Abstract
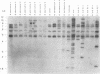
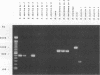
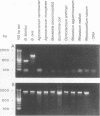
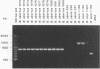
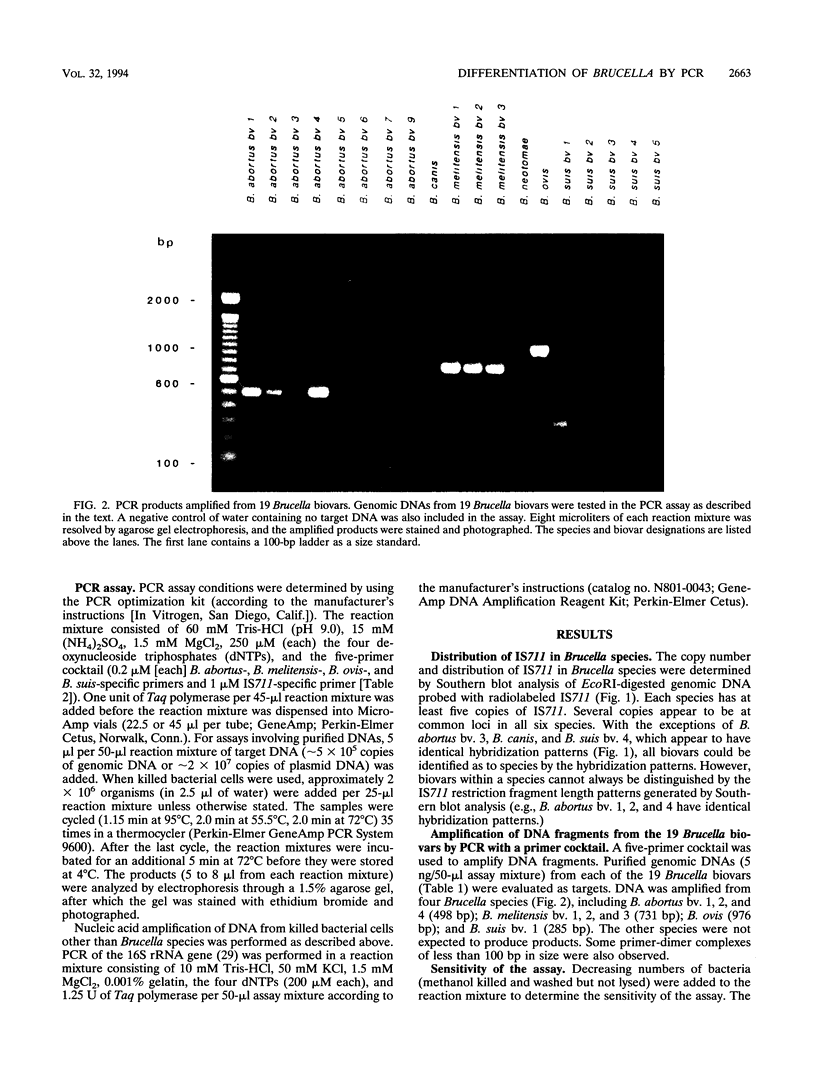
Several PCR assays which identify the genus Brucella but do not discriminate among species have been reported. We describe a PCR assay that comprises five oligonucleotide primers which can identify selected biovars of four species of Brucella. Individual biovars within a species are not differentiated. The assay can identify three biovars (1, 2, and 4) of B. abortus, all three biovars of B. melitensis, biovar 1 of B. suis, and all B. ovis biovars. These biovars include all of the Brucella species typically isolated from cattle in the United States, a goal of the present research. The assay exploits the polymorphism arising from species-specific localization of the genetic element IS711 in the Brucella chromosome. Identity is determined by the size(s) of the product(s) amplified from primers hybridizing at various distances from the element. The performance of the assay with U.S. field isolates was highly effective. When 107 field isolates were screened by the described method, there was 100% agreement with the identifications made by conventional methods. Six closely related bacteria (Agrobacterium radiobacter, Agrobacterium rhizogenes, Ochrobactrum anthropi, Rhizobium leguminosarum, Rhizobium meliloti, and Rhodospirillum rubrum) and two control bacteria (Bordetella bronchiseptica and Escherichia coli) tested negative by the assay.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allardet-Servent A., Bourg G., Ramuz M., Pages M., Bellis M., Roizes G. DNA polymorphism in strains of the genus Brucella. J Bacteriol. 1988 Oct;170(10):4603–4607. doi: 10.1128/jb.170.10.4603-4607.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baily G. G., Krahn J. B., Drasar B. S., Stoker N. G. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992 Aug;95(4):271–275. [PubMed] [Google Scholar]
- Cook D. R., Noble J. W. Isolation of Brucella suis from cattle. Aust Vet J. 1984 Aug;61(8):263–264. doi: 10.1111/j.1751-0813.1984.tb15539.x. [DOI] [PubMed] [Google Scholar]
- Fekete A., Bantle J. A., Halling S. M., Sanborn M. R. Preliminary development of a diagnostic test for Brucella using polymerase chain reaction. J Appl Bacteriol. 1990 Aug;69(2):216–227. doi: 10.1111/j.1365-2672.1990.tb01512.x. [DOI] [PubMed] [Google Scholar]
- Furrer B., Candrian U., Hoefelein C., Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991 May;70(5):372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Detilleux P. G., Tatum F. M., Judge B. A., Mayfield J. E. Deletion of the BCSP31 gene of Brucella abortus by replacement. Infect Immun. 1991 Nov;59(11):3863–3868. doi: 10.1128/iai.59.11.3863-3868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Tatum F. M., Bricker B. J. Sequence and characterization of an insertion sequence, IS711, from Brucella ovis. Gene. 1993 Oct 29;133(1):123–127. doi: 10.1016/0378-1119(93)90236-v. [DOI] [PubMed] [Google Scholar]
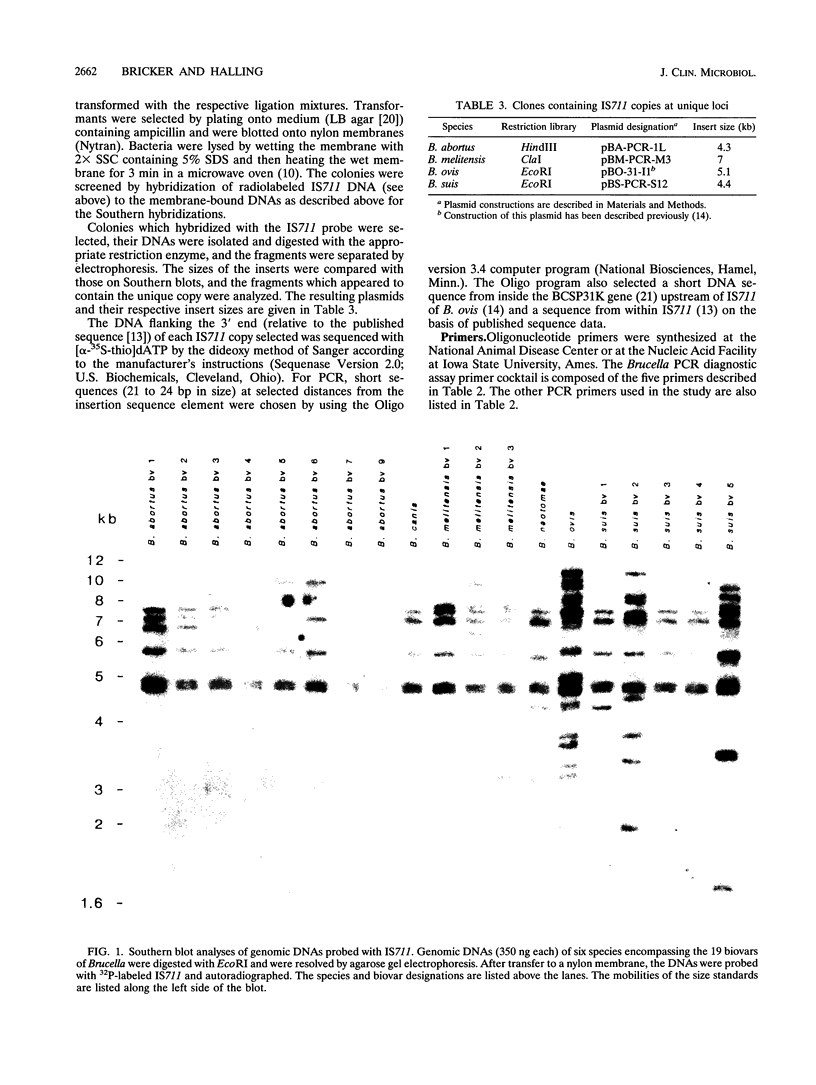
- Halling S. M., Zehr E. S. Polymorphism in Brucella spp. due to highly repeated DNA. J Bacteriol. 1990 Dec;172(12):6637–6640. doi: 10.1128/jb.172.12.6637-6640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington R., Jr, Brown G. M. Laboratory summary of Brucella isolations and typing: 1975. Am J Vet Res. 1976 Oct;37(10):1241–1242. [PubMed] [Google Scholar]
- Herman L., De Ridder H. Identification of Brucella spp. by using the polymerase chain reaction. Appl Environ Microbiol. 1992 Jun;58(6):2099–2101. doi: 10.1128/aem.58.6.2099-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A., Liesack W., Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992 Aug;30(8):1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J. E., Bricker B. J., Godfrey H., Crosby R. M., Knight D. J., Halling S. M., Balinsky D., Tabatabai L. B. The cloning, expression, and nucleotide sequence of a gene coding for an immunogenic Brucella abortus protein. Gene. 1988;63(1):1–9. doi: 10.1016/0378-1119(88)90540-9. [DOI] [PubMed] [Google Scholar]
- Norton J. H., Thomas A. D. Brucella suis infection in pregnant cattle. Aust Vet J. 1979 Nov;55(11):525–527. doi: 10.1111/j.1751-0813.1979.tb07016.x. [DOI] [PubMed] [Google Scholar]
- O'Hara M. J., Collins D. M., de Lisle G. W. Restriction endonuclease analysis of Brucella ovis and other Brucella species. Vet Microbiol. 1985 Aug;10(5):425–429. doi: 10.1016/0378-1135(85)90024-0. [DOI] [PubMed] [Google Scholar]
- Ouahrani S., Michaux S., Sri Widada J., Bourg G., Tournebize R., Ramuz M., Liautard J. P. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J Gen Microbiol. 1993 Dec;139(12):3265–3273. doi: 10.1099/00221287-139-12-3265. [DOI] [PubMed] [Google Scholar]
- Plikaytis B. B., Eisenach K. D., Crawford J. T., Shinnick T. M. Differentiation of Mycobacterium tuberculosis and Mycobacterium bovis BCG by a polymerase chain reaction assay. Mol Cell Probes. 1991 Jun;5(3):215–219. doi: 10.1016/0890-8508(91)90043-j. [DOI] [PubMed] [Google Scholar]
- Verger J. M., Grimont F., Grimont P. A., Grayon M. Taxonomy of the genus Brucella. Ann Inst Pasteur Microbiol. 1987 Mar-Apr;138(2):235–238. doi: 10.1016/0769-2609(87)90199-2. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991 Jan;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. G., Cooper J. E., Gilmour A. Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Appl Environ Microbiol. 1991 Jun;57(6):1793–1798. doi: 10.1128/aem.57.6.1793-1798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leek M. L., Becker H. N., Humphrey P., Adams C. L., Belden R. C., Frankenberger W. B., Nicoletti P. L. Prevalence of Brucella sp. antibodies in feral swine in Florida. J Wildl Dis. 1993 Jul;29(3):410–415. doi: 10.7589/0090-3558-29.3.410. [DOI] [PubMed] [Google Scholar]