Summary
MicroRNAs (miRNAs) are post transcriptional regulators of gene expression that are important for the control of a multitude of critical processes in mammalian cells. Increasing evidence supports that miRNAs also have important functions in viral replication and may be used by host cells to control viral infection. Expression of miRNAs has been reported for various groups of viruses including herpesviruses, small DNA viruses and retroviruses. The recent identification of target genes regulated by some of these viral miRNAs suggests that they may function in the control of lytic and latent viral replication, in the limitation of antiviral responses, in the inhibition of apoptosis, and in the stimulation of cellular growth. In this review, we summarize in brief recent findings on the antiviral activities of cellular miRNAs and the viral counter-responses to the cell’s RNAi restriction.
Keywords: microRNA, virus replication, innate immunity, post transcriptional regulation, viral gene expression, virus-host interaction
1. Introduction and approaches for analyzing viral and cellular microRNAs (miRNAs)
1.1 MiRNA biogenesis and function
MiRNAs have emerged as ubiquitous regulators of gene expression in metazoans, from plants to invertebrates to vertebrates. Increasingly, evidence supports that important cellular processes are regulated by miRNAs; these include cellular development, differentiation, proliferation, apoptosis, and metabolism [1], [2], [3]. Hence, it is not surprising that miRNAs may also play roles in the disease pathogenesis of cancers and infections [4], [5], [6], [7]. Here, we review in brief some recent findings on how the miRNA pathway is exploited by mammalian viruses and how host cell miRNAs may reciprocally influence viral replication.
Mature miRNAs are single-stranded oligoribonucleotides of 19–25nt in size that originate from larger RNA polymerase II (RNAP II) transcripts, termed primary miRNAs (pri-miRNA) [3], [8], [9]. Pri-miRNAs can reside in exons or introns of genes and contain a stem-loop structure with an imperfectly duplexed stem. During maturation, the stem-loop of a pri-miRNA is processed through the enzymatic activities of two type III RNAses, Drosha and Dicer. The processing starts in the nucleus where Drosha trims the pri-miRNA to a transcript of about 70nt, termed pre-miRNA, which is subsequently exported into the cytoplasm by exportin 5. In the cytoplasm, a second RNAse, Dicer processes the pre-miRNA into a ~22nt duplex mature miRNA. Thereafter, one strand of the mature miRNA (the “guide” strand) is captured into an Argonaute-containing effector complexes [RNA-induced silencing complex (RISC)] while the accompanying “passenger” strand is frequently degraded. The miRNA-guide strand in RISC serves for target recognition of protein-coding mRNAs via base complementarity with sequence elements, which mostly reside in their 3’ untranslated regions (UTR). The number of targeted sequences and their extent of complementarity with the miRNA-guide stand determine the magnitude and mechanism of suppression. MiRNAs, that are fully complementary to their mRNA targets, can induce direct endonucleolytic cleavage within the base-paired region leading to rapid decay of the entire transcript [10],. On the other hand, in a setting of mRNA-miRNA base-pairings that are imperfectly matched, downregulation functions best if the two sequences can base pair through a 7-nt miRNA “seed” sequence, defined as positions 2–8 from the 5’ end of the miRNA [11]. As yet, the suppression mechanism of protein production from mi-RISC-targeted mRNAs is not fully understood. Nonetheless, one plausible model for the mechanism by which miRNAs induce translational inhibition, deadenylation, and degradation is via the translocation of the targeted mRNA into P bodies, cytoplasmic structures, which do not contain ribosomes. Such translocation could be mediated by a P body component (GW182) binding to the Argonaute protein in the miRNA-RISC complex. Once within the P body, the targeted mRNA can be de-adenylated by resident de-adenylases, and then either decapped and degraded or held in stasis (i.e. spatially removed from the translational machinery) [11].
1.2 Identification of viral miRNAs and their targets
The most well-established method used to identify virus-encoded miRNAs is based on the experimental isolation of small RNAs from infected cells followed by cDNA cloning, sequencing and then in silico sequence analyses [9], [12], [13]. On the other hand, not infrequently, de novo computer-based miRNA gene prediction methods are used [14], [15] to initiate the identification of candidates miRNAs. Nevertheless, such candidate miRNAs need to be verified, if not by cloning, by direct detection with Northern blotting or real-time PCR.
Once a predicted miRNA is authenticated, understanding which mRNA(s) the miRNA targets is no simple feat. Bioinformatic computation is employed to search for miRNA-seed sequences in the 3’ untranslated regions of potential target mRNAs [9], [16]. While certain targeting rules support that a “seed” sequence complementarity between miRNA-mRNA is critical to physiological interactions, experimental realities are that imperfect complementarities frequently suffice for silencing [17], and that one miRNA can potentially modulate the expression of up to 100 discrete mRNAs [18]. Because a miRNA can affect a large number of mRNAs, a useful methodology is to screen for potential targets using micro array –based analyses of mRNA down-regulation [19]. To this end, miRNA-encoding vectors expressing a single or multiple miRNAs can be introduced into cultured cells, and resulting mRNA profiles can be analyzed. Alternatively, miRNA targeted mRNAs can be isolated from P-bodies by immunopurification [20]. Array-based and P-body derived data can then be individually validated by confirming that a miRNA can indeed regulate a putative mRNA target. One way to perform the confirmation is to position the 3’ UTR from the mRNA “targeted” by a miRNA into a chimeric transcript which contains a reporter gene. The ability of the miRNA when overexpressed to silence the reporter-containing transcript would be one criterion supportive of specific targeting.
1.3 Identification of cellular miRNAs and their viral targets
While it is well-accepted that mammalian viruses encode miRNAs that can regulate viral [12] [21] [22] [23] and cellular [24] [25] [26] functions, it remains debated whether cellular miRNAs physiologically regulate the replication of mammalian viruses [27]. Two methods have been used to shed light on this latter question. First, miRNA-processing enzymes such as Drosha and Dicer have been knocked down to reduce the processing of mature mammalian miRNAs [28], [29], [30]. When mammalian miRNAs levels were reduced, virus replication in such cells became more robust (see below). In a separate approach, when virus-encoded RNAi suppressors were overexpressed in mammalian cells, viral replication in such cells increased by 5- to 10- fold [31]. Taken together, these results suggest a mammalian miRNA/RNAi function that moderates the replication of viruses. Second, the role of individual cellular miRNA have been assessed using chemically modified antisense-oligoribonucleotides, or antagomirs, [32], [33]. This type of analysis is increasingly employed to inactivate individual cellular miRNAs in order to verify their roles in targeting viruses.
2. A selective overview of virus-encoded microRNAs
2.1 Herpesviruses
By far most of the extant viral miRNAs that have been identified come from herpesviruses. This is a family DNA viruses with large viral genomes approximating 150–230kb containing more than one hundred open reading frames. These viruses, besides their ability to lytically infect cells, can also establish latent infections in which viral gene expression is confined to a few specialized genes. Latency is closely related to the ability of these viruses to establish life-long persistence in their hosts. Many herpes miRNAs are expressed during latency, serving to allow the virus to control viral and cellular functions to minimize the exposure of viral proteins made in infected cells to the host’s immune system. It should be noted that while cellular miRNAs are frequently conserved across species (e.g. between mouse and human), miRNAs encoded by different herpes viruses are mostly not conserved with each other or with their host miRNAs. This lack of conservation could indicate a rapid evolution of herpes miRNA-genes. Alternatively, in instances where herpes-miRNAs serve to target host mRNAs, this may reflect the genetic variability of the 3'UTR target sites in cellular transcripts. Because 3’UTRs are not used for protein coding, they may experience less selective pressure for conservation and thus could diverge more rapidly from species to species.
2.2 Herpes simplex virus (HSV)
HSV belongs to the alpha subgroup of herpesviruses, which can persist in neurons. Although several miRNAs are predicted for HSV1 and HSV2 [15], [34], one virally-encoded miRNA has been verified [34]. No viral miRNAs have been identified yet in the alpha herpes varicella zoster virus [15].
2.3 Human cytomegalovirus (HCMV)
HCMV is the prototype of beta herpesviruses. CMVs are viruses with the largest genome size; they persist in haematopoetic cells like granulocytes and their progenitors. Nine HCMV miRNAs were cloned from lytically infected primary cells. They originate from sequences distributed across the viral genome. Three of these are transcribed from the complementary strand of known ORFs, five miRNAs are located in intergenic regions, and one within an intron [15]. Murine cytomegalovirus (MCMV), a close relative of HCMV, provides the most frequently used animal model for HCMV, which does not replicate in mice. 17–18 miRNAs were found to be encoded by MCMV; all of them are expressed during lytic replication. None of the MCMV miRNAs has significant homologies with HCMV-miRNAs [35], [36]. As yet, no microRNAs have been found encoded by the beta herpesviruses HHV-6 and HHV7 [15].
2.4 Epstein-Barr Virus (EBV)
EBV is a gamma herpesvirus. After a primary infection, EBV persists life-long in human B-lymphocytes [37]. The virus is capable of immortalizing normal human B-cells in vitro and is associated with several human malignant diseases including Hodgkin’s lymphoma, endemic Burkitt’s lymphoma and nasopharyngeal carcinoma. In malignant cells or in vitro transformed lymphoblastoid cells, a group of EBV latency-associated genes or subsets are expressed. The complete set of latency-associated genes contains latency associated membrane proteins (LMP) and Epstein-Barr virus associated nuclear antigens (EBNAs). The unspliced primary EBNA transcripts are the sources for the 3 miRNAs (BHRF1miRNAs) that are encoded in an intron [12], [38], [39]. In latent infection, EBV also expresses at least 14 BART miRNAs, which are encoded by a genomic region which also encodes the non-coding BamHI A rightward transcripts (BARTs) [12], [39], [40]. Seven of EBV’s microRNAs are closely related to microRNAs of an EBV-related monkey virus (Rhesus lymphocryptovirus) and provides a first example of microRNA conservation within the herpesvirus family [41].
2.5 Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8)
KSHV is another gamma herpesvirus linked to human malignancies such as Kaposi’s sarcoma, primary effusion lymphoma (PEL), and Castleman’s disease [42]. KSHV has 12 miRNAs within a 5-kb latency-associated region of the viral genome, which also encodes the transforming protein–coding Kaposin [15], [43]. The miRNAs are expressed in KSHV-positive primary effusion lymphoma-derived cells and endothelial cells. Rhesus Monkey Rhadinovirus (RRV), an animal virus model for KSHV, encodes several miRNAs in the same genomic location as the KSHV miRNAs. Nevertheless, KSHV and RRV miRNAs are unrelated in their primary sequences [44]. A related mouse gamma herpesvirus 68 (MHV68) expresses 9 miRNAs, which also cluster within a small (6-kb) region of the viral genome [15].
2.6 Small DNA Viruses
Viruses of the adenovirus and polyomavirus families have also been shown to express miRNAs. In the case of adenoviruses, the miRNA is processed from a non-coding cytoplasmic RNA polymerase III transcript, called virus-associated RNA (VAI). VAI confers resistance to cellular interferon-related defenses and contributes to viral replication. It was recently found that VAI RNA can be processed by Dicer into a miRNA that serves silencing function during adenovirus infection of cells [45]. Separately, a single miRNA (mirS1), which is derived from the 3’ UTR of the viral late transcript, was characterized for SV40 [21].
2.7 Retroviruses
The human immunodeficiency virus (HIV-1) was predicted earlier by computational analyses to potentially encode five candidate miRNA precursors [14], [46]. With one exception [47], to date these putative miRNAs have not reportedly been cloned successfully [15], [13]; however, some predicted moieties from HIV envelope Env [33], Nef reading frame [48], [49] and the 5’ non-coding TAR leader sequence [26] [50] have been detected and characterized by Northern blotting. The difficulties in cloning HIV-1 derived non-coding RNAs from infected cells [15] suggest that they exist in low abundance. Nonetheless, more recent in depth experiments accompanied by massively-parallel pyrosequencing have cloned and identified several HIV-1 small non-coding RNA (sncRNA) species (ML Yeung and KT Jeang, unpublished data). Additional studies are, however, needed to address the physiological relevance of these viral sncRNAs in HIV-1 infection [51].
3. Diverse functions provided by viral miRNAs
3.1 Regulation of viral transcripts
Frequently viral miRNAs are encoded by the opposite strand of a protein coding gene, which results in a perfect match of miRNA and the coding mRNA target. This complementarity holds obvious implications for the control of viral protein expression. Such control can affect the expression of regulatory proteins during the lytic or the latency/persistence phases of replication. For example, the latency associated membrane protein LMP-1 of EBV, a transforming protein, is under the control of viral miRNAs; a cluster of BART miRNAs targets the 3' UTR of the transcript and represses LMP1 protein expression. Functional consequences of this regulation affect LMP1-induced NF-κβ signaling and apoptosis resistance [52]. The regulation exerted by miRNAs could explain the discrepancy observed between LMP-1 protein expression and its mRNA expression, and the varying levels of LMP1 in nasopharyngeal carcinoma [52].
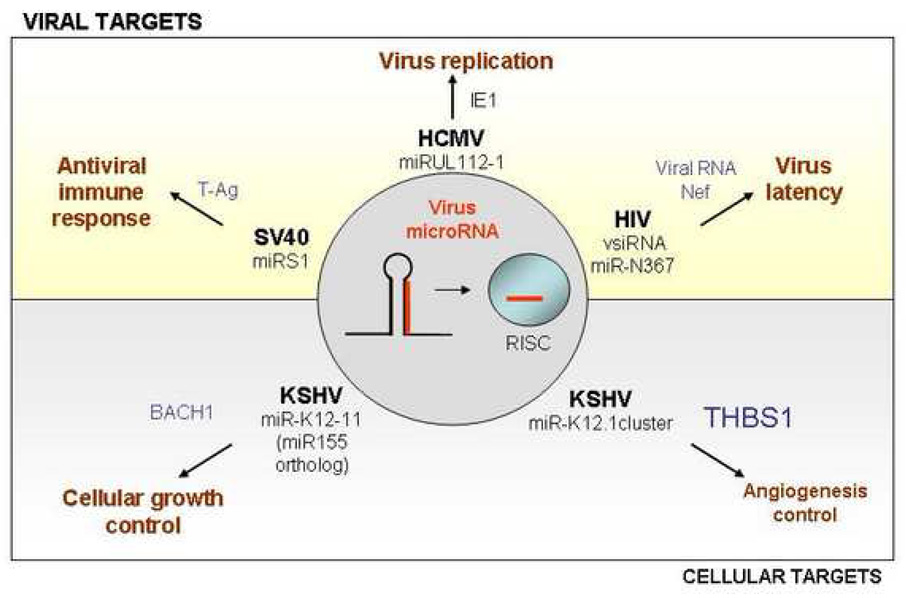
MiRNAs have also been shown to be involved in the control of regulatory proteins expressed at the beginning of viral replication (Figure 1). For instance, the expression of the immediate early IE1 (UL123, IE72) protein of human cytomegalovirus (HCMV), a transcription factor required for the expression of many viral genes, is targeted by virus-encoded miRNA, miR-UL112-1. The IE1 3'UTR target site is necessary and sufficient to direct miR-UL112-1-specific inhibition of expression in transfected cells [22] [23]. Expression of early viral protein synthesis is also limited in SV40 by miRNA control. A microRNA expressed late in infection targets the 3’UTR of the early transcript and reduces the expression of T antigen. This repression of T antigen is advantageous for the virus since it reduces sensitivity of infected cells to cytotoxic T cells [21].
Figure 1. Potential functions of viral miRNAs.
Like their cellular counterparts, viral miRNAs (red) are produced from hairpin-shaped RNA progenitors and are incorporated into the RNA-induced silencing complex (RISC). Viral miRNAs can act on both, the control of viral genes (top) or on cellular genes (bottom) by repressing their expression. The figure shows examples of miRNAs from different viruses, their targets (blue), and the proposed effects on antiviral immune response, viral replication and persistence, apoptosis and growth control (brown).
Similar to some herpesviruses, the role of viral miRNA in the life cycle of HIV remains incompletely clarified. The reported HIV-1 encoded small non-coding RNAs are capable, when over expressed, to reduce the levels of viral transcripts [33], [48], [26], and thus they could contribute to the maintenance and/or establishment of viral latency. Some of these viral sncRNAs lack classical targets in the 3’ UTR regions of genes and instead have complementarity with coding mRNA sequences. Thus, they may function by RNA destabilization, or as has been suggested for the Nef-miRNA and for the TAR-miRNA by suppression of long terminal repeat (LTR) transcription [48], [26].
3.2 Interference with host gene expression
Since miRNAs are not targets of the host’s immune response, they are ideal means for viruses to regulate the host cell during a latency phase. The 12 miRNAs encoded by KSHV are probably also part of its latency associated transcription program in transformed cells and in Kaposi’s sarcoma. Gene expression profiling in cells stably expressing all KSHV-encoded miRNAs identified eight genes which were down-regulated between 4- and 20-fold. One of these genes encodes thrombospondin 1 (THBS1) whose protein expression was decreased >10-fold [19]. THBS1 is a tumor suppressor and anti-angiogenic factor, which exerts its anti-angiogenic effect in part by activating the latent form of TGF-β (Figure 1). Thus KSHV-encoded miRNAs may contribute directly to pathogenesis by down-regulation of THBS1, a major regulator of cell adhesion, migration, and angiogenesis [19]. One KSHV miRNA, miR-K12-11, could in particular contribute to malignant transformation by the virus. It shares 100% seed sequence homology with the oncogenic hsa-miR-155, and accordingly is dubbed an ortholog of miR-155. MiR-155 has been shown to be up-regulated in lymphomas and to be critically important for B-cell development [53], [54]. Its overexpression in transgenic mice induces B -cell lymphoma [55]. Due to the homology in seed sequences, it is highly likely, that the viral miRNA is functionally equivalent to the oncogenic cellular microRNA. In support of this notion, miR-K12-11 and miR-155 regulate a common set of cellular targets including the transcriptional repressor BACH-1 [24] [25].
4. Actions of cellular miRNAs in viral infections
4.1 Cellular miRNAs influence the replication of mammalian viruses
If viruses use virally encoded miRNAs to influence the cell’s milieu, is there evidence that in return cellular miRNAs are employed by the host to reshape the course of viral infections? Several recent experimental findings are consistent with mammalian small non-coding RNAs acting to regulate the invading viruses. First, cellular short interfering RNAs (siRNAs) [56] and a separate class of small non-coding RNAs called piRNAs appear to work in germ and somatic cells to suppress the replication of mammalian endogenous retroviruses [57], [58]. Recently, a subset of Dicer-processed miRNAs has also been implicated in repressing retrotransposon activities in mouse ES cells [59]. Second, robust bioinformatics analyses coupled with experimental validation have revealed that discrete human miRNAs can indeed target various types of viruses including HIV-1 [60], [61], [62], [63], [64]. In most instances, the cellular miRNAs act to moderate viral replication in cells. Hence, human miR-32 has been reported to restrict the replication of primate foamy virus type 1 (PFV-1) [63], and the 3’ portion of HIV-1 mRNAs was shown to be redundantly targeted for repression by a cluster of human miRNAs including miR-28, miR-125b, miR-150, miR-223 and miR-382 [62]. Interestingly, in a rare exception, human miR-122 targets the 5’ UTR of HCV RNA; and rather than repressing replication, miR-122 appears to augment intracellular HCV production [65]. This latter phenomenon is incompletely understood since cells that lack miR-122 appear to support well HCV replication [66]. Third, three independent studies have documented that deliberate repression of mammalian Dicer (which reduces the overall processing of human miRNAs) increased the replication of HIV-1 [29], vesicular stomatitis virus (VSV) [30], [67], and influenza A virus [28]. One interpretation of these results is that the mammalian Dicer-miRNA pathway acts physiologically to censor the replication of viruses such as HIV-1, VSV, and influenza A.
4.2 Interferon-mediated antiviral activity through miRNAs
The interferon (IFN) signaling pathway is well-recognized as a cellular antiviral defense. One prototypic way for eliciting IFN is through exposure of cells to long double-stranded (ds) RNAs. This stimulation holds some similarity to the use of short dsRNAs to elicit RNAi. Interestingly, there is evidence that suggests partial overlap between the IFN and the RNAi pathways. For example, RNA-binding proteins that suppress the IFN effector, protein kinase R (PKR) ([68], [69], [70], also appears to be capable of suppressing RNAi [33,71], and cellular proteins (e.g. PACT, TRBP) that act in IFN’s antiviral mechanism [72], [73] are also important for miRNA processing and function [74], [75], [76], [77]. Recently, an even more direct link between IFN and RNAi has emerged. Interferon beta (IFNβ) was reported to modulate the expression of numerous cellular miRNAs. Intriguingly, eight of these IFNβ-induced miRNAs have sequence-predicted targets within the hepatitis C virus (HCV) genomic RNA. Moreover, the introduction of synthetic miRNA-mimics corresponding to these IFNβ-induced miRNAs recapitulated the antiviral effects of IFNβ on HCV replication and infection, while the neutralization of these IFNβ-induced miRNAs with antagomirs attenuated IFNβ’s antiviral effects against HCV [78]. These new findings support a convergence in the use of miRNAs by mammalian cells in IFN-dependent and IFN-independent defenses against viral infection.
4.3 Viral counter-responses to the cell’s RNAi restriction
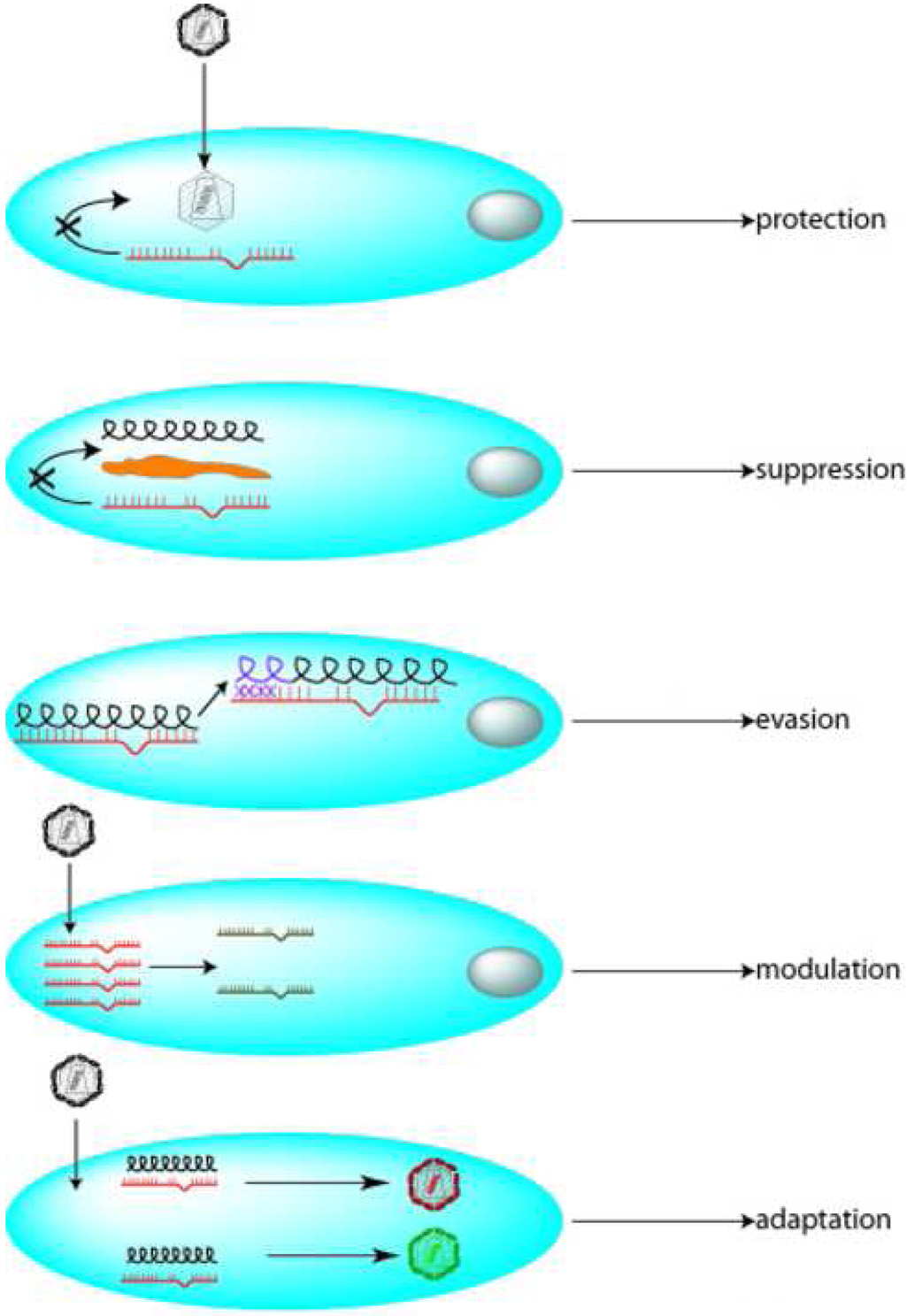
Above, we have outlined evidence for miRNAs being used as a part of the cell’s RNAi-restriction of virus replication. If cells thusly restrict viruses, how might the viruses respond? Viruses appear capable of responding in five different ways. These responses include protection against restriction, suppression of RNAi, mutational escape from RNAi, modulation of the cell’s miRNA/RNAi profile, and adaptation to restriction (Figure 2).
Figure 2. Five potential measures used by viruses to counter the cell’s RNAi restriction.
A virus particle is shown to infect a cell. In the first “protection” measure, viral RNAs could be sheathed by proteins so as to be protected against access by cellular RNAi. Second, viruses could encode RNA-binding proteins or RNA-decoys that sequester the cell’s RNAi effectors leading to “suppression” of interference. In a third way, viruses can mutate their nucleotide sequences so that previous complementarities present in si-/mi-RNA sequences are not longer valid. The mutations then allow for “evasion” by the viruses from RNAi. Fourth, the virus can change the expression profile of cellular miRNAs producing a “modulation” of miRNA levels. Finally, viruses (red) initially sensitive to restriction by a miRNA could evolve to a form (green) that can beneficially utilize the miRNA for its replication. HCV is a putative example of this “adaptation”.
First, viruses can shield their genomes to protect them against the cell’s RNAi restriction [79]. Second, viruses can transcribe small RNA-decoys that competitively occupy the cell’s RNAi machinery [80], [81], [82]) or synthesize RNA-binding proteins [33], [71], [83], [84] that sequester and neutralizes the cell’s antiviral siRNA/miRNA activities (Table 1). While RNA-binding proteins work well to suppress RNAi in plant cells, how well these proteins work in mammalian context has been debated [13]. Third, viruses can mutate their primary sequences or the secondary structure of their transcripts to evade complementarity driven siRNA/miRNA- restrictions [85], [86]. Finally, viruses may modulate the cell’s miRNA expression profile [29], [87], in order to suppress noxious miRNAs and to elevate propitious counterparts, or alternatively, viruses may learn to adapt miRNA-restriction to benefit viral replication [65]. As alluded to above, human hepatitis C virus is currently the one rare example of a virus that has apparently evolved an adaptation paradigm [65].
Table 1.
Selected examples of virus-encoded RNAi counter measures
5. Concluding remarks and future perspectives
About 8% of the human genome is composed of endogenous retroviruses (HERV) [88], [89]. HERVs contain repetitive genetic elements such as their long terminal repeat (LTR) sequences which in lower eukaryotic cells have been found to be processed into small repeat association short interfering RNAs (rasiRNAs; [90], [91]). While this brief review has primarily focused on mammalian cellular miRNAs, miRNAs encoded by mammalian viruses, and their relevant RNA-binding proteins, one should not lose sight of the fact that the universe of small RNA-based gene regulation is likely to be populated by other short non-coding RNAs including rasiRNA, piRNA [92], and additional yet discovered classes. Many mammalian viruses also encode perfectly matched sense and antisense transcripts [93], [94]. It remains to be explored how these RNAs form duplexes and whether these double stranded complexes are processed into small functional entities. In a relatively short period of time much has been achieved in characterizing small non-coding RNAs and their interactions with viruses; nevertheless, it is a good bet that much more remain to be discovered in the coming years.
Acknowledgements
Work in RG’s laboratory is supported by grants from Wilhelm Sander-Stiftung, EU (INCA, LSHC-CT-2005-018704) and by Deutsche Forschungsgemeinschaft (DFG-GRK1071, GR 1224/3-1). Work in KTJ’s laboratory is supported by intramural funding from NIAID, NIH; and by the Intramural AIDS Targeted Antiviral Program from the Office of the Director, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ralph Grassmann, Email: grassmann@viro.med.uni-erlangen.de.
Kuan-Teh Jeang, Email: kj7e@nih.gov.
References
- 1.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5:837. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 9.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 16.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Suppl:S8. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 17.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS. Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 22.Grey F, Meyers H, White EA, Spector DH, Nelson J. A Human Cytomegalovirus-Encoded microRNA Regulates Expression of Multiple Viral Genes Involved in Replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5453. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's Sarcoma-Associated Herpesvirus Encodes an Ortholog of miR-155. J Virol. 2007;81:12836. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung ML, Benkirane M, Jeang KT. Small non-coding RNAs, mammalian cells, and viruses: regulatory interactions? Retrovirology. 2007;4:74. doi: 10.1186/1742-4690-4-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol. 2007;88:2627. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- 29.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 30.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 31.de VW, Haasnoot J, van d, van VMT, Zorgdrager F, Paxton W, Cornelissen M, van KF, de HP, Berkhout B. Increased virus replication in mammalian cells by blocking intracellular innate defense responses. Gene Ther. 2008;15:545. doi: 10.1038/gt.2008.12. [DOI] [PubMed] [Google Scholar]
- 32.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNAs profile during lytic infection and show features of post-transcriptional regulation. J Virol. 2007 doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 38.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81:9967. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS. Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ensser A, Fleckenstein B. T-cell transformation and oncogenesis by gamma2-herpesviruses. Adv. Cancer Res. 2005;93:91. doi: 10.1016/S0065-230X(05)93003-0. [DOI] [PubMed] [Google Scholar]
- 43.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364:21. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 46.Bennasser Y, Le SY, Yeung ML, Jeang KT. MicroRNAs in human immunodeficiency virus-1 infection. Methods Mol Biol. 2006;342:241. doi: 10.1385/1-59745-123-1:241. [DOI] [PubMed] [Google Scholar]
- 47.Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86:751. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- 49.Omoto S, Fujii YR. Cloning and detection of HIV-1-encoded microRNA. Methods Mol Biol. 2006;342:255. doi: 10.1385/1-59745-123-1:255. [DOI] [PubMed] [Google Scholar]
- 50.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar A, Jeang KT. Insights into cellular microRNAs and human immunodeficiency virus type 1 (HIV-1) J. Cell Physiol. 2008 doi: 10.1002/jcp.21488. [DOI] [PubMed] [Google Scholar]
- 52.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 55.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006;13:763. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res Commun. 2005;337:1214. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 61.Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4(+) T lymphocytes. Nat Med. 2007;13:1241. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 63.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe Y, Kishi A, Yachie N, Kanai A, Tomita M. Computational analysis of microRNA-mediated antiviral defense in humans. FEBS Lett. 2007;581:4603. doi: 10.1016/j.febslet.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 65.Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 66.Kumar A. RNA interference: a multifaceted innate antiviral defense. Retrovirology. 2008;5:17. doi: 10.1186/1742-4690-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller S, Imler JL. Dicing with viruses: microRNAs as antiviral factors. Immunity. 2007;27:1. doi: 10.1016/j.immuni.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 2000;373:361. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 69.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1350. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase. PKR, Virology. 1995;213:413. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 71.Haasnoot J, de VW, Geutjes EJ, Prins M, de HP, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS. Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase. PKR, EMBO J. 1998;17:4379. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282:17649. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 77.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Westerhout EM, ter BO, Berkhout B. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006;3:57. doi: 10.1186/1742-4690-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein. TRBP, J Biol Chem. 2006;281:27674. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 83.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 2005;79:7371. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 Human immunodeficiency virus type 1. J. Virol. 2004;78:2601. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffiths DJ. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews1017. REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 91.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 2007;12:851. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2007 doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 93.Cavanagh MH, Landry S, Audet B, rpin-Andre C, Hivin P, Pare ME, Thete J, Wattel E, Marriott SJ, Mesnard JM, Barbeau B. HTLV-I antisense transcripts initiating in the 3'LTR are alternatively spliced and polyadenylated. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, Barbeau B. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J. Virol. 2007;81:10540. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, Huang Y, Cai X, Huang A. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res. 2008;133:250. doi: 10.1016/j.virusres.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 98.Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 2004;85:993. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- 100.Bucher E, Hemmes H, de HP, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 2004;85:983. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]