGCATTCGGAGTCAGAGTTCTTCTGTGCTGGCGGCTGAC; and EGF reverse CGCGGTACCTTAACGCAGTTCCCACC. Two independent PCR reactions were carried with the first using MVP1812 forward × MVP/EGF reverse primers with the MVP cDNA as the template and the second using MVP/EGF forward × EGF reverse primers using the synthetic EGF DNA as the template. Following gel purification, a second round of PCR was carried out using the products from the first two PCR reactions with the MVP1812 forward and EGF reverse primers. The product of this PCR reaction was purified on a Qiagen PCR column, digested with XhoI and KpnI, agarose gel purified, and ligated to similarily digested CP-MVP in pFastBac. The resultant construct was CP-MVP-EGF pFastBac. The dual expression vector, pFastBac dual (Invitrogen), was used to co-express two different MVPs (VSVG-MVP and CP-MVP-EGF). The VSVG-MVP was subcloned by digesting with EcoR1, end-repaired to blunt the 5′ overhang, followed by digestion with KpnI and ligated to SmaI/KpnI digested pFastBac Dual to create V-MVP pFastBac dual. The KpnI site in CP-MVP-EGF pFastBac was changed to an XbaI site. Then CP-MVP-EGF was subcloned by digesting with EcoR1 and XbaI, and ligated to EcoR1/XbaI digested V-MVP pFastBac dual to create V-MVP/CP-MVP-EGF pFastBac dual, where the p10 promoter controls expression of V-MVP and the polyhedrin (pH) promoter controls expression of CP-MVP-EGF. mCherry in pRSET-B was a kind gift of Dr. Roger Tsien at the University of California, San Diego41. The 236 amino acid coding region of mCherry was fused to minimal interaction domain (mINT) derived from VPARP (amino acids 1563-1724) by PCR ligation using the following primers: mCherry forward- CGCGGATCCATGGTGAGCAAGGGCGAGGA; mCherry-mINT reverse- TCCTGCCAGTGTTGTGTGCACTTGTACAGCTCGTCCATGCC; mCherry-mINT forward- GGCATGGACGAGCTGTACAAGTGCACACAACACTGGCAGGA; mINT reverse- GGGCTCGAGTTAGCCTTGACTGTAATGGAG. Two PCR reactions were carried out with the first using mCherry forward × mCherry-mINT reverse using the mCherry in pRSET as the template, the second used mCherry-mINT forward × mINT reverse using mINT in pET28 as the template. The PCR reactions were purified on a Qiagen column and a second round of PCR was carried out using mCherry forward × mINT reverse and both purified PCR products as the template. The resultant PCR product containing the fused mCherry-mINT was purified on a Qiagen column, digested with Bam H1 and Xho I, gel purified, and ligated to Bam H1/Xho I cut pFastBac to form mCherry-mINT pFastBac. Construction of GL-INT in pFastBac has been described previously.22
Expression and purification of recombinant vaults
Recombinant baculoviruses were generated according to the Bac-to-Bac protocol (Invitrogen). For vault purification, baculovirus-infected Sf9 insect cells were subjected to a standard protocol described previously.30 The protein concentration of purified vault proteins was determined using the BCA assay (Pierce) and their purity was analyzed by fractionating on SDS-PAGE gel followed by staining with Coomassie. All vaults samples were routinely analyzed by staining with uranyl acetate and viewed on an electron microscope as previously described.
CryoEM and single particle reconstruction
For the CP-MVP-VSVG reconstruction, 78 cryoelectron micrographs were collected on an FEI Tecnai-12 (120kV, LaB6) microscope with a Gatan UltraScan 1000 (2k × 2k) CCD camera at a magnification of 67,000 with a defocus value of about -1micron. A total of 273 particle images were selected and processed with Imagic43 imposing D8 symmetry, the final resolution was 38 Å at the FSC 0.5 threshold. The density slices were displayed with the AVS software package (Advanced Visual Systems, Waltham, MA) after filtering the CP-MVP and CP-MVP-VSVG reconstructions to the same resolution.
Antibodies
Primary antibodies used for Western blot analyses were anti-MVP rabbit polyclonal44, anti-MVP monoclonal 1023C (Santa Cruz), anti-VPARP rabbit polyclonal24, anti-VSVG monoclonal P5D4 (Sigma), anti-T7 mouse monoclonal (Novagen), anti-EGF rabbit polyclonal (Calbiochem), anti-EGF receptor clone LA22 mouse monoclonal (Upstate), and anti-phospho-EGFR (Tyr1173) clone 9H2, Alexa fluor 488 conjugate mouse monoclonal (Upstate). Secondary antibodies used were peroxidase-conjugated goat anti-rabbit IgG (BioRad), peroxidase-conjugated sheep anti-mouse IgG (Amersham Biosciences), 15nm gold labeled goat anti-mouse IgG (Amersham Biosciences), and Fluorescein (FITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch).
Bead binding assays
Lysates of Sf9 cells infected with the appropriate baculovirus were prepared by lysing cells in 50 mM Tris-Cl, pH 7.4, 75 mM NaCl, 0.5 mM MgCl2 containing 1% Triton X-100, protease inhibitor cocktail (Sigma), and 1 mM PMSF, followed by centrifugation at 20,000×g for 10min. The supernatants (S20) were used for the binding assays. Protein G Sepharose Fast Flow (GE Healthcare) or IgG-Sepharose 6 Fast Flow (Amersham Biosciences) beads were equilibrated in lysis buffer. S20 lysates from CP-MVP-VSVG infected cells were incubated with 2 μg of either anti-T7 monoclonal antibody or anti-VSVG monoclonal antibody and Protein G Sepharose beads overnight at 4°C. The beads were then washed with lysis buffer and boiled in SDS-PAGE sample buffer. Purified vaults or S20s from CP-MVP or CP-MVP-Z infected cells were incubated with IgG-Sepharose beads for 2h to overnight at 4°C. The beads were then washed with phosphate buffered saline (PBS) and boiled in SDS-PAGE sample buffer. The eluted proteins were loaded onto a 6% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane and probed with polyclonal anti-MVP antibodies followed by peroxidase-conjugated goat anti-rabbit IgG.
Binding analysis of the Z domain in purified vaults by ELISA
Purified vaults from CP-MVP-Z or CP-MVP-VSVG infected Sf9 cells were diluted to 2 μg/ml in Tris buffered saline (TBS) and applied to a 96-well dish by serially diluting from 100 ng to 12.5 ng per well (all assays were carried out minimally in duplicate). The plates were incubated at 4°C overnight to allow binding. Unoccupied sites were blocked with 5% BSA in TBS containing 0.05% Tween 20 (TBST) for 1 h at 25°C. After removal of the blocking solution the wells were washed 3 times for 5 min each at 25°C with TBST. Primary antibody [rabbit IgG (Sigma) at 0.5 μg/ml or mouse IgG (Sigma) at 1.67 μg/ml] diluted in TBST was applied for 1 h at 25°C. Following removal of the primary antibody the wells were washed as described above. Either HRP- conjugated goat anti-rabbit or HRP-conjugated sheep anti-mouse secondary antibody (diluted 1:2000) in TBST was applied for 1 h at 25°C. After washing a substrate-chromogen was added (DakoCytomation), and developed for 15 min. The reaction was stopped by the addition of 1N H2SO4. The absorbance was read directly at 405 on a Perkin Elmer HTS 7000Plus Bioassay reader.
The binding affinity of human, rabbit, and mouse IgG to the Z domain in purified vaults was compared by ELISA. Microtiter plates were coated with 50 ng per well of purified CP-MVP-Z recombinant vaults as described above. Z binding assays were performed with 1 μg/ml of human IgG1k (Sigma), 4 μg/ml of rabbit IgG (Sigma), or 32 μg/ml of mouse IgG (Sigma) in a two-fold dilution series. HRP-conjugated goat anti-human, HRP-conjugated goat anti-rabbit, or HRP-conjugated sheep anti-mouse were used as secondary antibodies (all diluted 1:2000) to detect bound complexes with an enhanced TMB chromogen (DakoCytomation), and developed for 15 min. The reaction was stopped by the addition of 1N H2SO4. The absorbance was read directly at 450 on a Perkin Elmer 1420 multilabel counter.
Bone marrow dendritic cells and flow cytometry
Immature bone marrow derived dendritic cells (DCs) were produced from mice as described previously45,46. Briefly, bone marrow cells were isolated from the fibias and tibias of mice and cultured with 20 ng GM-CSF for six days in RPMI-1640 supplemented with 10% heat-inactivated FCS (Gibco), 2 mM L-glutamine (Gibco), 50 μM of 2-ME (Gibco), 100U/ml penicillin (Gibco), streptomycin (Gibco), and 20ng/ml (200U/ml) recombinant mouse GM-CSF (Invitrogen Corp). At day 6, the cells were enriched by positive selection using magnetic microbeads conjugated to a cell surface molecule expressed on all murine DC, anti-mouse CD11c mAb (Miltenyi Biotec), following the manufacturers’ protocol. Purity of approximately 94% was achieved as assessed by flow cytometry. The cells were found to be immature by the lack of expression of MHC class II and other co-stimulatory molecules. The immature DC (1×106) were incubated with media, CP-MVP-Z/GL-INT vaults (500 μg) bound to rabbit IgG (250 μg) or CP-MVP-Z/GL-INT vaults (500 μg) bound to mouse IgG (250 μg) for 30 minutes at 37°C. These cells were collected and stained for CD11c-PE (phycoerythrin, emits fluorescence at 535 nm) a cell surface marker of murine DC and analyzed using a flow cytometer (FACS Calibur, Dickinson Corp). The flow cytometer is designed to measure the number of cells expressing a certain fluorochrome by using multiple filters that detect light emitted from the fluorochromes used in this study. A minimum of 10,000 gated events per sample were collected and plotted as dotplots.
Vault targeting to cells
A431, A549, and HeLa cells were grown and maintained in DMEM supplemented with 10% fetal bovine serum. Cells (4×104/well) were plated onto 12mm glass coverslips coated with poly-L-lysine in 4-well petri-dishes and incubated at 37°C (with 5%CO2) for 16h. Purified CP-MVP-Z/mCherry-mINT vaults (100 μg) were incubated with anti-EGFR antibody LA22 (1 μg) in PBS containing 0.05% Tween 20 (Fisher Scientific) at 4°C for 16 h with tumbling. The cells were serum starved for 1 h before the addition of antibody-bound vaults, followed by 1 h incubation at 4°C in DMEM (0.25ml containing 0.2% FBS per well) for specific membrane binding studies. They were washed three times with cold PBS and fixed in 4% paraformaldehyde. Cells were mounted in Vinol 205 and visualized using fluorescent microscopy (Axio Imager Z1 microscope, Carl Zeiss, Germany).
Cell proliferation assay
Recombinant human epidermal growth factor (rhEGF) was purchased from Promega. Cell proliferation assays were performed using CellTiter 96 AQueous assay kits (Promega) on HeLa cells following the manufacturers instructions. Briefly, rhEGF or V-MVP/CP-MVP-EGF vaults were serially diluted from 500 to ∼0.5 fM with one row left untreated (8 wells per dilution). Semi-confluent HeLa cells were detached using TrypLE Express (Invitrogen), counted, and 104 cells were plated per well in DMEM-HiGlu serum free media without phenol red (Invitrogen). The plates were incubate for ∼44 h in a humidified CO2 incubator at 37°C. The MTS/PMS solution (CellTiter 96 AQueous kit) was added and the plates were incubated in a humidified atmosphere for ∼40 min. The absorbance at 490 nm was read on a Perkin Elmer 1420 multilabel counter.
EGFR immunofluorescence
Cells were plated and grown as described above. The cells were serum starved for 1 h before the addition of anti-EGFR antibody LA22, followed by 1 h incubation at 4°C in DMEM (1 μg anti-EGFR antibody/0.25 ml containing 0.2% FBS per well). The cells were washed three times with cold PBS and incubated with FITC-conjugated AffiniPure Goat Anti-Mouse IgG (H+L) (1:200 dilution, Jackson ImmunoResearch) at RT for 1h. They were washed three times with cold PBS and fixed in 4% paraformaldehyde. Cells were mounted in Vinol 205 and visualized using fluorescent microscopy (Axio Imager Z1 microscope, Carl Zeiss, Germany). Serum starved cells were stimulated with either rhEGF (500 ng/ml, 15min, 37°C) as suggested by the manufacturer or V-MVP/CP-MVP-EGF (200 μg/ml, 1h, 4°C) or untreated. They were washed three times with cold PBS and fixed in 4% paraformaldehyde. Fixed cells were incubated with 3 μg/ml anti-phospho-EGFR (Tyr1173) clone 9H2 Alexa Fluor® 488 conjugate (Upstate) in PBS for 1 h. Cells were washed three times with PBS and mounted in Vinol 205. Fluorescent microscopy was used to observe the phosphorylated EGFR.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institutes of Health awards EB-004553 (to L. Rome & V. Kickhoefer) and AI-026328 (to K. Kelly); the National Science Foundation MCB-0210690 (to L. Rome) and MCB-9722353 (to P. Stewart); and a grant from the UCLA AIDS Institutes (to K. Kelly). M.Silvestry acknowledges support from the NIH Molecular Biophysics Training Grant at Vanderbilt (T32-GM008320). We thank H. Roseboro, R. Carlson, and J. Akana for excellent technical assistance.
REFERENCES AND NOTES
- 1.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Nanocrystal Targeting In Vivo. Proc Natl Acad Sci U S A. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brigger I, Dubernet C, Couvreur P. Nanoparticles in Cancer Therapy and Diagnosis. Adv Drug Deliv Rev. 2002;54:631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 3.Barratt G. Colloidal Drug Carriers: Achievements and Perspectives. Cell Mol Life Sci. 2003;60:21–37. doi: 10.1007/s000180300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manchester M, Singh P. Virus-Based Nanoparticles (VNPs): Platform Technologies for Diagnostic Imaging. Adv Drug Deliv Rev. 2006;58:1505–1522. doi: 10.1016/j.addr.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In Vivo Cancer Targeting and Imaging with Semiconductor Quantum Dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 6.Jain RK. Delivery of Molecular Medicine to Solid Tumors: Lessons from In Vivo Imaging of Gene Expression and Function. J Control Release. 2001;74:7–25. doi: 10.1016/s0168-3659(01)00306-6. [DOI] [PubMed] [Google Scholar]
- 7.Jain RK. Transport of Molecules across Tumor Vasculature. Cancer Metastasis Rev. 1987;6:559–593. doi: 10.1007/BF00047468. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, Barth RF, Yang W, Kawabata S, Zhang L, Green-Church K. Targeted Delivery of Methotrexate to Epidermal Growth Factor Receptor-Positive Brain Tumors by Means of Cetuximab (IMC-C225) Dendrimer Bioconjugates. Mol Cancer Ther. 2006;5:52–59. doi: 10.1158/1535-7163.MCT-05-0325. [DOI] [PubMed] [Google Scholar]
- 9.Park JW, Kirpotin DB, Hong K, Shalaby R, Shao Y, Nielsen UB, Marks JD, Papahadjopoulos D, Benz CC. Tumor Targeting Using Anti-Her2 Immunoliposomes. J Control Release. 2001;74:95–113. doi: 10.1016/s0168-3659(01)00315-7. [DOI] [PubMed] [Google Scholar]
- 10.Volpers C, Thirion C, Biermann V, Hussmann S, Kewes H, Dunant P, von der Mark H, Herrmann A, Kochanek S, Lochmuller H. Antibody-Mediated Targeting of an Adenovirus Vector Modified to Contain a Synthetic Immunoglobulin G-Binding Domain in the Capsid. J Virol. 2003;77:2093–2104. doi: 10.1128/JVI.77.3.2093-2104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada T, Iwasaki Y, Tada H, Iwabuki H, Chuah MK, VandenDriessche T, Fukuda H, Kondo A, Ueda M, Seno M, et al. Nanoparticles for the Delivery of Genes and Drugs to Human Hepatocytes. Nat Biotechnol. 2003;21:885–890. doi: 10.1038/nbt843. [DOI] [PubMed] [Google Scholar]
- 12.Biermann V, Volpers C, Hussmann S, Stock A, Kewes H, Schiedner G, Herrmann A, Kochanek S. Targeting of High-Capacity Adenoviral Vectors. Hum Gene Ther. 2001;12:1757–1769. doi: 10.1089/104303401750476258. [DOI] [PubMed] [Google Scholar]
- 13.Flenniken ML, Willits DA, Harmsen AL, Liepold LO, Harmsen AG, Young MJ, Douglas T. Melanoma and Lymphocyte Cell-Specific Targeting Incorporated into a Heat Shock Protein Cage Architecture. Chem Biol. 2006;13:161–170. doi: 10.1016/j.chembiol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Murphy EA, Majeti BK, Barnes LA, Makale M, Weis SM, Lutu-Fuga K, Wrasidlo W, Cheresh DA. Nanoparticle-Mediated Drug Delivery to Tumor Vasculature Suppresses Metastasis. Proc Natl Acad Sci U S A. 2008;105:9343–9348. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zitzmann S, Mier W, Schad A, Kinscherf R, Askoxylakis V, Kramer S, Altmann A, Eisenhut M, Haberkorn U. A New Prostate Carcinoma Binding Peptide (DUP-1) for Tumor Imaging and Therapy. Clin Cancer Res. 2005;11:139–146. [PubMed] [Google Scholar]
- 16.Arap W, Haedicke W, Bernasconi M, Kain R, Rajotte D, Krajewski S, Ellerby HM, Bredesen DE, Pasqualini R, Ruoslahti E. Targeting the Prostate for Destruction through a Vascular Address. Proc Natl Acad Sci U S A. 2002;99:1527–1531. doi: 10.1073/pnas.241655998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-Specific Targeting of Nanoparticles by Multivalent Attachment of Small Molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 18.Stella B, Arpicco S, Peracchia MT, Desmaele D, Hoebeke J, Renoir M, D’Angelo J, Cattel L, Couvreur P. Design of Folic Acid-Conjugated Nanoparticles for Drug Targeting. J Pharm Sci. 2000;89:1452–1464. doi: 10.1002/1520-6017(200011)89:11<1452::aid-jps8>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 19.Mazor Y, Barnea I, Keydar I, Benhar I. Antibody Internalization Studied Using a Novel IgG Binding Toxin Fusion. J Immunol Methods. 2007;321:41–59. doi: 10.1016/j.jim.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin Treatment of Cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 21.Suprenant KA. Vault Ribonucleoprotein Particles: Sarcophagi, Gondolas, or Safety Deposit Boxes? Biochemistry. 2002;41:14447–14454. doi: 10.1021/bi026747e. [DOI] [PubMed] [Google Scholar]
- 22.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, et al. Engineering of Vault Nanocapsules with Enzymatic and Fluorescent Properties. Proc Natl Acad Sci U S A. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha NL, Heuser JE, Chugani DC, Rome LH. Vaults. III. Vault Ribonucleoprotein Particles Open into Flower-Like Structures with Octagonal Symmetry. J Cell Biol. 1991;112:225–235. doi: 10.1083/jcb.112.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-Kd Vault Protein, VPARP, Is a Novel Poly(ADP-Ribose) Polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH. Vaults and Telomerase Share a Common Subunit, TEP1. J Biol Chem. 1999;274:32712–32717. doi: 10.1074/jbc.274.46.32712. [DOI] [PubMed] [Google Scholar]
- 26.Kickhoefer VA, Searles RP, Kedersha NL, Garber ME, Johnson DL, Rome LH. Vault Ribonucleoprotein Particles from Rat and Bullfrog Contain a Related Small RNA That Is Transcribed by RNA Polymerase III. J Biol Chem. 1993;268:7868–7873. [PubMed] [Google Scholar]
- 27.Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH. Vaults Are Up-Regulated in Multidrug-Resistant Cancer Cell Lines. J Biol Chem. 1998;273:8971–8974. doi: 10.1074/jbc.273.15.8971. [DOI] [PubMed] [Google Scholar]
- 28.van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA. Multiple Human Vault RNAs. Expression and Association with the Vault Complex. J Biol Chem. 2001;276:37715–37721. doi: 10.1074/jbc.M106055200. [DOI] [PubMed] [Google Scholar]
- 29.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the Major Vault Protein: Novel Roles in Signal Pathway Regulation and Immunity. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. Assembly of Vault-Like Particles in Insect Cells Expressing Only the Major Vault Protein. J Biol Chem. 2001;276:23217–23220. doi: 10.1074/jbc.C100226200. [DOI] [PubMed] [Google Scholar]
- 31.Mikyas Y, Makabi M, Raval-Fernandes S, Harrington L, Kickhoefer VA, Rome LH, Stewart PL. Cryoelectron Microscopy Imaging of Recombinant and Tissue Derived Vaults: Localization of the MVP N Termini and VPARP. J Mol Biol. 2004;344:91–105. doi: 10.1016/j.jmb.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Anderson DH, Kickhoefer VA, Sievers SA, Rome LH, Eisenberg D. Draft Crystal Structure of the Vault Shell at 9-Å Resolution. PLoS Biol. 2007;5:2661–2670. doi: 10.1371/journal.pbio.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA. Structural Domains of Vault Proteins: A Role for the Coiled Coil Domain in Vault Assembly. Biochem Biophys Res Commun. 2002;291:535–541. doi: 10.1006/bbrc.2002.6472. [DOI] [PubMed] [Google Scholar]
- 34.Poderycki MJ, Kickhoefer VA, Kaddis CS, Raval-Fernandes S, Johansson E, Zink JI, Loo JA, Rome LH. The Vault Exterior Shell Is a Dynamic Structure That Allows Incorporation of Vault-Associated Proteins into Its Interior. Biochemistry. 2006;45:12184–12193. doi: 10.1021/bi0610552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldsmith LE, Yu M, Rome LH, Monbouquette HG. Vault Nanocapsule Dissociation into Halves Triggered at Low pH. Biochemistry. 2007;46:2865–2875. doi: 10.1021/bi0606243. [DOI] [PubMed] [Google Scholar]
- 36.Esfandiary R, Kickhoefer VA, Rome LH, Joshi SB, Middaugh CR. Structural Stability of Vault Particles. J Pharm Sci. 2008 doi: 10.1002/jps.21508. [DOI] [PubMed] [Google Scholar]
- 37.Ng BC, Yu M, Ajaykumar G, Rome LH, Monbouquette HG, Tolbert SH. Encapsulation of Semiconducting Polymers in Vault Protein Cages. Nano Lett. 2008 doi: 10.1021/nl080537r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu M, Ng BC, Rome LH, Tolbert SH, Monbouquette HG. Reversible pH Lability of Cross-Linked Vault Nanocapsules. Nano Lett. 2008 doi: 10.1021/nl080536z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlen M. A Synthetic IgG-Binding Domain Based on Staphylococcal Protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 40.Braisted AC, Wells JA. Minimizing a Binding Domain from Protein A. Proc Natl Acad Sci U S A. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved Monomeric Red, Orange and Yellow Fluorescent Proteins Derived from Discosoma Sp. Red Fluorescent Protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 42.Wells A. EGF Receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 43.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A New Generation of the Imagic Image Processing System. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 44.Siva AC, Raval-Fernandes S, Stephen AG, LaFemina MJ, Scheper RJ, Kickhoefer VA, Rome LH. Up-Regulation of Vaults May Be Necessary but Not Sufficient for Multidrug Resistance. Int J Cancer. 2001;92:195–202. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1168>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, Kelly KA. Prostaglandin E2 Modulates Dendritic Cell Function During Chlamydial Genital Infection. Immunology. 2008;123:290–303. doi: 10.1111/j.1365-2567.2007.02642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An Advanced Culture Method for Generating Large Quantities of Highly Pure Dendritic Cells from Mouse Bone Marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.