Abstract
Fatty Acid Synthase (FASN) is the terminal enzyme responsible for fatty acid synthesis and is upregulated in tumors of various origins to facilitate their growth and progression. Because of several reports linking the fatty acid synthase and proteasome pathways, we asked whether FASN inhibitors could combine with bortezomib, the FDA-approved proteasome inhibitor, to amplify cell death. Indeed, bortezomib treatment augmented sub-optimal FASN inhibitor concentrations to reduce clonogenic survival, which was paralleled by an increase in apoptotic markers. Interestingly, FASN inhibitors induced accumulation of ubiquinated proteins and enhanced the effects of bortezomib treatment. In turn, bortezomib increased fatty acid synthesis, suggesting crosstalk between the pathways. We hypothesized that cell death resulting from crosstalk perturbation was mediated by increased unfolded protein response (UPR) signaling. Indeed, disruption of crosstalk activated and saturated the adaptation arm of UPR signaling, including eIF2α phosphorylation, activating transcription factor 4 (ATF4) expression, and X-box binding protein 1 (XBP-1) splicing. Furthermore, while single agents did not activate the alarm phase of the UPR, crosstalk interruption resulted in activated JNK and C-EBP homologous protein (CHOP)-dependent cell death. Combined, the data support the concept that the UPR balance between adaptive to stress signaling can be exploited to mediate increased cell death and suggests novel applications of FASN inhibitors for clinical use.
Keywords: fatty acid synthase, proteasome, ER stress, unfolded protein response, orlistat, C75
Introduction
Fatty Acid Synthase (FASN) is the central enzyme responsible for catalyzing the ultimate steps of fatty acid synthesis in mammalian cells (1, 2). Interestingly, high levels of FASN expression have been noted in many types of tumors. Accordingly, FASN expression levels correlate with advanced tumor stage and grade, poor patient prognosis and reduced overall- and disease-free survival (3–5). A functional correlation between FASN expression levels and tumor cell survival has been firmly established with the development of small molecule inhibitors of FASN that induce cell death specifically in tumor cells and reduce tumor growth in spontaneous and xenograft tumor models (6–10).
FASN inhibitors induce a number of anti-tumor effects including cell cycle arrest and cell death (3–5). Some of the effects of FASN inhibitors are mediated through key tumor signaling pathways. For instance, it has been demonstrated that pharmacological inhibition of FASN activity results in reduced Akt phosphorylation in multiple tumor cell lines (11, 12). Conversely, phosphoinositide 3-kinase (PI3K) and Akt can drive FASN expression in tumor cells (12, 13). The demonstration that reduced FASN activity negatively affects Akt activation identifies feedback between the two pathways. As a result, blocking both pathways results in amplified cell death (11, 12, 14). Synergy between FASN inhibitors and other chemotherapeutics has been noted in multiple cell lines (15–19). It has also be demonstrated that FASN overexpression protects breast cancer cells from chemotherapy-induced cell death (20). The demonstrated links between FASN, oncogenic pathways and ultimate response to chemotherapy underscore the potential of FASN inhibitors for clinical use and suggest novel strategies for targeting tumor cells to improve cell killing efficiency.
In prostate cancer FASN expression is stabilized by the ubiquitin-specific protease 2a (USP2A), a deubiquitinating enzyme (21). Treating prostate tumor cells with the proteasome inhibitor MG-132, also increases FASN expression, supporting evidence of FASN regulation by the proteasome (21). Inhibition of the proteasome can also stabilize nuclear SREBP-1 and increase FASN expression (22). Conversely, inhibiting FASN affects the proteasome pathway. Specifically, inhibition of FASN reduces expression of the E3 ubiquitin ligase Skp2 that is responsible for mediating the stability of key cellular proteins such as p27 (23). FASN inhibition also affects the expression profiles of several E2 ubiquitin conjugation enzymes and several E3 ubiquitin ligases (24). Combined, these data suggest a level of crosstalk between the fatty acid synthase and proteasome pathways.
Previous work from our laboratory demonstrated that inhibition of FASN activity induces endoplasmic reticulum (ER) stress and activation of the unfolded protein response (UPR) (25). Interestingly, bortezomib (Velcade), an FDA-approved inhibitor of the 26S proteasome, also activates the UPR in tumor cells (26–30). Given the connections between the proteasome and FASN, especially in prostate cancer, and that inhibitors of each induce ER stress, we hypothesized that FASN inhibitors and bortezomib could enhance prostate tumor cell death through UPR-driven mechanisms. The data presented herein demonstrate that functional crosstalk between the fatty acid synthase and a proteasome pathway in prostate tumor cell lines is the basis for UPR-mediated death by two clinically-relevant ER stressing agents.
Materials and Methods
Materials
The PC-3, DU145 and FS-4 cells were obtained from American Type Culture Collection (Manassas, VA). Cell culture medium and supplements were from Invitrogen (Carlsbad, CA). Antibodies against eukaryotic translation initiation factor 2α (eIF2α), phosphor-eIF2α, phosphorylated JNK, total JNK (Thr183/Tyr185), phosphorylated c-Jun (Ser63), Lamin A/C, α-tubulin, cleaved caspase-3 (Asp175), and cleaved PARP (Asp214) were from Cell Signaling Technologies (Beverly, MA). Antibody against fatty acid synthase was from BD Transduction Labs (San Diego, CA). Antibody against β-actin was from Sigma (St. Louis, MO). Antibodies against CHOP (R-20), ATF4 (CREB2 C-20), ubiquitin (FL-76), and XBP-1 (M-186) were from Santa Cruz Biotechnology, Incorporated (Santa Cruz, CA). TRIzol was from Invitrogen (Carlsbad, CA). Avian Myleoblastosis Virus (AMV) Reverse Transcriptase and Taq Polymerase were from Promega (Madison, WI). 14C-acetate was purchased from GE Healthcare (Piscataway, NJ). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA), except for those designed for siRNA which were synthesized by Dharmacon (Lafayette, Co). Orlistat was purchased from Roche (Nutley, NJ), bortezomib was purchased from Millenium Pharmaceuticals (Cambridge, MA) and JNK inhibitor II, SP600125 and JNK inhibitor V, AS601245, from Calbiochem (San Diego, CA). All other reagents were purchased from Sigma (St. Louis, MO), Calbiochem (San Diego, CA) or BioRad (Hercules, CA).
Cell culture and drug treatments
Prostate tumor cell lines were maintained in RPMI 1640 and FS-4 human foreskin fibroblasts were maintained in DMEM-high glucose, both supplemented with 10% fetal bovine serum at 37°C and 5% CO2. Cells were treated for the times and with drug concentrations as indicated. Orlistat was extracted from capsules in EtOH as described previously and stored at −80°C (8). Further dilutions were made in dimethyl sulfoxide (DMSO). Bortezomib was dissolved in DMSO and stored as individual 20 µmol/L aliquots at −20°C.
Immunoblot analysis
Cells were harvested after the indicated treatments, washed with ice-cold phosphate-buffered saline, and lysed in buffer containing 1% Triton X-100 to prepare for immunoblots. Lysis buffer was supplemented with protease, kinase, and phosphatase inhibitors 200 µmol/L phenylmethanesulphonylfluoride (PMSF), 5 µg/ml aprotinin, 5 µg/ml pepstatin A, 5 µg/ml leupeptin, 1 µmol/L sodium fluoride (NaF), 1 µmol/L sodium orthovanadate (Na3VO4), 50 µmol/L okadeic acid just before use. For nuclear proteins, such as ATF4, CHOP, and XBP-1, nuclear extracts were obtained by harvesting cells, washing with ice-cold PBS, then lysing cells using a harvest buffer containing 10 mmol/L HEPES (pH 7.9), 50 mmol/L NaCl, 500 mmol/L sucrose, 100 µmol/L EDTA, 0.5% Triton-X 100 with 1 mmol/L DTT, 10 mmol/L tetrasodium pyrophosphate, 100 mmol/L NaF, 17.5 mmol/L β-glycerophosphate, 1 µmol/L Na3VO4, 1 mmol/L PMSF, 4 µg/ml aprotinin, 2 µg/ml pepstatin A added just before use to separate the cytoplasmic and nuclei. The nuclei were pelleted in a swinging bucket rotor and then washed with buffer containing 10 mmol/L HEPES (pH 7.9), 10 mmol/L KCl, 100 µmol/L EDTA, and 100 µmol/L EGTA with 1 mmol/L DTT, 1 mmol/L PMSF, 1 µmol/L Na3VO4, 4 µg/ml aprotinin, and 2 µg/ml pepstatin added just before use. The nuclei were then lysed in a buffer containing 10 mmol/L HEPES (pH 7.9), 500 mmol/L NaCl, 100 µmol/L EDTA, 100 µmol/L EGTA, and 0.1% NP-40 with 1 mmol/L DTT, 1 mmol/L PMSF, 1 µmol/L Na3VO4, 4 µg/ml aprotinin, and 2 µg/ml pepstatin added just before use. Protein samples were electrophoresed through 10%, 12%, or 13.5% SDS-polyacrylamide gels and transferred to nitrocellulose, except for blots to detect phospho-eIF2α and eIF2α, which were transferred to Immobilon-P membrane (PVDF). Immunoreactive bands were detected by enhanced chemiluminescence (Perkin Elmer). Lamin A/C and β-actin were used as loading controls for immunoblots.
Quantification of Ubiquitin-Modified Proteins
Autoradiographs were scanned using a HP ScanJet4890. The digital files were then quantified using UN-SCAN-IT (Orem, UT). The average intensity of ubiquitin detection was calculated relative to vehicle treated samples for each blot. Then the values for three independent experiments were averaged and graphed on a logarithmic scale.
Fatty acid synthesis assays
To measure fatty acid synthesis, 1 × 105 cells per well were seeded in 24-well plates. Cells were treated with FASN inhibitors or bortezomib as indicated for two hours. 14C-acetate (1 µCi) was added to each well for an additional two hours. Cells were collected, washed and lipids were extracted and quantified by scintillation counting as previously described (8).
Clonogenic survival assays
PC-3 and DU145 cells were plated in 6-well plates at a density of 2000 cells per well 48 hours prior to each experiment. Fresh medium containing the indicated drugs was added at the indicated concentrations for sixteen hours. The media was then removed, the wells were washed and fresh medium was added. Plates were incubated until macroscopic colonies were formed. Visualization of colonies was performed as described previously (25). Colonies were quantified by counting.
Combination Index and statistical calculations
PC-3 cells were treated with a dose response of orlistat, bortezomib, or the combination and analyzed for clonogenic survival. Calculations were performed based on the Chou-Talalay method that calculates a combination index based on the equation: CI = (D1)/(Dx1) + (D2)/(Dx2) + (D1)(D2)/(Dx1)(Dx2) where (D1) and (D2) are the doses of the individual drugs that have ‘x’ effect when used in combination and (Dx1) and (Dx2) are the doses of the drugs having ‘x’ effect when used separately (31). A CI < 1 indicates synergism.
For clonogenic survival assays, survival of treated cells was normalized relative to vehicle treated cells and P-values between combined and single agent treated cells were determined by two-tailed student's t tests. For fatty acid synthesis assays, bortezomib treated cells were compared to vehicle treated cells and P-values were determined by two-tailed student's t test. For the ubiquitin-modification blots, the quantification of three separate blots was averaged for each treatment and then significance measured relative to untreated controls by two-tailed student’s t test.
Detection of XBP-1 splicing and GADD34 expression and suppression of CHOP expression with siRNA
Cells were exposed to the various drug treatments or transfected with siRNA for the indicated times. RTPCR was performed for XBP-1 splicing and GADD34 expression as described previously (25). To knockdown CHOP levels, an siGENOME SMARTpool siRNA oligonucleotide cocktail against CHOP (1, AAAUGAAGAGGAAGAAUCA; 2, GAAUCUGCACCAAGCAUGA; 3, CCAGCAGAGGUCACAAGCA; 4, GAGCUCUGAUUGACCGAAU) and one control siRNA against luciferase as a negative control (Luc sense, CUUACGUGAUACUUCGAUU) were designed and synthesized by Dharmacon. The individual siRNAs (83 nmol/L) were transfected into cells at plating with siPORT NeoFX transfection reagent (Ambion) according to manufacturers instructions. After 48 hours, transfection media was removed and fresh media containing indicated drug was added to cells. After indicated treatment times, cells were collected and nuclear protein was harvested for immunoblot analysis of CHOP and lamin A/C or tubulin.
Results
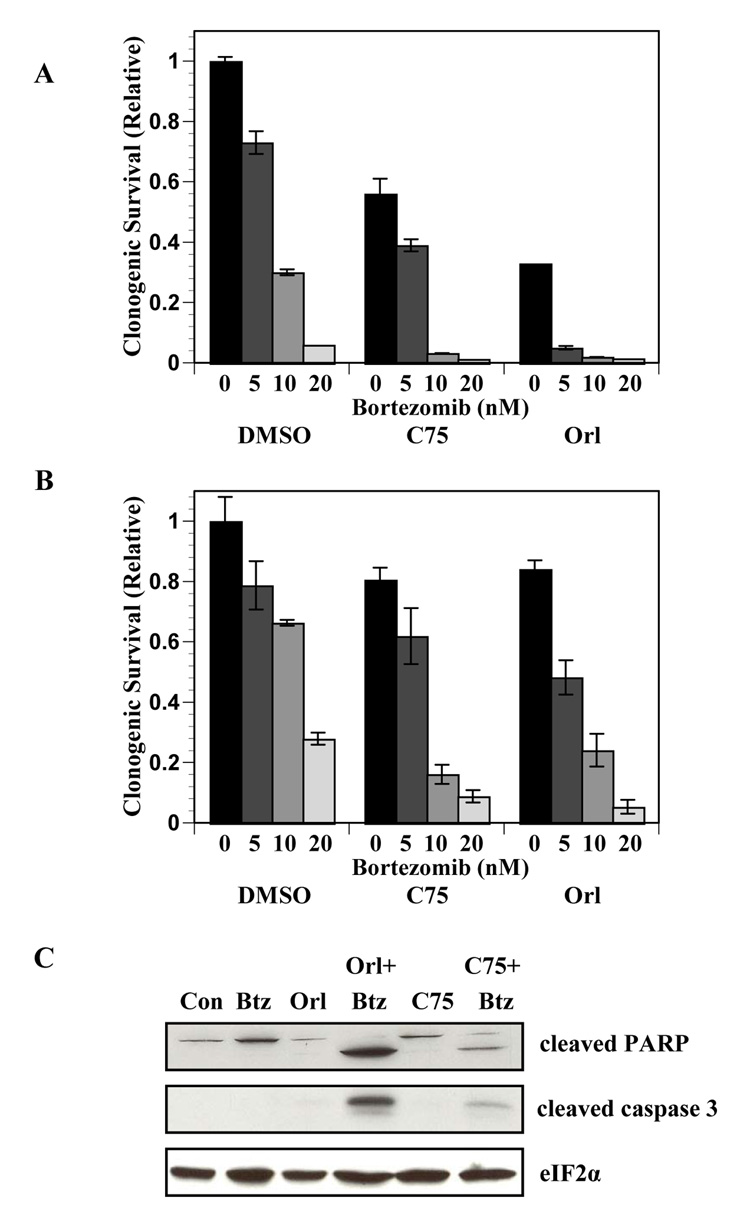
We hypothesized that the identified connections between the fatty acid synthesis and proteasome pathways would provide a novel strategy to target UPR activation and increase cell death in prostate tumor cells. To test this hypothesis PC-3 and DU145 cells were examined for clonogenic survival after treatment with orlistat or C75 and the proteasome inhibitor bortezomib (Velcade, PS-341). Clonogenic survival of PC-3 and DU-145 cells was reduced by bortezomib treatment in a dose-dependent manner (Figure 1A and B). Sub-optimal concentrations of FASN inhibitors were used to reduce cell killing by any of the single agents (data not shown). Clonogenic survival of PC-3 cells treated with orlistat and C75 was reduced by 60% and 30%, respectively (P < 0.001, Figure 1A). In DU145 cells, survival was only reduced by about 20% with each inhibitor. When the FASN inhibitors were combined with bortezomib, clonogenic survival was strikingly diminished compared to cells treated with the single agents (P ≤ 0.005, Figure 1A and Figure 1B). Although isobologram analyses were inconclusive, an analysis of the combination–index suggests that combining FASN inhibitors with bortezomib results in synergism (31). Consistent with previous findings, sub-optimal concentrations of orlistat and C75 did not result in cleavage of PARP or caspase-3. However, when FASN inhibitors were combined with bortezomib significant levels of both cleaved PARP and cleaved caspase-3 were detected (Figure 1C). Combined, the data showed that combining bortezomib and FASN inhibitors resulted in enhanced cell death.
Figure 1. FASN inhibitors combine with bortezomib to increase cell death.
Clonogenic survival assays were performed on PC-3 (A) and DU145 (B) cells. Cells were treated with the indicated concentrations of bortezomib in the presence of DMSO, C75 (10 µg/ml) or orlistat (25 µmol/L) for 16 hours. Clonogenic survival was determined relative to vehicle-treated controls. Statistical significance was determined by two tailed student’s t test of cells treated with the combination of FASN inhibitors and bortezomib compared to cells treated with FASN inhibitors alone (P ≤ 0.005). C, PC-3 cells treated with bortezomib (5 nmol/L), orlistat (25 µmol/L), C75 (10 µg/ml), or the combination of FASN inhibitors and bortezomib for 18 hours and collected for immunoblot analysis using antibodies specific for cleaved PARP, cleaved caspase 3, and total eIF2α. Images represent cropped immunoblots. Full length scans available in supplementary data figure 1.
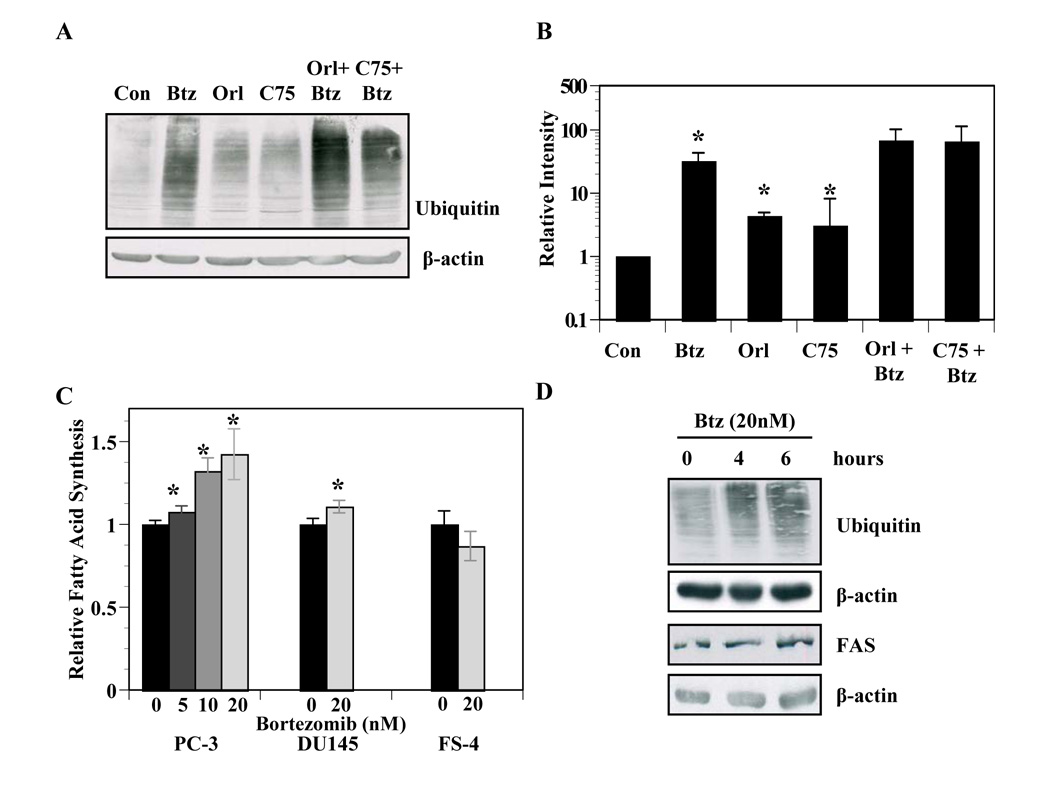
Given the established links between the fatty acid synthesis and proteasome pathways, we asked whether inhibition of FASN would affect the function of the proteasome. PC-3 cells were treated with FASN inhibitors, bortezomib, or FASN inhibitors and bortezomib together followed by immunoblot analysis of ubiquitin-modified proteins. As expected, bortezomib induced accumulation of ubiquitin-modified proteins (P ≤ 0.05, Figures 2A and 2B). Interestingly, PC-3 cells treated with orlistat or C75 also induced a modest but significant accumulation of ubiquitin-modified proteins (P ≤ 0.01). The co-treatment of FASN inhibitors with bortezomib also appeared to cause an additive accumulation of ubiquitin-modified proteins (Figures 2A, 2B), further suggesting that FASN activity contributes to the functionality of the proteasomal pathway. Next, we asked whether bortezomib affected fatty acid synthesis. Bortezomib induced a dose-dependent increase in fatty acid synthesis in PC-3 cells and a modest but significant increase in DU145 cells (P ≤ 0.05, Figure 2C). On the other hand, bortezomib had no effect on fatty acid synthesis in FS-4 human fibroblasts. The increased fatty acid synthesis was not associated with increased FASN protein levels (Figure 2D). Collectively, these data demonstrate crosstalk between the FASN and proteasome pathways.
Figure 2. Fatty acid synthesis and proteasome pathways exhibit crosstalk.
A, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib, (5 nmol/L), C75 (9 µg/ml), or the combination of bortezomib and orlistat or C75 for 16 hours and subjected to immunoblot analysis with antibodies specific for ubiquitin and β-actin. B, Three separate experiments of PC-3 cells treated as in A were quantified by measuring intensity of the exposure from ubiquitin antibody in each treatment as compared to vehicle treated lanes using UN-SCAN-IT (Orem, UT). Asterisks indicate statistical significance measured by student’s t test (P ≤ 0.05). C, PC-3 cells, DU145 cells and FS4 fibroblasts were incubated with the indicated concentrations of bortezomib for two hours followed by the addition of 14C-acetate (1 µCi) for two hours. Cells were collected, washed and lipids were extracted and quantified relative to vehicle-treated control as described in Experimental Methods. Asterisks indicate statistical significance by student’s t test (P ≤ 0.05). D, PC-3 cells were treated with vehicle or bortezomib (20 nmol/L) for 4 or 6 hours in duplicate. Cells were collected for immonoblot analysis using antibodies specific to fatty acid synthase, ubiquitin, or β-actin. Images represent cropped immunoblots. Full length scans available in supplementary data figure 2.
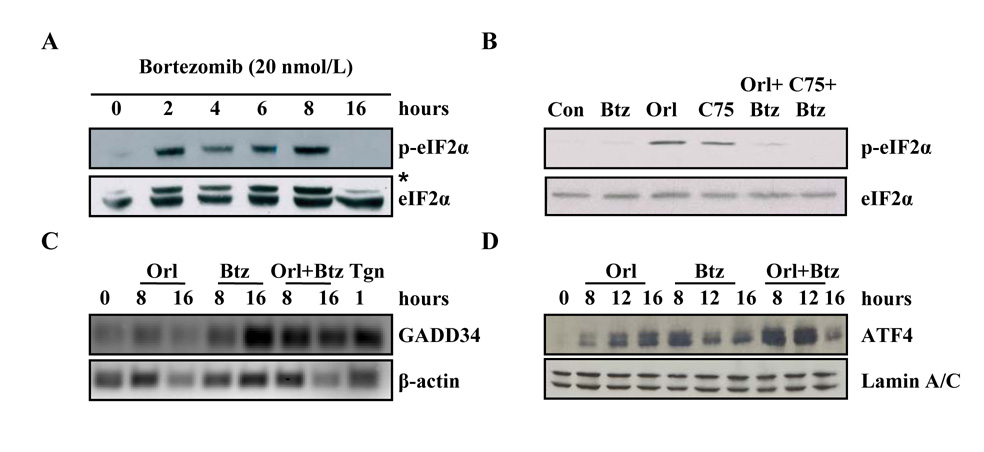
Previous work demonstrated that FASN inhibitors induce ER stress in tumor cells and others have shown that bortezomib induces ER stress (25, 28–30, 32). To characterize the role of the UPR in cells treated with the combination of FASN inhibitors and bortezomib, we first examined the phosphorylation status of eIF2α. Bortezomib induced a rapid and transient phosphorylation of eIF2α that diminished by sixteen hours (Figure 3A). Consistent with previous findings, phosphorylation of eIF2α was sustained through 16 hours in cells treated with FASN inhibitor (25). Disrupting the crosstalk between the proteasome and FASN resulted in nonphosphorylated eIF2α at sixteen hours, consistent with the lack of phosphorylated eIF2α with bortezomib alone at the same time point (Figure 3B).
Figure 3. Bortezomib and FASN inhibitors combine to saturate the PERK arm of UPR adaptation signaling.
A, PC-3 cells were treated with vehicle (DMSO) or 20 nmol/L bortezomib for the indicated times. Samples were resolved by SDS-PAGE, transferred to PVDF and the membrane was probed with antibodies specific for phospho-eIF2α and total eIF2α. B, PC-3 cells were treated with vehicle, bortezomib (5 nmol/L), orlistat (25 µmol/L), C75 (10 µg/ml), or the combination of bortezomib and orlistat or bortezomib and C75 for 16 hours and samples were subjected to immunoblot analysis and probed with antibodies specific for phosphor-eIF2α and total eIF2α. Asterisk denotes higher molecular weight nonspecific band. C, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (20 nmol/L), or the combination of bortezomib and orlistat for the indicated times. Total RNA was collected and semi-quantitative RT-PCR was performed using primers specific for GADD34 and β-actin. D, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (5 nmol/L), or the combination of bortezomib and orlistat. Nuclear fractions were isolated and prepared for immunoblot analysis and probed with antibodies specific for ATF4 and Lamin A/C. Images represent cropped immunoblots. Full length scans available in supplementary data figure 3.
In the UPR signaling cascade, growth arrest and DNA damage-inducible protein 34 (GADD34) is induced, mediating dephosphorylation of eIF2α to restore bulk translation (33, 34). To test whether single and combined agents induce GADD34 expression over the course of sixteen hours, semi-quantitative RT-PCR was performed with oligonucleotides specific for GADD34. PC-3 cells treated with orlistat showed slight GADD34 mRNA at eight and sixteen hours compared to vehicle treated controls (Figure 3C). Consistent with the timecourse of eIF2α phosphorylation in bortezomib treated cells, GADD34 mRNA expression was minimal at eight hours followed by robust expression at sixteen hours (Figure 3C). Inhibiting crosstalk between pathways induced GADD34 mRNA expression by eight hours to a level higher than seen with bortezomib or orlistat alone that was maintained through 16 hours, correlating with the lack of phosphorylated eIF2α at the same timepoint (Figure 3C). Together, these data showed that interrupting crosstalk between the proteasome and FASN resulted in acceleration of the eIF2α – GADD34 feedback loop indicative of increased UPR signaling and suggested that proteotoxicity may contribute to cell death.
Phosphorylation of eIF2α is a primary event in UPR signaling that activates downstream signals including the pro-survival activating transcription factor 4 (ATF4) (35, 36). Therefore, we tested whether interrupting crosstalk between proteasome and fatty acid synthesis pathways would increase ATF4 expression. Orlistat induced early expression of ATF4 at eight hours that was sustained through the sixteen hour time point (Figure 3D) consistent with robust phosphorylation of eIF2α (Figure 3B) and low levels of GADD34 (Figure 3C) in orlistat treated cells. Bortezomib treatment induced expression of nuclear ATF4 by eight hours that decreased at twelve and sixteen hours, consistent with phosphorylated eIF2α status and GADD34 expression (Figure 3). Interfering with the FASN – proteasome crosstalk induced ATF4 expression that was more robust than orlistat or bortezomib treated cells at eight hours but decreased by the sixteen hour time point (Figure 3D). Correspondingly, GADD34 levels were high and phosphorylated eIF2α levels were low at sixteen hours in combination treated cells (Figure 3C). Together, the time courses of eIF2α phosphorylation, GADD34, and ATF4 expression indicated that adaptation signals of the PERK arm of the UPR pathway was not only active in cells treated with FASN inhibitors and bortezomib, but was saturated when both pathways were inhibited.
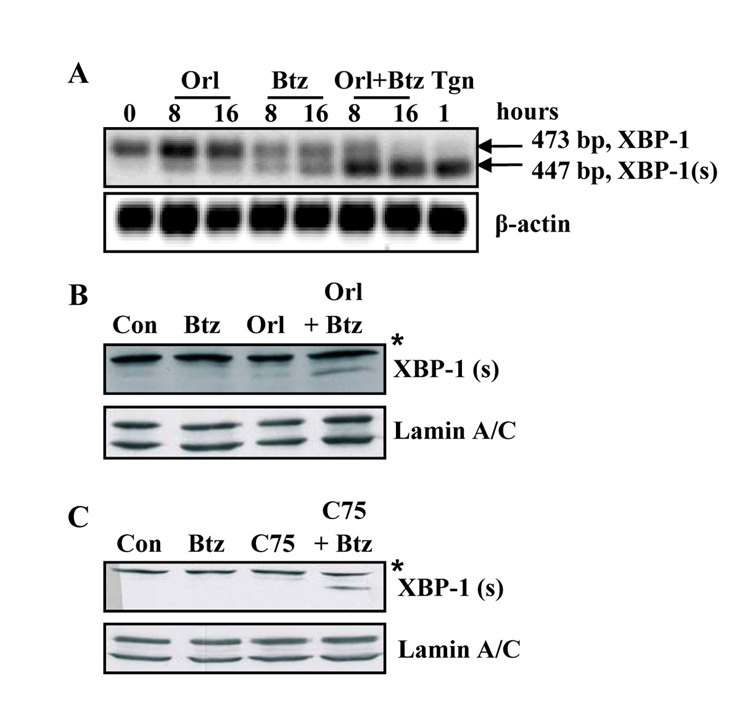
We next examined the IRE1-mediated processing of X-box binding protein 1 (XBP-1) in cells treated with orlistat, bortezomib or both. Sub-optimal concentrations of orlistat and bortezomib induced moderate splicing of XBP-1 mRNA by eight hours (Figure 4A). Treating cells with both agents caused and earlier and more robust processing of XBP-1 that was complete by sixteen hours (Figure 4A). Correspondingly, at these times and concentrations, accumulation of the XBP-1(s) transcription factor only occurred in cells treated with both FASN inhibitor and bortezomib (Figure 4B and C). Therefore, interfering with FASN and the proteasome simultaneously enhanced the adaptation response through IRE1-mediated processing of XBP-1 in prostate tumor cells.
Figure 4. Combining bortezomib with FASN inhibitors enhances IRE1-mediated XBP-1 mRNA processing.
A, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (5 nmol/L) or the combination of bortezomib and orlistat. Total RNA was collected with TRIzol and RT-PCR was performed with oligonucleotides specific for XBP-1 and β-actin. XBP-1 (us) is indicated by the 473 bp product and XBP-1 (s) is indicated by the 447 bp fragment. PC-3 cells were treated for one hour with thapsigargin (1 µmol/L) as a positive control. B, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (5 nmol/L), or the combination of bortezomib and orlistat. Nuclear lysates were collected as described in Experimental Methods for immunoblot analysis with antibodies specific for XBP-1 and Lamin A/C. Immunoblot indicates the 55 kD XBP-1 (s) protein. Asterisk denotes higher molecular weight nonspecific band. C, PC-3 cells were treated with vehicle, bortezomib (5 nmol/L), C75 (9 µg/ml), or the combination of bortezomib and C75. Asterisk denotes higher molecular weight nonspecific band. Images represent cropped immunoblots. Full length scans available in supplementary data figure 4.
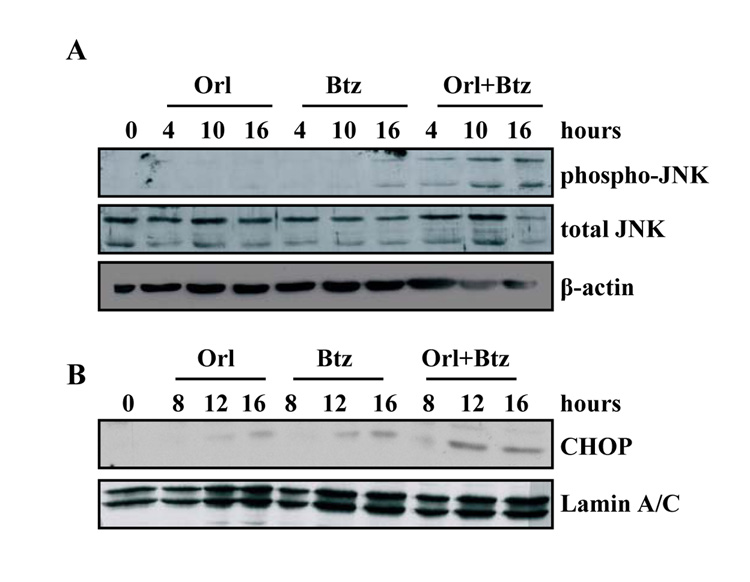
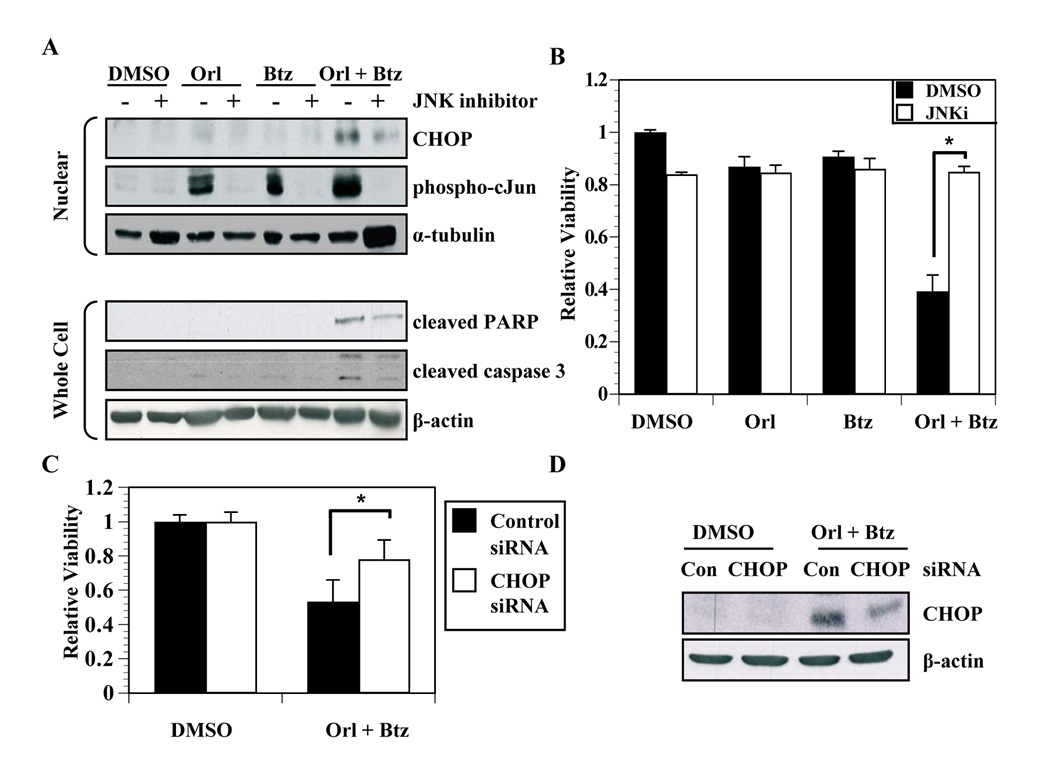
We next asked whether IRE1-activated signals characteristic of the alarm arm of the UPR pathway were upregulated in cells in which we inhibited fatty acid synthesis-proteasome crosstalk. The phosphorylation status of Jun N-terminal kinase (JNK) was examined by immunoblot (Figure 5A). Orlistat did not induce JNK phosphorylation, and bortezomib treatment induced only minimal JNK activation at sixteen hours (Figure 5A). Treating cells with both agents in combination, though, induced JNK phosphorylation within four hours that increased through sixteen hours. Similar results were observed in DU145 cells (data not shown). The C/EBP homologous protein (CHOP) transcription factor can be induced by JNK activation (37, 38). Orlistat only induced moderate CHOP expression at sixteen hours (Figure 5B). Similarly, at twelve hours, bortezomib induced minimal CHOP expression that slightly increased by sixteen hours (Figure 5B). On the other hand, combined inhibition of FASN and the proteasome pathways induced an earlier and more robust CHOP expression (Figure 5B). A JNK inhibitor was used to verify the role of JNK in mediating CHOP expression. Pharmacological inhibition of JNK blocked the phosphorylation of JNK and c-Jun (Figure 6A). More importantly, inhibition of JNK also reduced CHOP expression and cleavage of caspase-3 and PARP (Figure 6A). Consistent with these findings, trypan-blue exclusion assays also demonstrated that JNK inhibition protected cells from cell death induced by the combination of orlistat and bortezomib (P < 0.01, Figure 6B).
Figure 5. Combining bortezomib and FASN inhibitors induces JNK activation and CHOP expression.
A, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (20 nmol/L), or the combination of bortezomib and orlistat for the indicated times. Whole cell lysates were collected for immunoblot analysis of phospho-JNK, total JNK, and β-actin. B, PC-3 cells were treated with vehicle, orlistat (25 µmol/L), bortezomib (20 nmol/L), or the combination of bortezomib and orlistat for the indicated times. Nuclear lysates were collected as described in Experimental Methods for immunoblot analysis with antibodies specific for CHOP and Lamin A/C. Images represent cropped immunoblots. Full length scans available in supplementary data figure 5.
Figure 6. UPR-associated cell death is mediated by JNK activation and CHOP.
A, PC-3 cells were treated with vehicle, orlistat (50 µmol/L), bortezomib (20 nmol/L), or the combination of bortezomib and orlistat with and without JNK inhibitor (JNKi, 20 nmol/L). Nuclear lysates were collected as described in Experimental Methods for immunoblot analysis with antibodies specific for CHOP, phosphorylated c-Jun and α-tubulin. Whole cell lysates were analyzed with antibodies specific for cleaved PARP, cleaved caspase 3, and β-actin. B, PC-3 cells were treated with vehicle, orlistat (50 µmol/L), bortezomib (20 nmol/L), or the combination of bortezomib and orlistat with and without JNK inhibitor (20 nmol/L). Cells were collected and counted using a trypan-blue exclusion assay and the ratio of viable cells was calculated relative to vehicle treated cells. Statistical significance was determined using student’s t test (P < 0.01). C, PC-3 cells were transfected with a SMARTpool siRNA against CHOP or a control siRNA. After 48 hours, cells were treated with vehicle or the combination of bortezomib (20 nmol/L) and orlistat (50 µmol/L) for 18 hours. Cells were then collected and counted using trypan blue exclusion and the ratios of viable cells relative to vehicle treated controls were calculated from three independent experiments. Statistical significance between CHOP siRNA and control siRNA cells treated with orlistat and bortezomib was determined by two-tailed student’s t test (P < 0.01). D, Cells from C were analyzed by immunoblot with antibodies specific for CHOP and β-actin. Images represent cropped immunoblots. Full length scans available in supplementary data figure 6.
We next tested whether CHOP is an effector of JNK signaling that regulates cell death in this scenario. PC-3 cells were transfected with control or CHOP-targeted siRNA, and after 48 hours the cells were treated with vehicle, orlistat or with the orlistat-bortezomib combination. The siRNA-mediated knockdown of CHOP had no effect on orlistat or bortezomib induced cell death (not shown). However, siRNA-mediated knockdown of CHOP expression protected cells from the effects of the orlistat-bortezomib combination (P < 0.01, Figure 6C) at a level that was proportional with the reduction in CHOP levels (Figure 6D). Therefore, it appears that the JNK-CHOP axis is an important mediator of cell death in cells where the UPR is saturated. Collectively, these data demonstrate that blocking crosstalk between FASN and the proteasome shifts UPR balance from adaptation phase to the alarm phase, resulting in increased cell death via JNK-mediated CHOP expression.
Discussion
We have previously shown that inhibiting FASN induces ER stress and UPR signaling before cell death occurs and hypothesize that cell death induced by FASN inhibition is dependent on UPR activation (25). The data herein reveal a functional nexus between FASN and the proteasome. The connection of each of these pathways with ER homeostasis provides a unique therapeutic opportunity. Indeed, when inhibitors of both the proteasome and FASN pathways are combined, increased cell death occurs through the IRE1-JNK-CHOP arm of the UPR pathway.
Accumulating evidence links the proteasome and FASN pathways (21, 23, 24). The data herein suggests that the integrity of the proteasome pathway relies, at least in part, on functional fatty acid synthesis as FASN inhibitors cause the accumulation of ubiquinated proteins through an undetermined mechanism (Figure 2). It is possible that either the proteasome overall, or a component of the proteasome pathway, relies on the fatty acid synthesis pathway for proper function. Interestingly, the ER stressor tunicamycin can also induce the accumulation of ubiquitin-modified proteins and enhances the stability of E3 ligases gp78 and Hrd1 (39). Therefore, it is likely that the proper functioning of proteasome-mediated ER-associated degradation pathway is dependent upon global ER function.
Perhaps more interesting is that inhibiting the proteasome alone can increase fatty acid synthesis (Figure 2C). As the increased activity does not correlate with increased FASN protein levels, the data suggest a mechanism independent of SREBP-1 or FASN protein stabilization as others have demonstrated previously (21, 22). Induction of fatty acid synthesis by bortezomib could be due to several possibilities. Inhibition of the proteasome could affect turnover of a protein that modifies FASN or acetyl-CoA carboxylase (ACC). Because of the short time of treatment with bortezomib required to detect the increase of lipogenesis, a previously unidentified post-translational modification may occur to increase activity of either FASN or ACC. Alternatively, there could be increased incorporation of fatty acid into phospholipid. Previous studies have demonstrated that XBP-1(s) can increase phosphatidylcholine synthesis (40). However, significant splicing of XBP-1 was not observed within the same time period in which fatty acid synthesis increased (Figure 4). Therefore, it is unlikely that XBP-1(s) induces phospholipid synthesis in response to bortezomib treatment. Regardless, the crosstalk between FASN and proteasome pathways is the basis for amplification of UPR signaling and increased cell death when inhibitors of these pathways are combined. Increased UPR signaling in this sceneario, particularly that of the alarm arm, support a growing body of literature describing the ability for the UPR to induce cell death after adaptation signals have been saturated or are ineffective (41–43).
The activation of adaptation and stress signals is critical to the cellular balance between survival and cell death. PERK-dependent phosphorylation of eIF2α can act as a rheostat to fine tune signals that balance survival and death in a cell experiencing ER stress (44). If a cell is unable to sustain balanced levels of phosphorylated eIF2α, the cell dies due to proteotoxicity. If the cell has levels of phosphorylated eIF2α that are too high, the cell cannot translate enough protein to adapt and survive. As compared to the combination of FASN inhibitors with bortezomib and bortezomib alone, FASN inhibitors take the longest amount of time to induce cell death. Correspondingly, FASN inhibitors result in sustained phosphorylated eIF2α, therefore more cellular protection (Figure 3). Simultaneous inhibition of the proteasome and fatty acid synthesis pathways induces early and robust GADD34 expression to mediate the dephosphorylation of eIF2α (Figure 3). Coincidentally, the combined treatment quickly induces cell death, perhaps by proteotoxicity associated with decreased eIF2α phosphorylation.
The IRE1 arm of the UPR is another signaling pathway designed to mediate adaptation and alarm signals (41, 43, 45). IRE1 splices XBP-1 mRNA that is then translated to a stable ER-stress specific transcription factor that upregulates ER chaperones and other adaptation genes (46, 47). We show that FASN inhibitors and bortezomib both induce splicing of XBP-1, but that the combined treatment results in the accumulation of the XBP-1 transcription factor (Figure 4A–C). Therefore, blockade of FASN-proteasome crosstalk enhances adaptation signaling through the IRE1 arm of the UPR. Inhibiting both pathways also shifts the balance from adaptation to stress signaling downstream of IRE1. Activated IRE1 can also interact with tumor necrosis factor receptor adaptor factor 2 (TRAF2) to activate JNK and corresponding alarm signals, including expression of the ER stress pro-apoptotic protein CHOP (48, 49). Indeed, the combined treatment results in robust activation of JNK, as well as enhanced accumulation of CHOP, thereby confirming the hypothesis that increased ER stress shifts the UPR program to cell death. Consistent with other studies, the data presented here indicate that CHOP mediates cell death when FASN-proteasome crosstalk is interrupted (Figure 6) (50). That JNK and CHOP mediate death in this system is consistent with the notion that IRE1 guides cell fate in response to ER stress (43).
Altogether, the data indicate that rapid and robust UPR activation occurs when FASN and the proteasome are inhibited simultaneously. While the connection between the proteasome and the ER-associated degradation pathway has been established, the mechanism by which FASN mediates proteasome function remains to be determined. It will be important for future studies to determine the precise connections between these two important tumor support systems. Significant interest and opportunity lies in exploiting the UPR with therapeutic agents to augment tumor cell death. Collectively, these data provide insight into how FASN and the proteasome can be targeted to shift UPR balance from adaptation to stress signaling to affect increased tumor cell death.
Acknowledgments
Grant Support: This research was supported by the National Cancer Institute (5R01CA114104-02) and the Department of Defense (W81XWH-07-1-0024) to S.J.K. Views and opinions of, and endorsements by, the author(s) do not reflect those of the Department of Defense.
References
- 1.Jayakumar A, Tai MH, Huang WY, et al. Human fatty acid synthase: properties and molecular cloning. Proc Natl Acad Sci U S A. 1995;92:8695–8699. doi: 10.1073/pnas.92.19.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 3.Kridel SJ, Lowther WT, Pemble CWt. Fatty acid synthase inhibitors: new directions for oncology. Expert Opin Investig Drugs. 2007;16:1817–1829. doi: 10.1517/13543784.16.11.1817. [DOI] [PubMed] [Google Scholar]
- 4.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 5.Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Research. 2006;66:5977–5980. doi: 10.1158/0008-5472.CAN-05-4673. [DOI] [PubMed] [Google Scholar]
- 6.Funabashi H, Kawaguchi A, Tomoda H, et al. Binding Site of Cerulenin in Fatty Acid Synthetase. J Biochem (Tokyo) 1989;105:751–755. doi: 10.1093/oxfordjournals.jbchem.a122739. [DOI] [PubMed] [Google Scholar]
- 7.Kuhajda FP, Pizer ES, Li JN, et al. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc Natl Acad Sci U S A. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kridel SJ, Axelrod F, Rozenkrantz N, Smith JW. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Research. 2004;64:2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- 9.Alli PM, Pinn ML, Jaffee EM, McFadden JM, Kuhajda FP. Fatty acid synthase inhibitors are chemopreventive for mammary cancer in neu-N transgenic mice. Oncogene. 2005;24:39–46. doi: 10.1038/sj.onc.1208174. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Han WF, Landree LE, et al. Fatty acid synthase inhibition activates AMP-activated protein kinase in SKOV3 human ovarian cancer cells. Cancer Research. 2007;67:2964–2971. doi: 10.1158/0008-5472.CAN-06-3439. [DOI] [PubMed] [Google Scholar]
- 11.Wang HQ, Altomare DA, Skele KL, et al. Positive feedback regulation between AKT activation and fatty acid synthase expression in ovarian carcinoma cells. Oncogene. 2005;24:3574–3582. doi: 10.1038/sj.onc.1208463. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Shi Y, Giranda VL, Luo Y. Inhibition of the phosphatidylinositol 3-kinase/Akt pathway sensitizes MDA-MB468 human breast cancer cells to cerulenin-induced apoptosis. Mol Cancer Ther. 2006;5:494–501. doi: 10.1158/1535-7163.MCT-05-0049. [DOI] [PubMed] [Google Scholar]
- 13.Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, Swinnen JV. Role of the phosphatidylinositol 3'-kinase/PTEN/Akt kinase pathway in the overexpression of fatty acid synthase in LNCaP prostate cancer cells. Cancer Research. 2002;62:642–646. [PubMed] [Google Scholar]
- 14.Bandyopadhyay S, Pai SK, Watabe M, et al. FAS expression inversely correlates with PTEN level in prostate cancer and a PI 3-kinase inhibitor synergizes with FAS siRNA to induce apoptosis. Oncogene. 2005;24:5389–5395. doi: 10.1038/sj.onc.1208555. [DOI] [PubMed] [Google Scholar]
- 15.Menendez JA, Vellon L, Mehmi I, et al. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc Natl Acad Sci U S A. 2004;101:10715–10720. doi: 10.1073/pnas.0403390101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menendez JA, Vellon L, Lupu R. Antitumoral actions of the anti-obesity drug orlistat (XenicalTM) in breast cancer cells: blockade of cell cycle progression, promotion of apoptotic cell death and PEA3-mediated transcriptional repression of Her2/neu (erbB-2) oncogene. Ann Oncol. 2005;16:1253–1267. doi: 10.1093/annonc/mdi239. [DOI] [PubMed] [Google Scholar]
- 17.Menendez JA, Colomer R, Lupu R. Inhibition of tumor-associated fatty acid synthase activity enhances vinorelbine (Navelbine)-induced cytotoxicity and apoptotic cell death in human breast cancer cells. Oncol Rep. 2004;12:411–422. [PubMed] [Google Scholar]
- 18.Menendez JA, Lupu R, Colomer R. Inhibition of tumor-associated fatty acid synthase hyperactivity induces synergistic chemosensitization of HER −2/ neu -overexpressing human breast cancer cells to docetaxel (taxotere) Breast Cancer Res Treat. 2004;84:183–195. doi: 10.1023/B:BREA.0000018409.59448.60. [DOI] [PubMed] [Google Scholar]
- 19.Menendez JA, Vellon L, Colomer R, Lupu R. Pharmacological and small interference RNA-mediated inhibition of breast cancer-associated fatty acid synthase (oncogenic antigen-519) synergistically enhances Taxol (paclitaxel)-induced cytotoxicity. Int J Cancer. 2005;115:19–35. doi: 10.1002/ijc.20754. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Liu Y, Zhang J-T. A new mechanism of drug resistance in breast cancer cells: fatty acid synthase overexpression-mediated palmitate overproduction. Mol Cancer Ther. 2008;7:263–270. doi: 10.1158/1535-7163.MCT-07-0445. [DOI] [PubMed] [Google Scholar]
- 21.Graner E, Tang D, Rossi S, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 22.Hirano Y, Yoshida M, Shimizu M, Sato R. Direct Demonstration of Rapid Degradation of Nuclear Sterol Regulatory Element-binding Proteins by the Ubiquitin-Proteasome Pathway. J Biol Chem. 2001;276:36431–36437. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- 23.Knowles LM, Axelrod F, Browne CD, Smith JW. A fatty acid synthase blockade induces tumor cell-cycle arrest by down-regulating Skp2. J Biol Chem. 2004;279:30540–30545. doi: 10.1074/jbc.M405061200. [DOI] [PubMed] [Google Scholar]
- 24.Knowles LM, Smith JW. Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer. BMC Genomics. 2007;8:168–181. doi: 10.1186/1471-2164-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little JL, Wheeler FB, Fels DR, Koumenis C, Kridel SJ. Inhibition of fatty acid synthase induces endoplasmic reticulum stress in tumor cells. Cancer Research. 2007;67:1262–1269. doi: 10.1158/0008-5472.CAN-06-1794. [DOI] [PubMed] [Google Scholar]
- 26.Alberts B, Johnson A, Lewis J, et al., editors. Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. [Google Scholar]
- 27.Gardner RC, Assinder SJ, Christie G, et al. Characterization of peptidyl boronic acid inhibitors of mammalian 20 S and 26 S proteasomes and their inhibition of proteasomes in cultured cells. Biochem J. 2000;346:447–454. [PMC free article] [PubMed] [Google Scholar]
- 28.Obeng EA, Carlson LM, Gutman DM, et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawrocki ST, Carew JS, Pino MS, et al. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res. 2005;65:11658–11666. doi: 10.1158/0008-5472.CAN-05-2370. [DOI] [PubMed] [Google Scholar]
- 30.Fribley A, Zeng Q, Wang C-Y. Proteasome Inhibitor PS-341 Induces Apoptosis through Induction of Endoplasmic Reticulum Stress-Reactive Oxygen Species in Head and Neck Squamous Cell Carcinoma Cells. Mol Cell Biol. 2004;24:9695–9704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou T-C, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in Enzyme Regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 32.Kincaid MM, Cooper AA. ERADicate ER stress or die trying. Antioxid Redox Signal. 2007;9:2373–2387. doi: 10.1089/ars.2007.1817. [DOI] [PubMed] [Google Scholar]
- 33.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 35.Harding HP, Novoa I, Zhang Y, et al. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Molecular Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 36.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guyton KZ, Xu Q, Holbrook NJ. Induction of the mammalian stress response gene GADD153 by oxidative stress: role of AP-1 element. Biochem J. 1996;314:547–554. doi: 10.1042/bj3140547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urano F, Wang X, Bertolotti A, et al. Coupling of Stress in the ER to Activation of JNK Protein Kinases by Transmembrane Protein Kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y, Ballar P, Apostolou A, Doong H, Fang S. ER stress differentially regulates the stabilities of ERAD ubiquitin ligases and their substrates. Biochemical and Biophysical Research Communications. 2007;352:919–924. doi: 10.1016/j.bbrc.2006.11.121. [DOI] [PubMed] [Google Scholar]
- 40.Sriburi R, Bommiasamy H, Buldak GL, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 41.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 43.Lin JH, Li H, Yasumura D, et al. IRE1 Signaling Affects Cell Fate During the Unfolded Protein Response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novoa I, Zhang Y, Zeng H, et al. Stress-induced gene expression requires programmed recovery from translational repression. Embo J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 46.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 47.Lee A-H, Iwakoshi NN, Glimcher LH. XBP-1 Regulates a Subset of Endoplasmic Reticulum Resident Chaperone Genes in the Unfolded Protein Response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang XZ, Harding HP, Zhang Y, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. Embo J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 50.Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]