Abstract
The identification of the neutralization domains of hepatitis C virus (HCV) is essential for the development of an effective vaccine. Here, we show that the hypervariable region 1 (HVR1) of the envelope 2 (E2) protein is a critical neutralization domain of HCV. Neutralization of HCV in vitro was attempted with a rabbit hyperimmune serum raised against a homologous synthetic peptide derived from the HVR1 of the E2 protein, and the residual infectivity was evaluated by inoculation of HCV-seronegative chimpanzees. The source of HCV was plasma obtained from a patient (H) during the acute phase of posttransfusion non-A, non-B hepatitis, which had been titered for infectivity in chimpanzees. The anti-HVR1 antiserum induced protection against homologous HCV infection in chimpanzees, but not against the emergence of neutralization escape mutants that were found to be already present in the complex viral quasispecies of the inoculum. The finding that HVR1 can elicit protective immunity opens new perspectives for the development of effective preventive strategies. However, the identification of the most variable region of HCV as a critical neutralization domain poses a major challenge for the development of a broadly reactive vaccine against HCV.
Hepatitis C virus (HCV) is an important cause of morbidity and mortality worldwide (1–3). Infection with HCV becomes chronic in >80% of the cases and is a major cause of liver cirrhosis (4) and hepatocellular carcinoma (5). Although the development of a broadly reactive vaccine would be the most effective method for its control, concerns have been raised because of the high degree of genetic heterogeneity of HCV (6) and the lack of protective immunity against reinfection (7, 8) or superinfection (9, 10) documented both in humans and in chimpanzees. Viral isolate-restricted neutralizing antibodies against HCV have been demonstrated recently in infected individuals (11, 12), but their molecular target is presently unknown.
Several observations have suggested that the hypervariable region 1 (HVR1) could be involved in the neutralization of HCV. This assumption is based on the fact that the HVR1, which is located at the N terminus of the envelope glycoprotein 2 (E2) gene and consists of 34 amino acids spanning map position 384–414 (13), is the most variable region of the HCV genome (14, 15), contains linear epitopes that are recognized by patients’ antibodies (16–22) and mutates rapidly in vivo (23–26), suggesting that it is under the selective pressure of the host immune system. This hypothesis is further substantiated by the lack of variability in the HVR1 observed in an agammaglobulinemic patient over a period of 2.5 years (27). Recent data obtained in vitro suggest that antibodies, present in human sera and directed against the HVR1 as well as against the E2 protein of HCV, can prevent the binding of HCV to cells (28, 29). The potential importance of the HVR1 for HCV neutralization is also underscored by the analogy with the V3 loop of human immunodeficiency virus, which represents a principal neutralization domain and a major target of type-specific neutralizing antibodies (30).
To investigate whether the HVR1 of the E2 protein is a critical neutralization domain, in vitro neutralization of a pedigreed, in vivo-titrated HCV strain (H77) was attempted with a rabbit hyperimmune serum raised against a homologous synthetic HVR1 peptide. The residual infectivity was evaluated by inoculation of HCV-seronegative chimpanzees.
MATERIALS AND METHODS
Source of HCV.
The virus stock of HCV used for all the inocula was derived from a plasmapheresis unit obtained during the early acute phase of posttransfusion non-A, non-B hepatitis (on July 12, 1977) from a patient (H) who underwent open heart surgery (31). This plasma, designated H77, contained 106.5 50% chimpanzee-infectious doses (CID50) of HCV per ml, as shown by titration studies in chimpanzees (32) and was devoid of antibodies to HCV by all available tests. This viral stock has been used successfully to transmit non-A, non-B (NANB) hepatitis to chimpanzees in other studies (7, 14, 32).
Synthesis of the HVR1 Peptides and Generation of the Hyperimmune Sera.
Two synthetic peptides (HVR1-A and HVR1-B; see Fig. 1) of 21 and 31 amino acids, respectively, corresponding to the sequence of the HVR1 of H77 were generated. The two sequences were obtained independently in our laboratory by direct sequencing, following PCR amplification of cDNA from the same sample (H77). One of the two sequences was previously reported (23). Peptides HVR1-A and HVR1-B were conjugated to tetanus antitoxin either by glutaraldehyde (HVR1-A) or by carbodiimide (HVR1-B) coupling. Hyperimmune sera were generated by three sequential intradermal multiple site injections in rabbits of the peptide conjugate (500 μg) in Freund’s adjuvant according to a standard immunization protocol for rabbits (0, 1, and 6 months). The hyperimmune sera, designated LMF87 (anti-HVR1-A) and LMF92 (anti-HVR1-B), respectively, were tested in ELISA both against the homologous peptide (unconjugated) and against the recombinant E1/E2 complex from the H77 isolate expressed in vaccinia virus in BHK21 cells. The recombinant clone expressed in vaccinia had an HVR1 sequence identical to that of the HVR1-B peptide. As a control, the hyperimmune sera were tested against cell extracts of BHK21 infected with negative control recombinant vaccinia virus.
Figure 1.

Alignment of the amino acid sequences (in single-letter code) predicted from a portion of the E1 and E2 genes of HCV, strain H77, spanning map positions 317–508 (13). H77 denotes the sample obtained from a plasmapheresis unit collected from patient H during the acute phase of posttransfusion NANB hepatitis (31), which represents the standard viral stock used for all the inocula, which had been previously titrated in chimpanzees (32). H77-1 and H77-2 denote two distinct sequences obtained independently in our laboratory by direct sequencing, following PCR amplification of cDNA from the same sample (H77). One of the two sequences was previously reported (23). The HVR1 of the E2 protein, spanning map position 384–414 (13), is shown in boldface type. The boxed sequences denote the sequence of the peptides (HVR1-A and HVR1-B) used for generating hyperimmune rabbit sera. Matches are indicated by vertical bars and changes are indicated by colons.
Source of Antibodies for Neutralization.
As a potential source of neutralizing antibodies, we used the hyperimmune rabbit serum (LMF87, anti-HVR1) which was positive by ELISA against the homologous peptide and the recombinant E1/E2 protein complex, expressed in vaccinia systems. As negative controls, we used the preimmune rabbit serum that was negative by ELISA for all detectable antibodies to HCV, and plasma from a normal blood donor, negative for all antibodies to HCV (11), whose blood repeatedly failed to transmit hepatitis after transfusion (H.J.A., unpublished data). As a positive control, we used plasma obtained from patient H 2 years after primary infection, designated H79, which was previously shown to contain neutralizing antibodies against H77 (11). The H79 plasma was positive for antibodies to core, E1, E2, NS3, NS4, and NS5 of HCV (11).
In Vitro Neutralization Test.
The neutralization test was performed as described (11). Each antiserum was diluted 1:5 in PBS (pH 7.4) and then heat-inactivated at 56°C for 30 min before use. One vial of a dilution (in fetal bovine serum) of the challenge virus containing 3200 CID50 was further diluted 1:5 in ice-cold PBS (pH 7.4), and then one additional dilution was made in cold PBS with 20% fetal bovine serum to yield samples containing 64 CID50. The in vitro neutralization test was done by mixing the virus inoculum (64 CID50 in 1 ml) with one of the inactivated antisera (1 ml). The virus/antiserum mixtures were incubated overnight at 4°C. Each mixture (2 ml) was then inoculated intravenously into one HCV-seronegative chimpanzee.
Chimpanzees.
Five chimpanzees were included in this study. The animals were caged individually and maintained under conditions that met all relevant requirements for the use of primates in an approved facility. None of the chimpanzees included in this study had been previously exposed to HCV, and none of them had signs of active or past HCV infection, as measured by PCR and antibody testing. At the time of the study, all chimpanzees were negative for hepatitis B surface antigen and had normal hepatic enzyme levels. Weekly serum samples were monitored for alanine aminotransferase (ALT). Serum HCV RNA was determined in serial serum samples obtained at intervals of 1, 2, or 4 weeks, during an observation period of 24 weeks after the virus challenge. All serial samples were tested with a set of nested primers derived from the 5′ noncoding region of the HCV genome (7). Serum HCV RNA from selected samples obtained from each chimpanzee 2 weeks after inoculation was amplified with a set of primers that span part of the E1 and E2 genes (7), including the HVR1, and the PCR products were sequenced both directly and after molecular cloning. Weekly serum samples were also tested for antibodies to HCV (anti-HCV).
Anti-HCV Testing.
Antibodies against structural and nonstructural proteins of HCV (anti-HCV) were assayed in chimpanzee sera using a second generation ELISA according to the manufacturer’s instructions (Ortho Diagnostics).
RNA Extraction and PCR.
Total RNA extracted from 100 μl of serum or plasma using the guanidinium/phenol/chloroform method (33) was reverse-transcribed in a volume of 20 μl, and the resulting cDNA was amplified in a 100-μl reaction volume (33). PCR was performed using two sets of nested primers (7). The first, derived from the 5′ noncoding region (7), was used to investigate the course of HCV viremia, and the second, derived from the E1 and E2 genes (7), including the HVR1, for the comparative sequence analysis. The sensitivity, specificity, and details of our nested PCR technique, have been reported (7, 33). To reduce the risk of contamination, all the proper precautions were taken (7, 33). In addition, for each test sample, a negative control was tested in parallel throughout the entire procedure, starting from RNA extraction.
Sequencing Analysis.
PCR products amplified with the set of primers that span part of the E1 and E2 genes of the HCV genome, obtained from the H77 inoculum and from a selected sample from each chimpanzee, were purified by GeneClean (Bio 101). Double-stranded PCR fragments were directly sequenced by the dideoxynucleotide chain termination method with phage T7 DNA polymerase (Sequenase: United States Biochemical), as described (34). The amplified PCR products were also cloned into pGEM-T vector systems (Promega), and the molecular clones were sequenced using the Applied Biosystems model 373 automated DNA sequencer with a modified Sanger method. A total of 104 molecular clones from the H77 inoculum and 9 or 10 clones from each chimpanzee, on a sample obtained 2 weeks after inoculation, were sequenced.
RESULTS
Generation of Rabbit Hyperimmune Sera Against the HVR1 and in Vitro Neutralization.
We generated rabbit hyperimmune sera directed against two synthetic peptides corresponding to the HVR1 of HCV, strain H77, a pedigreed virus obtained during the early acute phase of post-transfusion NANB hepatitis from a patient (H) who underwent open heart surgery (31). Two separate aliquots of the H77 plasma were PCR-amplified with a set of primers spanning a portion of the E1 and E2 genes (7), including the HVR1 domain, and the DNA products were analyzed by direct sequencing (Fig. 1). Comparative analysis of the two predicted amino acid sequences of 191 residues showed only a single amino acid change (S → N at position 391), located within the HVR1 (Fig. 1). Based on these data, two synthetic peptides, representing the two sequences of the HVR1 of H77, were generated, and each of them was separately used to immunize two rabbits. One peptide (HVR1-A), bearing the S residue at position 391, was shorter (aa 390–410); the other (HVR1-B), bearing the N residue at position 391, was longer (aa 384–414). The hyperimmune sera, designated LMF87 (anti-HVR1-A) and LMF92 (anti-HVR1-B), respectively, were both reactive in an ELISA test against the homologous peptide (at a titer of 1:650,000), but only LMF87, directed against the shorter peptide (HVR1-A), recognized the recombinant E1/E2 protein complex derived from the H77 virus and expressed in a vaccinia system (at a titer of 1:70,000). Thus, the LMF87 anti-HVR1 serum was selected for in vitro neutralization experiments.
The chimpanzees and sera used in the experiments of HCV neutralization in vitro are indicated in Table 1. As negative controls, we used the respective preimmune rabbit serum and plasma from a normal blood donor, which were both negative for detectable antibodies to HCV. As a positive control, we used plasma obtained from patient H 2 years after primary infection, designated H79, which was previously shown to contain neutralizing antibodies against H77 (11). The protocol used for the neutralization test has been reported (11). The challenge stock was prepared from the standard H77 inoculum which contained 106.5 CID50 of HCV per milliliter, as determined by in vivo titration in chimpanzees (32). This inoculum had been used successfully for other challenge studies in chimpanzees (7, 11, 32). The in vitro neutralization test was performed by mixing the virus inoculum (64 CID50 in 1 ml) with the relevant serum (1 ml), previously heat-inactivated at 56°C for 30 min. The virus/serum mixtures were incubated overnight at 4°C and then inoculated i.v. into seronegative chimpanzees (Table 1). The dose of H77 virus used in this study (64 CID50) had successfully infected five of five chimpanzees in a previous study (11).
Table 1.
In vitro neutralization of HCV, as measured in chimpanzees: animals and antisera used
| Chimpanzee no. | Sera reacted with 64 CID50 of HCV,
strain H77 and inoculated into indicated chimpanzee
|
|
|---|---|---|
| Serum (final concentration) | Anti-HCV antibodies* detected in indicated serum | |
| 1480 | Preimmune rabbit serum (1:10) | None |
| 1479 | Normal human plasma (1:10) | None |
| 1484, 1486 | Hyperimmune rabbit anti-HVR1 (1:10) | Homologous HVR1 peptide, recombinant E1/E2 complex |
| 1475, 1442 | H79 human plasma (1:10) | Core, E1, E2, NS3, NS4, NS5 |
Anti-HCV antibodies directed against HVR1 (synthetic peptide with sequence of hypervariable region 1), NS3 (nonstructural 3 protein), NS4 (nonstructural 4 protein), NS5 (nonstructural 5 protein, detected by ELISA).
Course of HCV Infection in Chimpanzees After Antibody-Mediated in Vitro Neutralization.
After antibody-mediated in vitro neutralization, tests for residual infectivity were performed by intravenous inoculation of the serum/virus mixture into seronegative chimpanzees, the most sensitive model presently available to evaluate the infectivity of HCV (35). The two control animals (nos. 1480 and 1479) developed classical hepatitis C (Fig. 2A). Serum HCV RNA, as measured by PCR, was detected in the first bleeding, 1 week postinoculation, and remained persistently detectable throughout the observation period. Both animals seroconverted. In contrast, of the two animals whose inoculum contained the H77 virus mixed with the rabbit hyperimmune anti-HVR1 serum, one (no. 1484) did not show any virologic, biochemical or serologic evidence of HCV infection (Fig. 2B). Serum HCV RNA was never detected by PCR in any of the weekly samples tested throughout the observation period of 24 weeks; neither alterations in serum ALT levels nor antibody seroconversion occurred at any time after challenge. The second animal (no. 1486) developed acute hepatitis C, characterized by the appearance of HCV viremia within 1 week postinoculation and by elevation of serum ALT values and antibody seroconversion. The infection became chronic (Fig. 2B). Of the two chimpanzees whose inoculum contained the virus mixed with the H79 plasma, one (no. 1475) developed acute hepatitis, whereas the other (no. 1442), which was previously described (11), exhibited no signs of HCV infection or biochemical alterations (Fig. 2C). Thus, both the hyperimmune anti-HVR1 serum and the H79 plasma conferred protection from HCV infection in one of the two animals tested.
Figure 2.
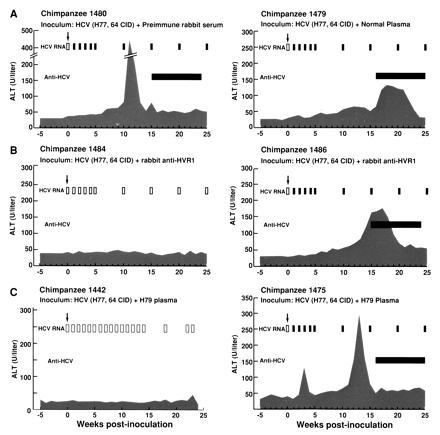
Course of HCV infection in chimpanzees after antibody-mediated in vitro neutralization. Details of the in vitro neutralization test, of the source of HCV, and of the antisera used in this study are provided in Materials and Methods. Neutralization of HCV was attempted with the preimmune rabbit serum and with an anti-HCV negative human plasma (A), with the hyperimmune rabbit anti-HVR1 serum (B), or with the H79 human plasma (C). The upper arrows indicate the time of challenge. The shaded areas indicate the values of serum ALT. Normal ALT values in chimpanzees range between 6 and 38 units/liter. Open bars indicate negative results for serum HCV RNA by PCR and solid bars indicate positive results. The horizontal bar indicates the time during which serum was positive for antibodies to HCV, as detected by second generation ELISA assay. Chimpanzee 1442 was described previously (11).
Sequence Analysis of the H77 Virus Used for Inoculation.
To interpret the results of the neutralization experiments, we performed an extensive sequence analysis of the H77 virus that was used for inoculation. RNA from the H77 viral stock was extracted, reverse transcribed, and amplified with a set of primers that span a portion of the E1/E2 region, including the HVR1 (7). The PCR products, 573 nt long, were purified and analyzed by both direct sequencing and sequencing of molecular clones. From the H77 viral stock, a total of 104 molecular clones was analyzed. The predicted amino acid sequences of the HVR1 are shown in Fig. 3. At least 19 different viral strains were simultaneously present within the H77 virus stock, confirming that HCV circulates in vivo as a complex quasispecies. One variant was predominant, representing 70/104 clones (67%). The sequence of this variant was identical, except for a single amino acid change (S → N at position 391), to the synthetic peptide HVR1-A which had been used to generate the hyperimmune serum (LMF 87) selected for the in vitro neutralization studies, as well as to one of the two sequences obtained by direct sequencing (Fig. 1). Other strains were represented by only six, five, four, and two clones, respectively. At least 11 strains were represented by a single clone (Fig. 3).
Figure 3.

Predicted amino acid sequence alignment of 31 amino acids (in single-letter code) representing HVR1 of the E2 gene, spanning map position 384–414, of 104 molecular clones obtained from HCV, strain H77, which was used for inoculation. The sequence obtained by direct sequencing is indicated at the top for comparison. The standard viral stock used for all the inocula (H77) was obtained from patient H during the acute phase of posttransfusion NANB hepatitis (31). The remaining sequence of the 191 amino acids spanning map position 318–508, which is not shown, is available upon request.
Comparative Sequence Analysis of the Viral Quasispecies Present in the H77 Inoculum and the Viruses Recovered from the Infected Chimpanzees.
We performed a comparative sequence analysis of the viral strains present in the H77 inoculum and the viruses recovered from the chimpanzees that developed HCV infection after challenge. From each chimpanzee, both direct sequencing and sequencing of 9 or 10 molecular clones were performed on a sample obtained 2 weeks after inoculation (Fig. 4). Remarkably, in the animal whose inoculum contained the virus mixed with the hyperimmune rabbit anti-HVR1 serum (no. 1486), neither the sequence of the 10 molecular clones nor the direct sequence was identical to the predominant clone against which the hyperimmune serum was directed. The viruses emerging from this animal were identical to two of the minor variants: 7 of the 10 clones corresponded to the second variant (6%) in the H77 quasispecies; the remaining 3 clones, as well as the directly determined sequence, corresponded to a variant represented by only 2% of the H77 quasispecies (Figs. 3 and 4). When compared with the sequence of the predominant strain of H77 virus used for challenge (Figs. 3 and 4), each of the two minor variants exhibited 4 amino acid changes over the 31 amino acids of the HVR1. In contrast, viruses identical to the predominant clone were consistently recovered from all the other chimpanzees, regardless of the serum used for neutralization. Altogether, these data indicate that in animal no. 1486 the rabbit hyperimmune anti-HVR1 serum was able to neutralize the predominant clone against which it had been raised, but was ineffective against some of the minor variants that emerged in vivo. Comparative sequence analysis was also performed in animal no. 1475, which developed acute hepatitis despite the presence in the inoculum of the H79 plasma, previously shown to contain neutralizing antibodies (11). The analysis demonstrated that the virus emerging in vivo was identical to the predominant clone (67% of the H77 viral quasispecies). Only a single amino acid change was detected in two of the nine clones. Similarly, the sequence obtained by direct sequencing corresponded to that of the predominant clone. Because this animal exhibited a biphasic pattern of serum ALT levels (Fig. 2C), raising the possibility that the second peak of hepatitis might be due to the emergence of a neutralization escape mutant, additional direct sequence analysis was performed over a period of 6 months after inoculation. Comparison of the sequences obtained 8, 12, 13, 18, and 25 weeks, respectively, postinoculation demonstrated 100% identity with the direct sequence obtained at week 2 postinoculation (Fig. 4). Remarkably, none of the minor variants was ever detected in this chimpanzee, which suggests that the in vitro neutralization was incomplete, and that HCV infection in this animal was initiated by a very limited number of viral particles of the predominant strain that were not effectively neutralized. These data support the hypothesis that the neutralizing immune response elicited by HCV in the host is weak, and further demonstrate the extreme sensitivity of the animal model for evaluating HCV infectivity (35).
Figure 4.
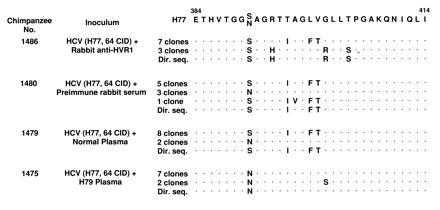
Comparative sequence analysis of the sequence of the H77 virus used for inoculation and the viruses recovered from the chimpanzees 2 weeks after inoculation. The two sequences indicated for H77 were obtained independently by direct sequencing (Dir. seq.). For each chimpanzee the results of both direct sequencing and sequencing of 9 or 10 molecular clones are indicated.
DISCUSSION
Our study provides evidence that the HVR1 of the E2 protein of HCV, the most variable region of the entire viral genome, is a critical neutralization domain. We showed that a hyperimmune rabbit serum raised against a synthetic peptide representing the HVR1 protected chimpanzees from homologous HCV infection, but not from the emergence of neutralization escape mutants that were already present in the complex viral quasispecies. These results indicate that the synthetic peptide had elicited neutralizing antibodies, but that such antibodies were type-specific and thus ineffective against some of the minor variants present in the inoculum. In fact, the rabbit anti-HVR1 hyperimmune serum was directed against the predominant clone present in the H77 plasma, which contained a mixed viral population (quasispecies) of at least 19 different variants. Thus, this study provides an in vivo model for the emergence of neutralization escape mutants, a strategy whereby HCV can evade the host’s immune surveillance and establish persistent infection in >80% of infected individuals (4). Neutralization escape mutants emerged in only one of the two animals whose inoculum contained the virus mixed with the hyperimmune anti-HVR1 serum, whereas the other chimpanzee was completely protected. A possible explanation for this finding is the different infectivity titer of each of the 19 viral variants present in the H77 inoculum. In fact, the two variants recovered from this animal represented only 6% and 2%, respectively, of the viral population of the H77 used for inoculation. Thus, the viral inoculum of 64 CID50 contained only ≈4 and 1 CID50, respectively, of these variants. These represent borderline titers for in vivo infectivity, which would not be expected to infect 100% of the animals, based on Poisson distributions of the variant viruses.
The observation that a hyperimmune serum against the HVR1 protected chimpanzees from homologous HCV infection provides direct evidence that anti-HVR1 antibodies can, in the absence of other virus-specific immune responses, prevent HCV infection. The HCV-neutralizing activity mediated by anti-HVR1 antibodies may be the experimental correlate of an in vivo protective immune response. Protection against the homologous strain of HCV was demonstrated in chimpanzees vaccinated with the recombinant putative E1/E2 proteins of HCV (36). The finding that HVR1 is a neutralization domain of HCV constitutes an important step for understanding the immune response elicited by HCV in the host, as well as for devising effective preventive strategies for the control of HCV infection. However, the fact that a major target of the neutralizing antibodies is the HVR1, the most variable region of the HCV genome, raises a new set of problems that need to be addressed for the development of a broadly reactive vaccine against HCV. For a vaccine to be practical in humans, it should provide protection against the great variety of HCV isolates that infect the human population. The high degree of genetic heterogeneity of HCV (6), which is reflected in the complex quasispecies nature of this virus (37), indicates that it might be difficult to realize effective preventive strategies. Nevertheless, our report demonstrates that it is indeed possible to elicit a protective humoral immune response against homologous HCV infection. However, because there is no obvious strategy to suppress the genetic variation of HCV, the finding that the HVR1 is a neutralization domain poses a major challenge that must be taken into account for any viable vaccination strategy for the control of HCV infection.
Footnotes
Abbreviations: HCV, hepatitis C virus; HVR1, hypervariable region 1; E1, envelope glycoprotein 1; E2, envelope glycoprotein 2; CID50, 50% chimpanzee-infectious dose; NANB, non-A, non-B; ALT, alanine aminotransferase.
References
- 1.IARC Working Group, World Health Organization: Hepatitis C Virus. IARC Monogr. 1994;59:165–221. [PMC free article] [PubMed] [Google Scholar]
- 2.Kiyosawa K, Tanaka E, Sodeyama T, Furuta S. Intervirology. 1994;37:101–107. doi: 10.1159/000150363. [DOI] [PubMed] [Google Scholar]
- 3.Bellentani S, Tiribelli C, Saccoccio G, Sodde M, Fratti N, De Martin C, Cristianini G The Dyonysos Study Group. Hepatology. 1994;20:1442–1449. doi: 10.1002/hep.1840200611. [DOI] [PubMed] [Google Scholar]
- 4.Alter H J, Seeff L B. In: Viral Hepatitis: Scientific Basis and Clinical Management. Zuckerman A Z, Thomas H C, editors. Edinburgh: Churchill Livingstone; 1994. pp. 467–498. [Google Scholar]
- 5.Nishioka K, Watanabe J, Furuta S, Tanaka E, Iino S, Suzuki H, Tsuji T, Yano M, Kuo G, Choo Q-L, Houghton M, Oda T. Cancer (Philadelphia) 1991;67:429–433. doi: 10.1002/1097-0142(19910115)67:2<429::aid-cncr2820670218>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Bukh J, Miller R H, Purcell R H. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 7.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, Purcell R H. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 8.Lai M E, Mazzoleni A P, Argiolu F, De Virgilis S, Balestrieri A, Purcell R H, Farci P. Lancet. 1994;343:388–390. doi: 10.1016/s0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- 9.Kao J K, Chen P J, Lai M Y, Chen D S. Gastroenterology. 1992;105:583–587. doi: 10.1016/0016-5085(93)90737-w. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto H, Mishiro S, Tokita H, Tsuda F, Miyarawa Y, Mayumi M. Hepatology. 1994;20:1131–1136. [PubMed] [Google Scholar]
- 11.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiner A J, Brauer M J, Rosemblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q-L, Houghton M. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 15.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 16.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore E E, Peterson D A, Mishiro S. Virology. 1993;195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 18.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. J Virol. 1994;68:4776–4784. doi: 10.1128/jvi.68.8.4776-4784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiya H, Kato N, Ootsuyama Y, Nakazawa T, Yamauchi K, Shimotohno K. Int J Cancer. 1994;57:664–670. doi: 10.1002/ijc.2910570509. [DOI] [PubMed] [Google Scholar]
- 21.Lesniewski R R, Boardway K M, Casey J M, Desai S M, Devare S G, Leung T K, Mushahwar I K. J Med Virol. 1993;40:150–156. doi: 10.1002/jmv.1890400213. [DOI] [PubMed] [Google Scholar]
- 22.Scarselli E, Cerino A, Esposito G, Silini E, Mondelli M U, Traboni C. J Virol. 1995;69:4407–4412. doi: 10.1128/jvi.69.7.4407-4412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata N, Alter H J, Miller R H, Purcell R H. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto H, Kojima M, Okada S-I, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ito Y, Mishiro S. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 25.Higashi Y, Kakumu S, Yoshioka K, Wakita T, Mizokami M, Ohba K, Ito T, Ishikawa T, Takayanagi M, Nagai Y. Virology. 1993;197:659–668. doi: 10.1006/viro.1993.1641. [DOI] [PubMed] [Google Scholar]
- 26.Kumar U, Brown J, Monjardino J, Thomas H C. J Infect Dis. 1993;167:726–730. doi: 10.1093/infdis/167.3.726. [DOI] [PubMed] [Google Scholar]
- 27.Kumar U, Monjardino J, Thomas H C. Gastroenterology. 1994;106:1072–1075. doi: 10.1016/0016-5085(94)90770-6. [DOI] [PubMed] [Google Scholar]
- 28.Zibert A, Schreier E, Roggendorf M. Virology. 1995;208:663–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]
- 29.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q-L, Chien D, Pileri P, Houghton M, Abrignani S. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letvin N L. N Engl J Med. 1993;329:1400–1405. doi: 10.1056/NEJM199311043291908. [DOI] [PubMed] [Google Scholar]
- 31.Koziol D E, Holland P V, Alling D W, Melpolder J C, Solomon R E, Purcell R H, Hudson L M, Shoup F J, Krakauer H, Alter H J. Ann Intern Med. 1986;104:488–495. doi: 10.7326/0003-4819-104-4-488. [DOI] [PubMed] [Google Scholar]
- 32.Feinstone S M, Alter H J, Dienes H, Shimizu Y K, Popper H, Blackmore D, Sly D, London W T, Purcell R H. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 33.Farci P, Alter H J, Wong D, Miller R H, Shih J W, Jett B, Purcell R H. N Engl J Med. 1991;325:98–104. doi: 10.1056/NEJM199107113250205. [DOI] [PubMed] [Google Scholar]
- 34.Bachman B, Luke W, Hunsmann G. Nucleic Acids Res. 1990;18:1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farci P, Purcell R H. In: Viral Hepatitis: Scientific Basis and Clinical Management. Zuckerman A, Thomas H C, editors. Edinburgh: Churchill Livingstone; 1993. pp. 241–267. [Google Scholar]
- 36.Choo Q-L, Kuo G, Ralston R, Weiner A J, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghton M, Muchmore E. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


