Abstract
Bacterial endospores derive much of their longevity and resistance properties from the relative dehydration of their protoplasts. The spore cortex, a peptidoglycan structure surrounding the protoplasm, maintains, and is postulated to have a role in attaining, protoplast dehydration. A structural modification unique to the spore cortex is the removal of all or part of the peptide side chains from the majority of the muramic acid residues and the conversion of 50% of the muramic acid to muramic lactam. A mutation in the cwlD gene of Bacillus subtilis, predicted to encode a muramoyl-l-alanine amidase, results in the production of spores containing no muramic lactam. These spores have normally dehydrated protoplasts but are unable to complete the germination/outgrowth process to produce viable cells. Addition of germinants resulted in the triggering of germination with loss of spore refractility and the release of dipicolinic acid but no degradation of cortex peptidoglycan. Germination in the presence of lysozyme allowed the cwlD spores to produce viable cells and showed that they have normal heat resistance properties. These results (i) suggest that a mechanical activity of the cortex peptidoglycan is not required for the generation of protoplast dehydration but rather that it simply serves as a static structure to maintain dehydration, (ii) demonstrate that degradation of cortex peptidoglycan is not required for spore solute release or partial spore core rehydration during germination, (iii) indicate that muramic lactam is a major specificity determinant of germination lytic enzymes, and (iv) suggest the mechanism by which the spore cortex is degraded during germination while the germ cell wall is left intact.
The ability of endospores produced by Gram-positive eubacteria (i.e., Bacillus and Clostridium) to survive harsh physical and chemical treatments and to remain in a dormant state for long periods is important in the food preservation industry and represents an interesting biological problem. The protoplast of the spore is relatively dehydrated (even when suspended in H2O) in comparison to that of a vegetative cell, resulting in metabolic dormancy, and this dehydration is responsible in large part for the heat and hydrogen peroxide resistance properties of the spore (1–3). The mechanisms by which this dehydration is achieved and maintained are not clear and remain an outstanding question in the field. Maintenance of protoplast dehydration is clearly dependent on the integrity of a surrounding peptidoglycan wall and the unique structure of this peptidoglycan has been postulated to confer on it a role in attaining protoplast dehydration (4, 5). The spore peptidoglycan is comprised of two contiguous structures, an inner layer called the germ cell wall and a thicker outer layer called the cortex. The germ cell wall appears to have a structure similar to that of vegetative cell wall peptidoglycan (6) and serves as the initial cell wall during spore outgrowth. The vegetative peptidoglycan of Bacillus subtilis is composed of glycan chains of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) residues (7). Each NAM residue carries a peptide side chain, approximately 40% of which are involved in interstrand peptide cross-links to form a network of peptidoglycan chains (7, 8). The cortex peptidoglycan has two unique structural modifications (9, 10) (Fig. 1). Approximately one-half of the muramic acid in the cortex carries no peptide side chain and is cyclized to form muramic lactam (MAL). These lactam residues are found in a highly regular distribution, alternating with NAM residues within the glycan strands. An additional 25% of the NAM has only an l-alanine side chain (9–11). The peptides on the remaining NAM residues are cross-linked at a frequency of 13–19% (9–11). Although the introduction of MAL is a general property of spore peptidoglycan and has been found in the spores of all Bacillus and Clostridium species examined (9), the function of the MAL residues, aside from the resulting decrease in peptidoglycan cross-linking, is unclear. However, it is the low degree of cortex peptidoglycan cross-linking that has given rise to hypotheses concerning a potential role of the cortex in achieving spore protoplast dehydration (4, 5). Loosely cross-linked peptidoglycan exhibits a significant change in volume upon alteration of its ionic environment (12). Therefore, the cortex could potentially have a mechanical activity that results in a decrease in the spore protoplast volume with an accompanying dehydration. During spore germination the cortex is rapidly degraded by autolysins (peptidoglycan lytic enzymes), which exhibit specificity for the cortex structure and which are held in an inactive state within the dormant spore; subsequently, the spore protoplast takes up water and releases solutes, and metabolic activity resumes.
Figure 1.
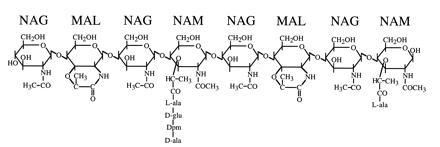
Structure of B. subtilis spore peptidoglycan. This structure was originally determined by Warth and Strominger (9, 10). NAM carries side chains of l-alanine, the tetrapeptide l-ala-γ-d-glu-diaminopimelic acid-d-ala, or the tripeptide l-ala-γ-d-glu-diaminopimelic acid (not shown). Approximately 50% of the muramic acid residues have been converted to MAL, which is found with great regularity at every second muramic acid position. The glycan chains can be cross-linked via the peptide side chains; peptide cross-links are between the ɛ-amino group of diaminopimelic acid of one peptide and the carboxyl-terminal d-alanine of another.
A gene of B. subtilis, cwlD, which appears to encode an autolysin of the N-acetylmuramoyl-l-alanine amidase class, was recently described (13). A null mutation in this gene resulted in the production of dormant spores that were able to carry out the first steps of germination, loss of refractility, and release of dipicolinic acid (DPA, a chelator which is concentrated in the spore protoplast during sporulation), but were unable to degrade the cortex and initiate growth (13). It was postulated that the product of cwlD was a germination-specific peptidoglycan lytic enzyme or alternatively, involved in spore peptidoglycan synthesis (13). We have further examined the properties of the spores produced by a cwlD mutant, including a detailed analysis of the spore peptidoglycan structure. Our analyses indicate that the germination/outgrowth defect in these spores is the result of a complete absence of MAL in the spore peptioglycan and that this structural modification has no influence on spore dehydration or heat resistance.
MATERIALS AND METHODS
Bacterial Strains, Spore Production, and Spore Germination.
All bacterial strains were derivatives of strain PS832, a prototrophic revertant of B. subtilis 168. Mutations were moved into this background by transformation (14). Sporulation was carried out in 2× SG (15) medium at 37°C. Spores were purified by gentle agitation at 4°C for 5–7 days with frequent washing in H2O. DPA was determined as described (16). For some experiments spore coats were permeabilized to lysozyme (decoating) as described (8). Spores were heat activated in H2O at 70°C for 30 min prior to germination in 10 mM Tris·HCl, pH 8.0/8 mM l-alanine at 37°C. Spore protoplast wet density was determined using metrizoic acid gradients as described (3, 17). Dormant spores were permeabilized prior to density determination such that the metrizoic acid could permeate the spore coats and cortex (17); this was found to be unnecessary for density determination of germinated spores. For determination of heat resistance identical samples of purified spores were heated in H2O at 90°C for various lengths of time. To control for increased germination as a result of heat activation all samples were maintained at an elevated temperature for a minimum of 30 min; the time of heating reported is the time at 90°C, the remainder of the time was at 70°C. The spores were then permeabilized as described above followed by five washes in H2O. The recovery of spores was determined at this point by measurement of optical density at 600nm (OD600nm). The washed spores were germinated in 0.3 M sucrose, 10 mM Tris·HCl (pH 7.5), 10 mM MgSO4, 30 mM CaCl2, 8 mM l-alanine, and 25 μg/ml lysozyme for 15 min at 37°C (18). The germinated spores were then diluted in the same solution lacking lysozyme and plated for determination of colony-forming units.
Spore Peptidoglycan Structure Analysis.
Hexosamines and diaminopimelic acid contents were assayed as described (3). A complete description of the reversed-phase high-pressure liquid chromatographic analysis of spore peptidoglycan structure will be published elsewhere (11). Briefly, lytic enzymes were inactivated and small molecules were extracted from purified spores by heating in 5% trichloroacetic acid at 95°C for 6 min (8, 19). The extracted spores were treated with trypsin and then heated in 1% sodium dodecyl sulfate for 15 min at 100°C to remove the majority of the protein. The spore peptidoglycan was digested to completion with a muramidase (Mutanolysin, Sigma), insoluble material (mainly residual spore coat proteins) was removed by centrifugation, and the soluble muropeptides were collected. The muropeptides were reduced with NaBH4 and separated on a Hypersil ODS column (Keystone Scientific, Bellefonte, PA) in a 50-mM NaPO4 buffer system with a gradient from pH 4.31, 0% methanol to pH 4.95, 20% methanol. Purified muropeptides were identified and quantified using amino acid/amino sugar analyses and matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
RESULTS
Phenotypic Characterization of cwlD Mutant Spores.
We moved the previously constructed cwlD::cat insertion mutation into our standard laboratory strain background by transformation and verified that it resulted in the phenotype originally described (13). The mutant strain produced as many spores per unit of culture as the wild type (based upon microscopic observation). Purified wild-type and cwlD mutant spore preparations contained equivalent amounts of DPA (15 μg/OD600 nm unit). When equal numbers of purified spores (based on optical density) were plated, the cwlD spores formed colonies at an efficiency of 5 × 10−4 with respect to the wild type. The plating efficiency was unaffected by heat activation (70°C, 30 min), chemical permeabilization of spore coats, and incubation under germinating conditions prior to plating (30 min at 37°C with 4 mM l-alanine). This plating efficiency of the cwlD mutant is higher than that previously observed (13), possibly due to the different strain background, but still represents a major defect. Spore germination is accompanied by a significant decrease in optical density due to the loss of spore refractility. During germination in liquid medium we observed that the decrease in optical density was slightly delayed in the cwlD mutant relative to the wild type, but after 90 min each spore preparation had lost >70% of its initial optical density. As observed previously (13), release of DPA from the germinating spores was also slightly delayed in the cwlD mutant but was equivalent to the wild type after 30 min. It was seen earlier that release of reducing sugars (due to peptidoglycan degradation) was dramatically reduced during germination of cwlD spores (13). We measured total hexosamine (NAG plus NAM) release during germination and found that the wild type released 37% of the total spore hexosamine after 90 min, whereas the cwlD mutant spores had released <3%.
Transmission electron microscopy of dormant and germinated spores revealed significant differences between the wild-type and cwlD mutant. As observed many times previously, micrographs of dormant wild-type spores reveal a densely staining spore protoplasm and almost a complete lack of staining of the spore cortex (Fig. 2A). In dormant spores of the cwlD mutant the protoplast has a similar appearance but the cortex shows a moderate staining (Fig. 2B). The appearance of wild-type spores changes dramatically following germination. The staining of the protoplast is less dense and the cortex area is significantly stained (Fig. 2C). The total size of the wild-type spores increased significantly upon germination (Fig. 2 A and C); this swelling is easily observable under the light microscope. In addition, the wild-type spore nucleoplasm acquired a fibrous appearance upon germination (Fig. 2C). In contrast, cwlD mutant spores appear almost unchanged following germination (Fig. 2D); the staining of the cortex is similar in the dormant and germinated spores, the staining density of the protoplast has not changed to the same degree as that observed in the wild type, and the nucleoplasm is not apparent.
Figure 2.
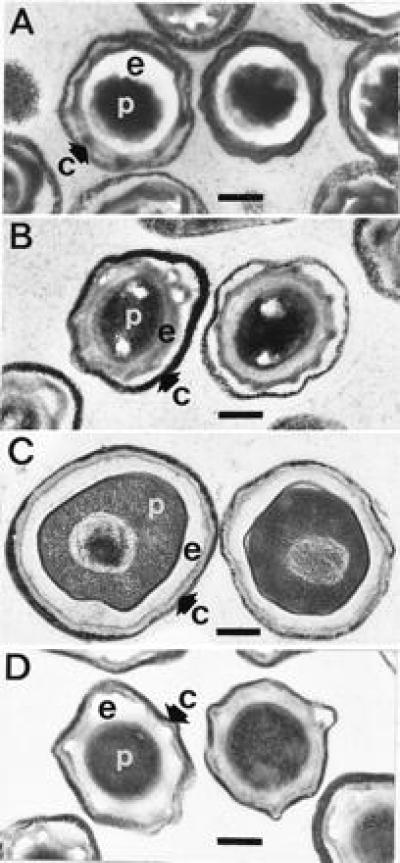
Transmission electron microscopy of dormant and germinated wild-type and cwlD mutant spores. Dormant spores or spores that had been exposed to germinants for 60 min at 37°C were fixed with glutaraldehyde, postfixed, stained, and photographed as described (20). (A and B) Wild-type and cwlD mutant dormant spores, respectively. (C and D) Wild-type and cwlD mutant germinated spores, respectively. c, spore coats and exosporium; e, spore cortex peptidoglycan; p, spore protoplast. (Bar = 0.25 μm.)
We determined the wet density of the spore protoplasts, which we interpret as water content, by equilibrium density centrifugation (17). The cwlD mutant spores exhibited a slight but reproducible decrease in protoplast wet density (1.366 ± 0.006, n = 5) relative to the wild type (1.372 ± 0.006, n = 5) (The errors indicate differences between spore preparations; in each case where wild-type and cwlD spores were prepared and analyzed side-by-side the cwlD spores had a slightly lower protoplast wet density.) Lindsay and colleagues (17) had indicated that the wet densities of the dormant spore protoplasts correspond to protoplast water contents of 37% and 35% (percent wet weight as H2O) for the mutant and wild type, respectively. Sixty minutes after exposure to germinants the protoplast wet density of the wild-type spores had dropped to 1.228 ± 0.009 (n = 3), whereas that of the cwlD mutant spores was 1.263 ± 0.002 (n = 3). We were unable to convert these wet densities to definitive protoplast water contents due to the lack of a published value for the dry density of germinated protoplast contents. However, calculations indicated that the wild-type germinated spores contained 12% more of their wet weight as water than did the cwlD germinated spores.
The slight difference in protoplast wet density between the wild-type and cwlD mutant dormant spores would not be expected to result in a significant difference in heat resistance (1, 2). To learn if the alteration in spore peptidoglycan structure had an effect on heat resistance it was necessary to determine conditions under which the block to outgrowth in the cwlD mutant spores could be overcome. Previous work indicated that spores treated with lysozyme partially recovered the ability to form colonies (0.01% of the spores) (13). We were able to obtain a high plating efficiency with cwlD mutant spores (0.2–0.5 relative to the wild type per OD600nm unit) following chemical permeabilization of the spore coats and lysozyme treatment in an isotonic solution. Following heat treatment for various lengths of time, spores were permeabilized, washed, lysozyme treated, and plated. Changes in optical density were used to control for physical losses during washes. These corrections were similar for all spore preparations. We found that the cwlD mutant spores were indistinguishable from those of the wild type in terms of heat resistance (D90 ≈6 min).
Spore Peptidoglycan Structure.
We have recently developed a procedure for the analysis of spore peptidoglycan structure using reversed-phase high-pressure liquid chromatography (11). The method involves complete digestion of the peptidoglycan with a muramidase (which cleaves between NAM and NAG but not between MAL and NAG) and quantitative analysis of the muropeptides that are liberated. Analyses of peptidoglycan from dormant wild-type spores (11) are briefly described here for the purpose of comparison. These analyses produced results (Fig. 3A, Table 1) consistent with a previous description of spore peptidoglycan structure in which ≈50% of the muramic acid had been converted to muramic acid (9, 10). The muramidase cleaved to produce disaccharides (NAG-NAM; peaks 1, 2, and 3; Table 1, Fig. 3A), tetrasaccharides (NAG-MAL-NAG-NAM; peaks 5, 10, and 13; Table 1, Fig. 3A), and hexasaccharides (NAG-MAL-NAG-MAL-NAG-NAM; peaks 18 and 19; Table 1, Fig. 3A). The NAM residues were substituted with either l-alanine, a tripeptide, or a tetrapeptide (Table 1). Several species of saccharide multimers cross-linked via their peptide side chains (peaks 8, 9, 14, 17, and 20; Table 1, Fig. 3A) were also identified. Quantitative analysis of the digestion products obtained from four spore preparations indicated that 50 ± 1% of the NAM residues had been converted to MAL, 25 ± 1% carried l-alanine side chains, and the remainder had peptide side chains. The diaminopimelic acid residues in 13 ± 1% of these peptide side chains were in peptide cross-links.
Figure 3.
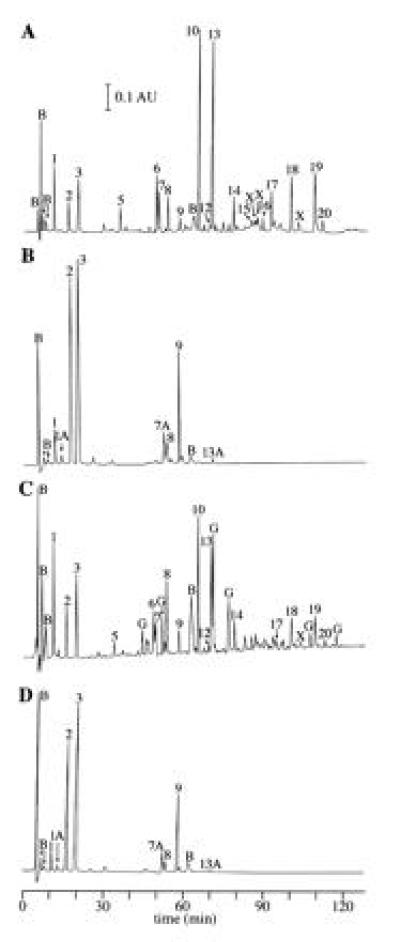
Reversed-phase high-pressure liquid chromatography separation of spore peptidoglycan muropeptides. Spore peptidoglycan was muramidase digested, reduced, and chromatographed as described. (A and B) Muropeptides derived from wild-type and cwlD mutant dormant spores, respectively. (C and D) Muropeptides derived from wild-type and cwlD mutant-germinated spores, respectively. Peaks are numbered as in ref. 11 and have been identified as shown in Table 1. Novel peaks derived from cwlD mutant spores (1A, 7A, and 13A) were identified as described in Table 2. Peaks labeled B are buffer components. Peaks 6, 7, 12, 15, 16, and those labeled X are the results of reduction of MAL in other identified compounds (9, 11); corrections were made for this reduction in the quantitative analysis of the spore peptidoglycan structure (11) (Table 1). Peaks labeled G are unidentified products of germination lytic enzyme activity.
Table 1.
Muropeptides produced by muramidase digestion of spore peptidoglycan
| Peak† | Structure‡ | Muropeptides,
nmol*
|
|||
|---|---|---|---|---|---|
| wt dorm | wt ger | cwlD dorm | cwlD ger | ||
| 1 | DS-TriP | 1.3 | 2.3 | 2.4 | 2.8 |
| 1A | DS-TriP-gly | 0 | ND | 1.2 | 1.2 |
| 2 | DS-Ala | 1.3 | 1.8 | 32.8 | 32.9 |
| 3 | DS-TP | 2.3 | 2.5 | 47.5 | 47.3 |
| 5 | TS-TP§¶ | 0.8 | 0.5 | 0 | 0 |
| 7A | DS-TriP-gly-DS-TP | 0 | 0 | 1.9 | 1.7 |
| 8 | DS-TriP-DS-TP | 0.4 | 0.8 | 0.7 | 0.7 |
| 9 | DS-TP-DS-TP | 0.3 | 0.2 | 5.2 | 5.3 |
| 10 | TS-TP¶ | 12.7 | 5.3 | 0 | 0 |
| 13 | TS-Ala¶ | 20.0 | 8.8 | 0 | 0 |
| 13A | DS-TP-DS-TP-DS-TP | 0 | ND | 0.2 | 0.1 |
| 14 | DS-TP-TS-TP¶ | 0.7 | 0.6 | 0 | 0 |
| 17 | TS-TP-TS-TP¶ | 0.9 | 0.2 | 0 | 0 |
| 18 | HS-TP¶ | 2.3 | 1.3 | 0 | 0 |
| 19 | HS-Ala¶ | 4.0 | 1.6 | 0 | 0 |
| 20 | TS-TP-HS-TP¶ | 0.4 | 0.2 | 0 | 0 |
Recovery of spore peptidoglycan was variable due to physical losses inherent in the purification procedure; however, recovery of muropeptides from muramidase digests was quantitative. These values are nanomoles of compound detected per 100 nmol of muramic acid in a single analysis of each spore type [except for wild-type germinated spores where this is nanomoles of compound detected per 49 nmol of muramic acid to account for muramic acid solubilized during germination (25% of total) and new, unidentified compounds (26% of UV-absorbing material). We make the assumption that the average extinction coefficient of these unidentified compounds is similar to that of the identified compounds.]. Values have been corrected for reduction of MAL and the number of MAL residues per molecule (11). Values of zero should be interpreted as less than 0.1 nmol. wt, wild type; dorm, dormant spores; ger, germinated spores; ND, not determined.
Peaks are numbered as in Fig. 3 and ref. 11.
Structures were determined using amino acid/amino sugar analyses and mass spectrometry (11). DS, disaccharide; TS, tetrasaccharide; HS, hexasaccharide; Ala, l-alanine side chain on NAM; TriP, tripeptide side chain on NAM; TP, tetrapeptide side chain on NAM. Where two saccharide moieties are indicated they are cross-linked via their peptide side chains.
This molecule contains an unidentified chemical modification (11).
These molecules contain MAL residues.
Analysis of peptidoglycan from cwlD spores produced the surprising result that 92% of the UV-absorbing material was in the identified disaccharide monomer and cross-linked disaccharide peaks (Table 1 and Fig. 3B) indicating that there was no MAL present in the cwlD mutant spore peptidoglycan. The identities of these disaccharide peaks were confirmed by co-chromatography with known compounds in a different gradient system (11) (not shown). Minor peaks that chromatographed in the positions of the tetrasaccharides containing MAL seen in the wild type did not co-chromatograph with the tetrasaccharides in the alternative gradient system (not shown). Several novel peaks (Fig. 3B) were identified using amino acid/amino sugar analyses and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. These molecules are: a disaccharide with a tripeptide side chain containing alanine, glutamic acid, diaminopimelic acid (Dpm), and an additional glycine residue (DS-TriP-gly) (peak 1A, Fig. 3, Table 2); DS-TriP-gly-DS-TP (peak 7A, Fig. 3, Table 2) (peak 7A contained two other minor compounds that were removed by chromatography on a different gradient system and were not identified); and the cross-linked trimer of DS-TP, DS-TP-DS-TP-DS-TP (peak 13A, Fig. 3, Table 2). We suspect that the glycine is attached to the ɛ-amino group of the Dpm, in the position of a glycine cross-bridge within a peptide cross-link. The fact that DS-TriP-gly is present in much lower quantities than DS-TriP-gly-DS-TP suggests that the former is derived from the latter by autolytic activity. Quantitative analysis of the identified peaks derived from five spore preparations revealed that in this altered spore peptidoglycan 33 ± 3% of the NAM had l-alanine side chains and the peptides on the remaining 67 ± 3% were cross-linked at a frequency of 12 ± 1%. Thus, the number of cross-links relative to total muramic acid was 8.0% in the cwlD mutant and 3.4% in the wild type.
Table 2.
Structures of unique muropeptides recovered from cwlD mutant spores
| Peak*
|
|||
|---|---|---|---|
| 1A | 7A | 13A | |
| NAG† | 1 (1) | 1 (2) | 1 (3) |
| NAM† | 1.0 (1) | 1.0 (2) | 1.2 (3) |
| Alanine† | 1.1 (1) | 1.7 (3) | 2.2 (6) |
| Glutamic acid† | 1.2 (1) | 1.4 (2) | 1.4 (3) |
| Dpm† | 1.2 (1) | 1.4 (2) | 1.4 (3) |
| Glycine† | 1.0 (1) | 0.6 (1) | 0 |
| Predicted structure | DS-TriP-gly | DS-TriP-gly-DS-TP | DS-TP-DS-TP-DS-TP |
| (M-H)− calc. m/z‡ | 926.9 | 1849.8 | 2788.8 |
| (M-H)− obs. m/z‡ | 926.5 | 1851.0 | 2787.8 |
Peaks are numbered as in Fig. 3 B and D. All nanomole values determined using amino acid and amino sugar analyses (11) are normalized to NAG = 1. Values in parentheses are predicted molar ratios.
In nanomoles.
(M-H)− is the deprotonated molecular ion observed in the negative ion mode. Calc. m/z is the mass/charge predicted from the amino acid/amino sugar analyses. Below m/z 2000 monoisotopic mass values are given; above m/z 2000 average mass values are given. Obs. m/z is the matrix-assisted laser desorption ionization time-of-flight mass spectrometry measured value.
Amino acid analyses of total spores indicated that the cwlD and wild type had equivalent amounts of protein per OD600nm unit. However, there was 2-fold more diaminopimelic acid in the cwlD mutant spores (not shown). This increase in diaminopimelic acid is consistent with the larger percentage of NAM residues substituted with peptide. This increase is insufficient to account for our measurements of total hexosamines per OD600nm unit of spores, which demonstrated that the cwlD mutant spores contained 29% more than the wild type (not shown). This discrepancy may be: (i) within the combined errors of the various measurements, (ii) due to the lack of characterization of several of the muropeptides present in the cwlD mutant spore peptidoglycan digest, or (iii) due to differential recovery of total hexosamine from wild-type and cwlD mutant spores. We are most confident of the diaminopimelic acid measurements due to the low number of manipulations required and the precision of the amino acid analyzer.
High-pressure liquid chromatographic structural analyses were carried out on the peptidoglycan from spores pelleted by low speed centrifugation 60 min after the initiation of spore germination. At this point in germination 25% and ≤3% of the total hexosamine found in dormant wild-type and cwlD mutant spores, respectively, had become soluble. Measurements of DPA release and protoplast wet densities indicated that ≥90% of the spores in these cultures had initiated germination. During this germination period there was no significant change in the muropeptides obtained from cwlD mutant spores (Fig. 3 B versus D, Table 1, and data not shown from three additional spore preparations). However, the peptidoglycan of wild-type spores underwent significant changes during germination (Fig. 3 C versus A). Quantitative analysis of the identified peaks derived from three spore preparations revealed two major peptidoglycan structural changes during germination of wild-type spores. The first was an increase in the percentage of hexosamines found in disaccharide muropeptides from 7 ± 1% to 17 ± 7% of the total, accompanied by decreases in tetrasaccharides and hexasaccharides. The second structural change was an increase in the percentage of NAM residues with tripeptide side chains from 6.5 ± 0.4% to 16 ± 5%, accompanied by decreases in l-alanine side chains and MAL residues. Germination was carried out in the absence of significant added nutrients so we believe that minimal new peptidoglycan synthesis took place during the time frame of the experiment. However, we cannot rule out the possibility that the apparent decrease in MAL and l-alanine side chains was simply due to synthesis of new vegetative wall-type peptidoglycan. Several new compounds appeared in the muramidase digest (Fig. 3C). These new peaks were not identified and constituted 26% of the total UV-absorbing material.
Effects of Other Autolysins on Spore Peptidoglycan Structure.
We analyzed the spore peptidoglycan structures produced by several other mutant strains lacking autolytic enzymes. We first moved mutations effecting these genes into our standard laboratory strain background by transformation. We detected no structural changes associated with losses of the products of cwlA (21, 22), cwlB (lytC) (23, 24), cwlC (25), or cwlG (lytD) (26, 27) (data not shown). As expected, we also found that the heat resistance of these mutant spores was equal to that of the wild type (not shown).
DISCUSSION
The most surprising conclusion to be drawn from our results is that normal spore dehydration and heat resistance can be obtained in the complete absence of MAL. Previous work showed a correlation between spore MAL content and degree of heat resistance (28), but in that case MAL content was probably a measure of total peptidoglycan content (29). Our results indicate that the observed correlation was certainly between total spore peptidoglycan content and heat resistance. The effect of MAL content on peptidoglycan flexibility has not been examined so it is difficult to predict what effect the absence of MAL might have on the postulated role of a spore cortex peptidoglycan mechanical activity in spore protoplast dehydration (4, 5). The fact that we have now identified two different types of major spore peptidoglycan structural alterations that have little or no effect on spore protoplast dehydration (this work and refs. 3 and 11) is suggestive that the cortex does not contribute to the process of dehydration but is simply required to maintain dehydration achieved by other means.
What then is the function of MAL synthesis in the spore peptidoglycan? Our results suggest that MAL serves as a specificity determinant for spore germination lytic enzymes. In the absence of MAL the spores were completely unable to initiate spore cortex peptidoglycan degradation (this work and ref. 13). In wild-type spores it appeared that MAL-containing peptidoglycan was preferentially degraded during germination. These results suggest that there is preferential synthesis of MAL in the cortex peptidoglycan relative to the germ cell wall. If MAL is the major specificity determinant of germination lytic enzymes then the cortex can be degraded without damage to the germ cell wall. Characterization of the germination lytic enzymes will be required to verify this prediction.
The most recent model of the process of spore germination (30) predicted that initiation of cortex hydrolysis was an early step required for rehydration of the protoplast. The ability of cwlD mutant spores to achieve significant protoplast rehydration in the absence of cortex degradation suggests that this is not true. Our results suggest the breakdown of a permeability barrier, allowing the release of solutes and uptake of water, is one step in protoplast rehydration while partial degradation of the cortex, increasing its flexibility, and allowing swelling of the spore, is another. Such a model for spore germination was previously suggested (31). It is possible that full protoplast rehydration in the wild type does not require any cortex hydrolysis and that the cwlD mutant does not achieve full rehydration simply because its cortex is less flexible, prohibiting swelling of the protoplast. Alternatively, the cwlD mutant cortex might be more flexible than that of the wild type, allowing a degree of rehydration in the mutant that might only be achieved following cortex hydrolysis in the wild type. Studies of strains lacking the cortex lytic enzymes involved in spore germination may resolve this issue.
The sequence similarity between the predicted cwlD product and demonstrated N-acetyl-muramoyl-l-alanine amidases (13) suggest that this activity is part of the pathway of MAL synthesis. It is probable that all NAM is incorporated into glycan strands with pentapeptides and that these are subsequently cleaved to tetrapeptides, tripeptides, l-alanine, or to produce MAL. The absence of MAL in the cwlD mutant spores is accompanied by increases in both l-alanine and peptide side chains, therefore the question remains whether the cleavage to l-alanine is on the pathway to MAL synthesis. Muropeptides containing glycine have not been previously detected in B. subtilis so their presence in the cwlD mutant spores was unexpected. This observation raises the possibility that this peptidoglycan modification is introduced during synthesis of wild-type spore peptidoglycan and is subsequently removed during spore maturation. Structural analysis of the spore peptidoglycan of another mutant strain (dacB) has shown the presence of the products of an N-acetyl-muramoyl-l-alanine amidase activity that cleaves entire tetrapeptides from glycan strands (11). Analysis of the spore peptidoglycan produced by a dacB cwlD double mutant and in vitro assays of the substrate specificity of the cwlD product will determine whether the amidase activity observed in dacB mutant spores is a function of the cwlD product. It is also not clear whether MAL synthesis is a direct outcome of the action of the cwlD product or whether another enzyme is required to complete the process. The cwlD mutant spore peptidoglycan may be an ideal substrate for assay of activities required for MAL synthesis and production of MAL-containing muropeptides can be easily detected using our reversed-phase high-pressure liquid chromatography analytical method. Such a system might also be useful for addressing the process by which the high degree of regularity of MAL synthesis within a glycan strand is maintained.
Acknowledgments
We thank J. Sekiguchi for providing strains and George Korza for performing amino acid and amino sugar analyses. This work was supported by Grants GM19698 (to P.S.) and RR1088 (to C.E.C.) from the National Institutes of Health. J.H. was the recipient of a fellowship from the Academy of Finland.
Footnotes
Abbreviations: NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid; MAL, muramic lactam; DPA, dipicolinic acid.
References
- 1.Beaman T C, Greenamyre J T, Corner T R, Pankratz H S, Gerhardt P. J Bacteriol. 1982;150:870–877. doi: 10.1128/jb.150.2.870-877.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakashio S, Gerhardt P. J Bacteriol. 1985;162:571–578. doi: 10.1128/jb.162.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popham D L, Illades-Aguiar B, Setlow P. J Bacteriol. 1995;177:4721–4729. doi: 10.1128/jb.177.16.4721-4729.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis J C, Snell N S, Burr H K. Science. 1960;132:544–545. doi: 10.1126/science.132.3426.544. [DOI] [PubMed] [Google Scholar]
- 5.Warth A D. In: Fundamental and Applied Aspects of Bacterial Spores. Dring G J, Ellar D J, Gould G W, editors. London: Academic; 1985. pp. 209–225. [Google Scholar]
- 6.Tipper D J, Linnet P E. J Bacteriol. 1976;126:213–221. doi: 10.1128/jb.126.1.213-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warth A D, Strominger J L. Biochemistry. 1971;10:4349–4358. doi: 10.1021/bi00800a001. [DOI] [PubMed] [Google Scholar]
- 8.Popham D L, Setlow P. J Bacteriol. 1993;175:2767–2769. doi: 10.1128/jb.175.9.2767-2769.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warth A D, Strominger J L. Proc Natl Acad Sci USA. 1969;64:528–535. doi: 10.1073/pnas.64.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warth A D, Strominger J L. Biochemistry. 1972;11:1389–1396. doi: 10.1021/bi00758a010. [DOI] [PubMed] [Google Scholar]
- 11.Popham, D. L., Helin, J., Costello, C. E. & Setlow, P. (1996) J. Bacteriol. 178, in press. [DOI] [PMC free article] [PubMed]
- 12.Ou L-T, Marquis R E. J Bacteriol. 1970;101:92–101. doi: 10.1128/jb.101.1.92-101.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anagnostopoulos C, Spizizen J. J Bacteriol. 1961;81:74–76. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leighton T J, Doi R H. J Biol Chem. 1971;254:3189–3195. [PubMed] [Google Scholar]
- 16.Nicholson W L, Setlow P. In: Molecular Biological Methods for Bacillus. Harwood C R, Cutting S M, editors. Chichester, England: Wiley; 1990. pp. 391–450. [Google Scholar]
- 17.Lindsay J A, Beaman T C, Gerhardt P. J Bacteriol. 1985;163:735–737. doi: 10.1128/jb.163.2.735-737.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitz-James P C. J Bacteriol. 1971;105:1119–1136. doi: 10.1128/jb.105.3.1119-1136.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquis R E, Bender G R. Can J Microbiol. 1990;36:426–429. [Google Scholar]
- 20.Setlow B, Hand A R, Setlow P. J Bacteriol. 1991;173:1642–1653. doi: 10.1128/jb.173.5.1642-1653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda A, Sekiguchi J. J Gen Microbiol. 1990;136:2209–2216. doi: 10.1099/00221287-136-11-2209. [DOI] [PubMed] [Google Scholar]
- 22.Foster S J. J Gen Microbiol. 1991;137:1987–1998. doi: 10.1099/00221287-137-8-1987. [DOI] [PubMed] [Google Scholar]
- 23.Lazarevic V, Margot P, Soldo B, Karamata D. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda A, Sekiguchi J. J Bacteriol. 1991;173:7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda A, Sekiguchi J. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid M H, Mori M, Sekiguchi J. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 27.Margot P, Mauël C, Karamata D. Mol Microbiol. 1994;12:535–545. doi: 10.1111/j.1365-2958.1994.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 28.Imae Y, Strominger J L. J Bacteriol. 1976;126:907–913. doi: 10.1128/jb.126.2.907-913.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickus G G, Warth A D, Strominger J L. J Bacteriol. 1972;111:625–627. doi: 10.1128/jb.111.2.625-627.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster S J, Johnstone K. Biochem J. 1986;237:865–870. doi: 10.1042/bj2370865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatani Y, Tanida I, Koshikawa T, Imagawa M, Nishihara T, Kondo M. Microbiol Immunol. 1985;29:689–699. doi: 10.1111/j.1348-0421.1985.tb00873.x. [DOI] [PubMed] [Google Scholar]


