Abstract
The ability to genetically modify mesenchymal stem cells (MSCs) seeded inside synthetic hydrogel scaffolds would offer an alternative approach to guide MSC differentiation and to study molecular pathways in three dimensions than protein delivery. In this report, we explored gene transfer to infiltrating MSCs into matrix metalloproteinase (MMP) degradable hydrogels that were loaded with DNA/poly(ethylene imine) (PEI) polyplexes. DNA/PEI polyplexes were encapsulated inside poly(ethylene glycol) (PEG) hydrogels crosslinked with MMP degradable peptides via Michael Addition chemistry. A large fraction of encapsulated polyplexes remained active after encapsulation (65%) and the mechanical properties of the hydrogels were unchanged by the encapsulation of the polyplexes. Cells were seeded inside the hydrogel scaffolds using two different approaches, clustered and homogeneous. The viability of MSCs was similar in hydrogels with and without polyplexes. Transgene expression was characterized with time using a secreted reporter gene and showed different profiles for clustered and homogeneously seeded cells. Clustered cells resulted in cumulative transgene expression that increased through the 21-day incubation, while homogeneously seeded cells resulted in cumulative transgene expression that plateaued after 7-days of culture. The use of hydrogel scaffolds that allow cellular infiltration to deliver DNA may result in long lasting signals in vivo, which are essential for the regeneration of functional tissues.
Introduction
Tissue regeneration aims to promote the healing of diseased or injured tissue through the use of a biodegradable scaffold that supports cellular infiltration and contains bioactive signals that guide invading cells through tissue formation [1, 2]. During morphogenesis, extracellular matrix (ECM) proteins, cytokines, growth factors, cell-cell contacts and mechanical stimuli provide signals to cells that result in cell fates that, when orchestrated, result in tissue or organ formation [3]. The goal when designing scaffolds for tissue regeneration is to design an environment that remodels or changes as the cells infiltrate and proliferate so that all stages of tissue formation can be guided and that the end product is a completely natural tissue. Although peptides and growth factors are generally utilized as the bioactive signals in tissue engineering scaffolds, we and others have investigated the use of DNA delivery as an alternative or complementary approach to introduce bioactive signals into tissue engineering scaffolds [4–9]. In this approach the cells themselves produce the protein signals through the delivery of DNA that encodes for the desired protein.
Gene delivery can be achieved with the use of modified viruses (viral delivery) or polymers (non-viral delivery) that encapsulate or condense plasmid DNA into particles that can transport the DNA inside the cell. Although viral delivery is generally more efficient than non-viral approaches, it has limitations due to its potential immunogenicity and insertion mutagenesis [10]. Because of the mentioned safety concerns, non-viral approaches have been investigated. Poly(ethylene imine) (PEI) is a cationic polymer that has been widely utilized for non-viral gene delivery. PEI is able to condense DNA through electrostatic interactions between the positively charged amines in the PEI and the negatively charged phosphates on the DNA, forming particles in the range of 50 to 200 nm [11]. DNA/PEI polyplexes enter the cell through endocytosis and are believed to be able to escape the endosome through endosomal buffering (the proton sponge effect, [12, 13]). PEI has been successfully used in vitro and in vivo delivering DNA or siRNA to the brain [14, 15], lungs [16–19], abdomen [20], and tumors [21–23].
DNA delivery from tissue engineering hydrogel scaffolds is a versatile approach to promote the expression of tissue inductive factors locally to be used as signals to promote tissue formation. Naked DNA, as well as complexed DNA, has been incorporated into hydrogel scaffolds including collagen [5], pluronic-hyaluronic acid [24], PEG-poly(lactic acid)-PEG [25], engineered silk elastin [26], fibrin [9, 27], and PEG-hyaluronic acid [28] hydrogels. However, gene transfer efficiency remains a major limitation and gene transfer to MSCs seeded in three-dimensions has not previously been investigated. We are interested in investigating gene transfer to MSCs in matrix metalloproteinase (MMP) degradable hydrogels due to their potential use to transfect MSC-like cells in vivo as they infiltrate the hydrogel scaffold. MMP-degradable hydrogels have been shown to allow cell migration through proteolytic degradation [29, 30] and to support cell growth in vivo [31, 32] and in vitro [29, 30, 33–35]. Thus we believe that these hydrogels are interesting materials to explore the delivery of genes to MSCs as they infiltrate the hydrogel scaffold.
In this report, we explored the delivery of PEI/DNA polyplexes from MMP degradable PEG hydrogels to MSCs grown inside the hydrogel scaffold. The end goal of these studies was to develop methods for the assessment of gene transfer to infiltrating cells as would occur when an acellular DNA loaded scaffold is implanted in vivo. We hypothesized that cells infiltrating into a hydrogel scaffold containing DNA/PEI polyplexes would result in the cells being transfected multiple times as they encountered new polyplexes during their migration into the scaffold. We used previously established MMP-degradable hydrogels [30–35] and developed methods to incorporate DNA/PEI polyplexes through the bulk of the matrix with high activity and low aggregation.
Methods
Materials
Peptides GCRDGPQGIWGQDRCG (MMPxl) and Ac-GCGWGRGDSPG-NH2 (RGD) were obtained from NEOMPS (Strasburg, France). Bovine plasma thrombin was purchased from Sigma-Aldrich (St. Louis, MO). Human fibrinogen (plasminogen depleted) was purchased from Enzyme Research Laboratories (South Bend, IN, USA). gWIZ secreted alkaline phosphatase mammalian expression vector (pSEAP, Genlantis, San Diego, CA) and green fluorescent protein-luciferase expression vector (pEGFP-LUC, BD Biosciences, Palo Alto, CA) were expanded using an endotoxin free Giga Prep kit from Qiagen following the manufacturer’s instructions. Linear poly(ethylene imine) (25 kg/mole, PEI) was purchased from Polysciences (Warrington, PA). All other products were purchased from Fisher Scientific unless noted otherwise.
Cell culture and zymography
Mouse bone marrow cloned mesenchymal stem cells (D1, CRL12424) were purchased from ATCC (Manassas, VA, USA) and cultured in DMEM (Sigma-Aldrich) medium supplemented with 10% bovine growth serum (BGS, Hyclone, Logan, Utah) and 1% penicillin/streptomycin (Invitrogen, Grand Island, NY) at 37 °C and 5% CO2. The cells were split using trypsin following standard protocols. To determine the extent to which D1 cells express matrix metalloproteinases, a gelatin zymogram was used (Bio-Rad, Hercules, CA). D1 cell mediums at passages 3, 6 and 10 were collected and assayed for total protein concentration using the Bradford assay (Pierce, Rockford, IL). 27 µg of total protein were loaded in each lane and the gel was run in 1x Tris-Glycine SDS running buffer at 110 V. The gel was then placed in renaturing buffer (2.5% Triton-100) for 30-min at room temperature and then incubated in the developing buffer (0.5–1% tris(hydroxymethyl)aminomethane, 1–2% sodium chloride) for 30-min at room temperature with further development in fresh developing buffer at 37 °C overnight. The gel was stained with commassie blue R-250 for 30-min (0.5% w/v, Thermo Scientific, Rockforld, IL) and de-stained in a methanol, acetic acid, and water solution (50:10:40) leaving clear bands where protease activity was present. Human active matrix metalloproteinase 2 (MMP-2, 4 ng/well, Calbiochem, Gibbstown, NJ), complete DMEM medium and BSA were run in the gel for comparison.
PEG-VS synthesis
Four-armed PEG-OH (20 kDa, Nektar, Aerogen Ltd., Galway, IE) was modified with divinyl sulfone as previously described with minor modifications [33]. Briefly, PEG-OH was dissolved in tetrahydrofluran (THF, Aldrich/Fluka, Basel, Switzerland) under inert atmosphere and heated to reflux in a Soxhlet apparatus filled with molecular sieves for 3–4-hours. The solution was allowed to cool to the touch and sodium hydride (NaH, Fluka, Basel, Switzerland), at 5-fold molar excess over OH groups, was added followed by the addition of divinyl sulfone, at 30-fold molar excess over OH groups. The reaction was carried out at room temperature (RT) under argon atmosphere with constant stirring for 3-days. Afterwards, the reaction solution was neutralized with acetic acid, filtered, concentrated and precipitated in ice-cold diethyl ether (Acros Organics/Chemie Brunschwig AG, Basel, Switzerland). Precipitation was repeated three times to remove unreacted divinyl sulfone. The final product was dried under vacuum and stored under argon at −20 °C. The degree of functionalization was confirmed with 1H NMR (in CDCl3): 3.6ppm (PEG backbone), 6.1 ppm (d, 1H, CH2), 6.4 ppm (d, 1H, CH2), and 6.8ppm (dd, 1H, -SO2CH). The degree of end group conversion, as shown by NMR, was found to be 99%.
PEG hydrogel formation and DNA/PEI polyplex encapsulation
PEG hydrogels were formed by Michael-type addition of bis-cysteine containing MMPxl peptides or bis-thiol containing PEG (HS-PEG-SH, 3400 g/mole, Nektar, Huntsville, AL) onto four-armed PEG–VS functionalized with cell adhesion peptides (RGD peptides) (Scheme 1). Lyophilized aliquots of the crosslinker (0.91 mg MMPxl or 3.280 mg HS-PEG-SH) were diluted in 10 µL of 0.3 M TOEA (pH=8.0) buffer immediately before mixing with a mixture of DNA/PEI polyplexes and PEG-VS. DNA/PEI polyplexes were formed by mixing plasmid DNA with PEI in DI water, vortexing for 15-s and incubating for 15-min at room temperature before adding 6.5 µL 3 M TOEA (final volume 65 µL) to the complex solution. A lyophilized aliquot of PEG-VS (6.5 mg) was dissolved in 25 µL of 0.3 M TOEA, mixed with 65 µL of DNA/PEI polyplex solution and 10 µL of MMPxl. The gel precursor solution was then placed either as a 100 µL drop or three 30 µL drops between two Teflon plates for 30-min at 37 °C to allow for gelation. The final gel was swelled in DMEM medium before being placed inside 96-well plates for long-term culture.
Scheme 1.
PEG hydrogels were formed through Michael addition of cysteine-containing matrix metalloproteinase sensitive peptides (MMPxl) with 4-armed PEG-vinyl sulfone pre-modified with cell adhesion peptides (PEG-RGD). Polyplexes were encapsulated into hydrogel matrix by mixing with the precursor solution prior to gelation. Cells were seeded as single cells or a cluster of cells (shown) inside the hydrogel matrix. As the cells infiltrate the scaffold, they encounter polyplexes and are transfected.
To visualize the distribution of the polyplexes inside the PEG hydrogels, gels were formed using the same protocol as described above except rhodamine labeled DNA (MirusBio Label IT nucleic acid TM rhodamine labeling kit) was used to form the polyplexes and phosphate buffered saline (PBS) was used to swell the gel. For these experiments, HS-PEG-SH crosslinker was used. The swelled gel was imaged using an inverted confocal microscope (Leica TCS SP MP) using a 40X objective over a 20 µm thick section of PEG gel.
DNA/PEI polyplex release kinetics and activity
In order to determine the extent of release of the encapsulated polyplexes and their activity post encapsulation, gels were formed using the protocols indicated above and with MMPxl crosslinker. To test the release kinetics, the gels were swelled in PBS for 2-hours and then the swelling solution was collected and the gels were placed in 200 µL of releasing solution (PBS, 0.25% trypsin/EDTA, or D1 conditioned media). At the indicated time points, 100 µL of the medium was removed and an additional 100 µL of fresh releasing medium was added. Following the final release medium collection, the gels were incubated with 0.25% Trypsin/EDTA to result in complete release of the DNA from the gel as a result of gel degradation. The DNA concentration in the samples was measured using HOECHT dye (H33258). In a typical measurement, 100 µL heparin (10 mg/mL in PBS) was incubated with 10 µL the released sample for 10-min at room temperature to displace the DNA from DNA/PEI complex. 10 µL of the above solution was then mixed with 100 µL H33258 assay solution (0.1 µg/mL H33258 in TNE buffer (0.2 M NaCl, 10 mM Tris, 1.3 mM EDTA)) and the fluorescence light intensity was read using a fluorometer equipped with a UV filter (Turner Biosystems, Sunnyvale, CA). The readout was analyzed using a standard curved measured using free DNA. The presence of heparin/PEI was not found to affect the binding efficiency of the HOECHT dye to DNA nor did the heparin/PEI interfere with the fluorescence reading. The data was plotted as a % release by dividing the DNA released at a given time point by the total DNA amount in the gel.
To determine the activity of the encapsulated DNA/PEI polyplexes, a PEG gel was prepared and swelled as indicated above using DNA encoding for luciferase (pEGFP-LUC). After swelling in PBS, the gel was degraded through incubation with 100 µL 0.25% trypsin at 37 °C for 30-min. The released DNA/PEI polyplexes (1 µg in 150 mM NaCl) were then added to plated D1 cells (50,000 cells in 24-well plates) and incubated for 48-hours. After the incubation time, the cells were washed with PBS and lysed using 200 µL of lysis buffer. The luciferase activity was measured using a luciferase assay kit (Promega, Madison, WI) as detailed below. For comparison, transfection using freshly prepared complexes supplemented with free PEG-VS, 0.25% trypsin/EDTA, or a combination of both was used.
Hydrogel Rheology
Plate-to-plate rheometry was used to determine if the presence of DNA/PEI polyplexes affected the mechanical properties of the gel. The gels were made as detailed above and cut to a size of 1.8 cm in diameter so that when compressed by 200 µm the gel filled the entire moving plate of the rheometer (2.0 cm in diameter). Compression of the gel was necessary to avoid slipping. The storage (G') and loss modulus (G") were measured using an AR2000 rheometer (TA Instruments, New Castle, DE) under constant strain of 0.05 and frequency from 0.1 to 10 Hz.
Cell encapsulation
D1 cells were encapsulated using two protocols: homogeneous encapsulation, resulting in single cells throughout the hydrogel, and clustered encapsulation, resulting in a single cluster of cells inside a fibrin clot (Scheme 1). The cells were used between passages 3 and 10. For the homogeneously plated cells, the cells were encapsulated during gelation by mixing 400,000 cells in a 30 µL gel. The cells were mixed with the PEG-VS/polyplex solution before the crosslinker was added. For the clusters of cells, fibrin clots were formed by resuspending D1 cells (1.8×106 cells) in 60 µL of a 2 mg/mL fibrinogen and 2 U/mL thrombin solutions in DMEM. The clusters were made by dropping 5 µL of the suspension onto a Teflon plate and incubating at 37 °C for 20-min. The clusters were incorporated into the PEG gel by placing them inside 30 µL of gel precursor solution. The resulting gel was pressed between two Teflon plates and gelled at 37°C for 30-min. The gel was swelled in DMEM for two hours and cultured for up to 20-days in DMEM supplemented with 10% BGS and 1% P/S. At the indicated time points, cells were imaged with an inverted fluorescence microscope or 25 µL medium was removed for alkaline phosphatase detection as detailed below. Cell migration rates out of fibrin clusters were determined by measuring the distance between the cluster-gel edges to the growing cell front. Measurements from 5 different locations in one gel were averaged. At day 13, the cells were stained for the actin network and the cell nuclei using phalloidin-488 (Invitrogen) and HOECHT dye. For staining, hydrogels were washed once with PBS before fixing with 4% paraformaldehyde for 4-min at room temperature. The hydrogels were then washed twice with PBS and permeated with 0.1% Triton X-100 in PBS followed by another PBS wash. The hydrogels were then incubated in a solution of BSA (1% in PBS) for 1-hour before addition of a solution containing Alexa488-phalloidin (25 µL stock in 200 µL of 1%BSA PBS) and HOECHT dye (1 µg/ml in 1%BSA PBS) and incubation for a 1-hour. Three final washes were made with 0.05% Tween-20 PBS solution after which imaging was conducted using a Nikon inverted microscope (Nikon TE2000U).
Reporter gene quantification/observation
Three types of reporter genes were employed: green fluorescent protein (GFP), luciferase (LUC), and secreted alkaline phosphatase (SEAP). Green fluorescent protein was observed using a fluorescence microscope (Nikon TE2000U) at predetermined time points. For fluorescence imaging the gels were cultured in medium without phenol red. Luciferase activity was determined in cell lysate using a luciferase assay kit following the manufacturer’s protocol and using a modulus luminometer (Turner Biosystems, Sunnyvale, CA) with a 10-s integration time. The luciferase activity was normalized using the total protein content of the lysate, which was measured using the Bradford assay (PIERCE, Rockford, IL). The SEAP activity was determined using Phospha-Light™ SEAP reporter gene assay system (Applied Biosystems, Bedford, MA) following the manufacturer’s instructions with a 20-min incubation with the substrate and using luminescence reader (Turner Biosystems, Sunnyvale, CA) with a 1-s integration time. The measured RLUs were related to mass of SEAP protein (pg) using a standard curve, RLU = 931.42X0.99 pg R2 = 0.999, made with recombinant SEAP protein and plotted using a log-log scale. To determine if the SEAP protein diffuse out from the PEG hydrogel into the cell culture medium freely, D1 cells were transfected with pSEAP/PEI polyplexes for 24-hours and released with trypsin. 150,000 cells were either encapsulated in 100 µL MMP degradable PEG hydrogel homogeneously or plated in the 24-well plate for another 24-hours. The total SEAP secreted to the medium was measured and compared.
Live/dead cell staining
Cell viability in the PEG gel was studied with the LIVE/DEAD® viability/cytotoxicity kit (Molecular Probes, Eugene, OR). Briefly, 1 µL of ethidium homodimer-1 and 0.25 µL of calcein AM from the kit were diluted with 500 µL DMEM without phenol red to make the staining solution. Each gel was stained with 150 µL of the staining solution for 30-min at room temperature in the dark and imaged with an inverted fluorescence microscopy (Nikon TE2000U).
Statistical analysis
Statistical analyses were done using the statistical package Instat (GraphPad Software, La Jolla, CA). For multiple comparisons, the means of triplicate independent samples were compared using the Tukey multiple comparisons analysis with the alpha level indicated in the figure legend.
Results
Characterization of DNA loaded PEG hydrogels
DNA/PEI polyplexes were encapsulated inside MMP-degradable PEG hydrogels by mixing the polyplexes with the gel precursors prior to crosslinking. To ensure that the polyplexes were homogeneously distributed throughout the hydrogel and to determine the maximum loading density of DNA without significant aggregation of the polyplexes, fluorescently labeled polyplexes were incorporated inside the hydrogel and visualized with confocal fluorescence microscopy. DNA/PEI polyplexes formed with 15, 30 and 50 µg of Rho-DNA with an N/P ratio of 7.5 and with 15 µg Rho-DNA with an N/P ratio of 15 were incorporated into a PEG hydrogel (100 µL, Figure 1). Polyplexes dispersed uniformly for the 15 µg and 30 µg/100 µL DNA loading densities (Figure 1A, B, D). However, when the concentration of polyplexes was raised to 50 µg/100 µL significant aggregation was observed (Figure 1C).
Figure 1.
Distribution of DNA/PEI polyplexes inside the PEG hydrogel. DNA labeled with TM-rhodamine was used to form the polyplexes prior to encapsulation inside the gel. Polyplexes made with 15 µg DNA/100 µL gel at N/P = 7.5 (A), 30 µg DNA/100 µL gel at N/P = 7.5 (B), 50 µg DNA/100 µL gel at N/P = 7.5 (C) and 15 µg DNA/100 µL gel at N/P = 15 (D) were encapsulated inside the hydrogel scaffold. Confocal microscopy was used to take the images using a 40x objective over a 20 µm thick section. Scale bar equals 10 µm.
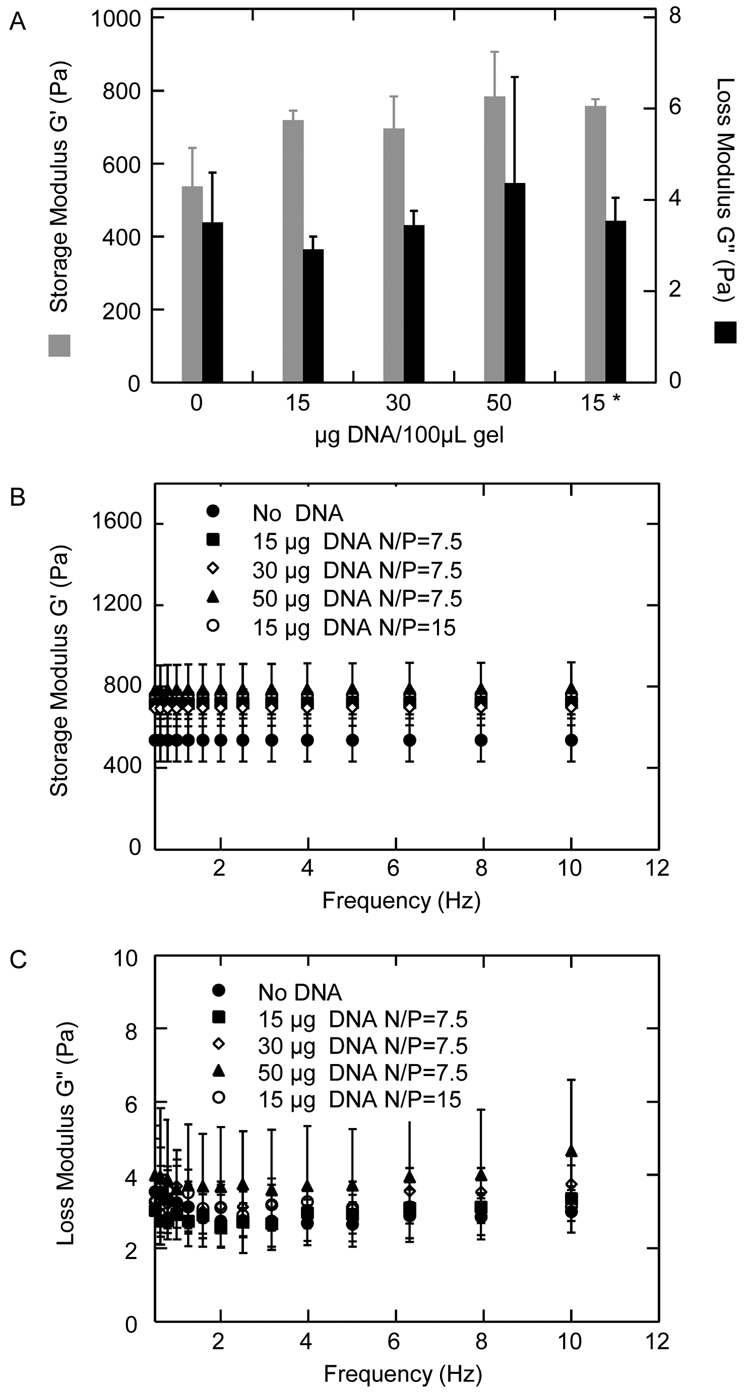
To ensure that the incorporation of the polyplexes did not significantly alter the mechanical properties of the hydrogels, the storage and loss moduli (G' and G"), which indicate the elastic and viscous properties, respectively, were measured with a rheometer. PEG hydrogels were formed with encapsulated DNA/PEI polyplexes (15–50 µg DNA/100 µL gel, N/P=7.5 or 15) and the resulting gels were tested for their mechanical properties (Figure 2). There is no statistical difference in G' and G" between hydrogels not containing DNA and hydrogels containing polyplexes formed with up to 30 µg DNA (p > 0.05, Figure 2A). Hydrogels formed with 50 µg of DNA polyplexes resulted in a statistical increase in G' when compared to hydrogels with no DNA (p < 0.05) (Figure 2A). The finding that the G' and G" did not cross at any measured frequency (0.1 to 10 Hz, Figure 2B, C) and that the storage and loss modulus were frequency independent (Figure 2B, C) were consistent with hydrogel rheology. The loss tangent values (ratio of G" to G') were lower than 0.006 for all the hydrogels tested indicating that the hydrogels were highly elastic.
Figure 2.
Storage (G') and loss modulus (G") of PEG hydrogel with and without DNA/PEI polyplexes measured using plate-to-plate rheometry. Polyplexes made with 15 µg DNA/100 µL gel at N/P = 7.5, 30 µg DNA/100 µL gel at N/P = 7.5, 50 µg DNA/100 µL gel at N/P = 7.5 and 15 µg DNA/100 µL gel at N/P = 15 (* in Figure A) were encapsulated inside the hydrogel scaffold. G' and G" were measured under constant strain of 0.05 and frequency from 0.1 to 10 Hz. Overall G' and G" (A), and G' (B) and G" (C) over the entire frequency sweep are shown.
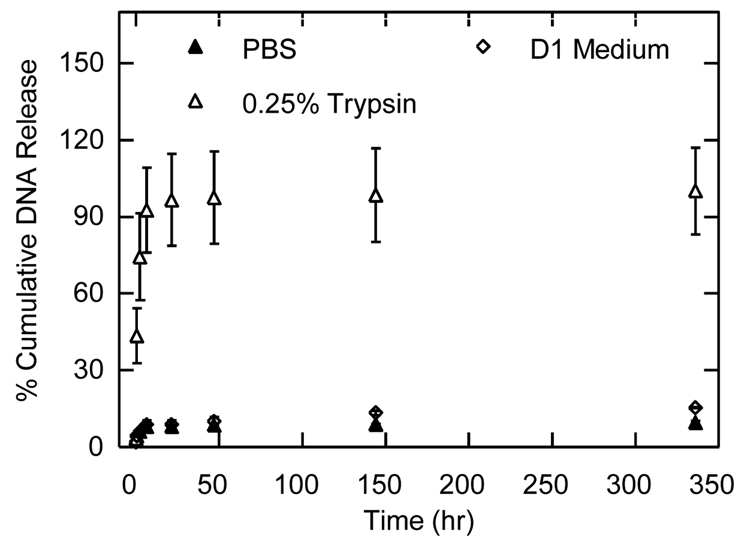
The release kinetics of the encapsulated polyplexes were tested in PBS, 0.25% trypsin, and D1 conditioned medium (Figure 3). Cumulative release in PBS and D1 conditioned media measured for 14-days was less than 15% of the total encapsulated DNA polyplexes, indicating that the polyplexes were not free to diffuse and remained entrapped within the hydrogel. In contrast, release in a trypsin containing solution resulted in a burst release of DNA polyplexes with 74.4% release after 2.5-hours of incubation. The gel degraded completely after 6-hours of incubation in trypsin.
Figure 3.
Cumulative release kinetics of DNA/PEI polyplexes encapsulated inside MMP-degradable PEG hydrogels. Hydrogels (100 µL) containing 15 µg of DNA complexed with PEI at an N/P of 7.5 were placed in PBS, trypsin or D1 conditioned medium. At the indicated time points the releasing medium was analyzed for DNA content. Data is shown as a percent of the total DNA found after complete gel degradation.
Activity of the encapsulated polyplexes
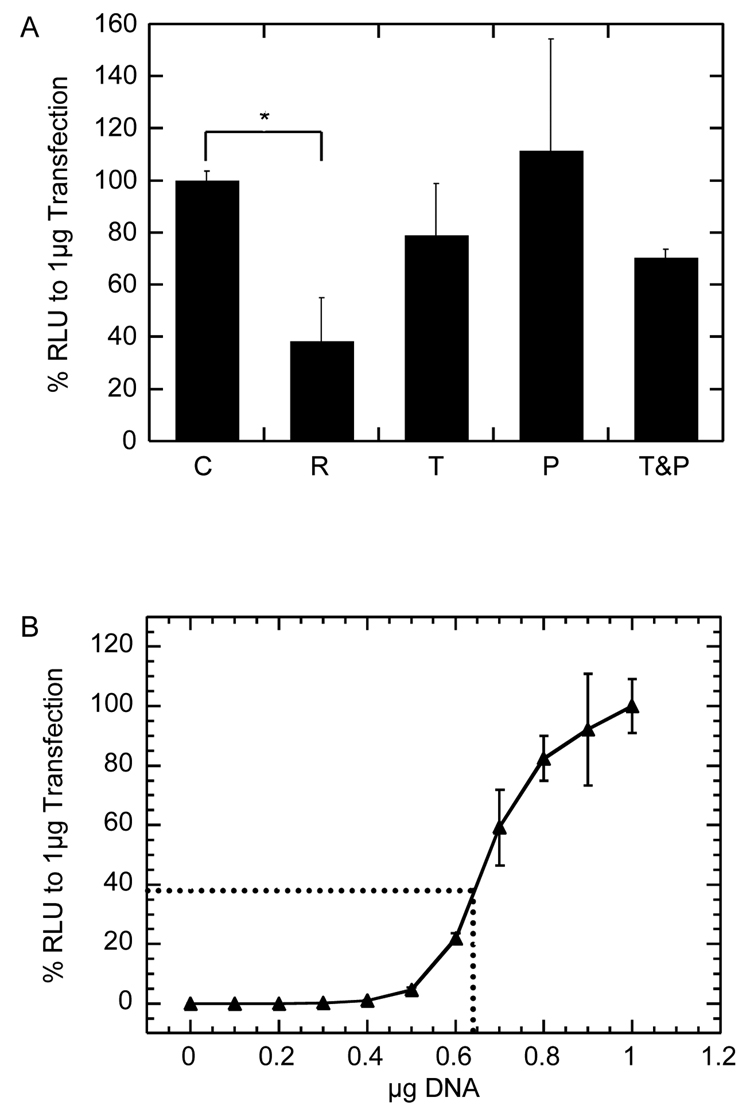
The activity of the encapsulated polyplexes is an important characterization step and was measured by degrading the gel and releasing the encapsulated polyplexes and then using those complexes for bolus transfection of plated D1 cells. Transfections were done using 1 µg of DNA from the released polyplexes in 24-well plates. Although collagenases can be used to degrade the PEG hydrogel, we found that the presence of collagenase I or collagenase IV in cell lysate resulted in severe reduction of luciferase activity (data not shown). In contrast, the use of trypsin to degrade the gel resulted in no statistically significant decrease of luciferase activity (p > 0.05, Figure 4A). Gene transfer mediated by trypsin-released polyplexes resulted in 38 ± 3.5% of the expected RLU/mg of protein (compared to transfection mediated by fresh polyplexes, Figure 4A). To determine if free PEG or the combination of PEG and trypsin contributed to the reduced activity of the released polyplex, transfections were performed using fresh polyplexes in the presence of PEG-VS or PEG-VS with trypsin at the same concentration found in the degraded hydrogels. Transfection efficiency was not decreased in the presence of PEG-VS or PEG-VS with trypsin, indicating that polyplex encapsulation was responsible for the loss of activity. Since the amount of luciferase activity is not linear with respect to DNA concentration, a dose-response curve for DNA concentration versus luciferase expression (RLU) was measured and normalized to the luciferase expression for 1 µg DNA (Figure 4B). This curve shows a sigmoidal dependence of DNA concentration to luciferase expression (RLU) and indicates that a 38% of luciferase activity corresponds to a transfection being performed with 65% of the DNA (0.65 µg) (Figure 4B dotted line). Since we observe 38% of the activity achieved with fresh polyplexes, we conclude that at least 65% of the encapsulated DNA/PEI polyplexes remain active.
Figure 4.
Activity of pEGFP-LUC/PEI polyplexes encapsulated inside MMP-degradable hydrogels (A). The activity of released polyplexes (R) was normalized to that of fresh polyplexes (C), and compared to fresh polyplexes with trypsin added (T), fresh polyplexes with PEG added (P), and fresh polyplexes with both trypsin and PEG added (T&P). Transfection using freshly prepared complexes supplemented with free PEG-VS, 0.25% trypsin/EDTA, or a combination of both at the same concentration found in the degraded hydrogels. Dose response curve of DNA/PEI polyplexes transfection efficiency normalized to the RLU found with 1 µg DNA (B). The dotted line represents polyplexes that had 37% of the RLU activity and corresponds to 65% of the DNA being present. The * symbol indicates statistical significance at a level of p<0.05 calculated using multiple comparisons.
D1 cell migration and viability inside DNA/PEI loaded PEG gels
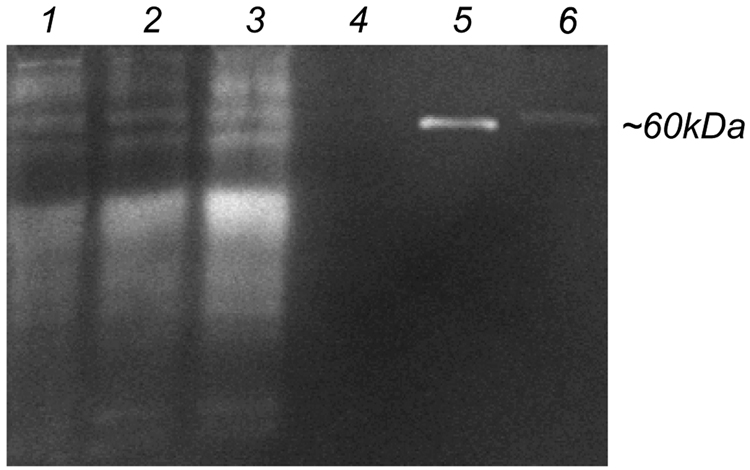
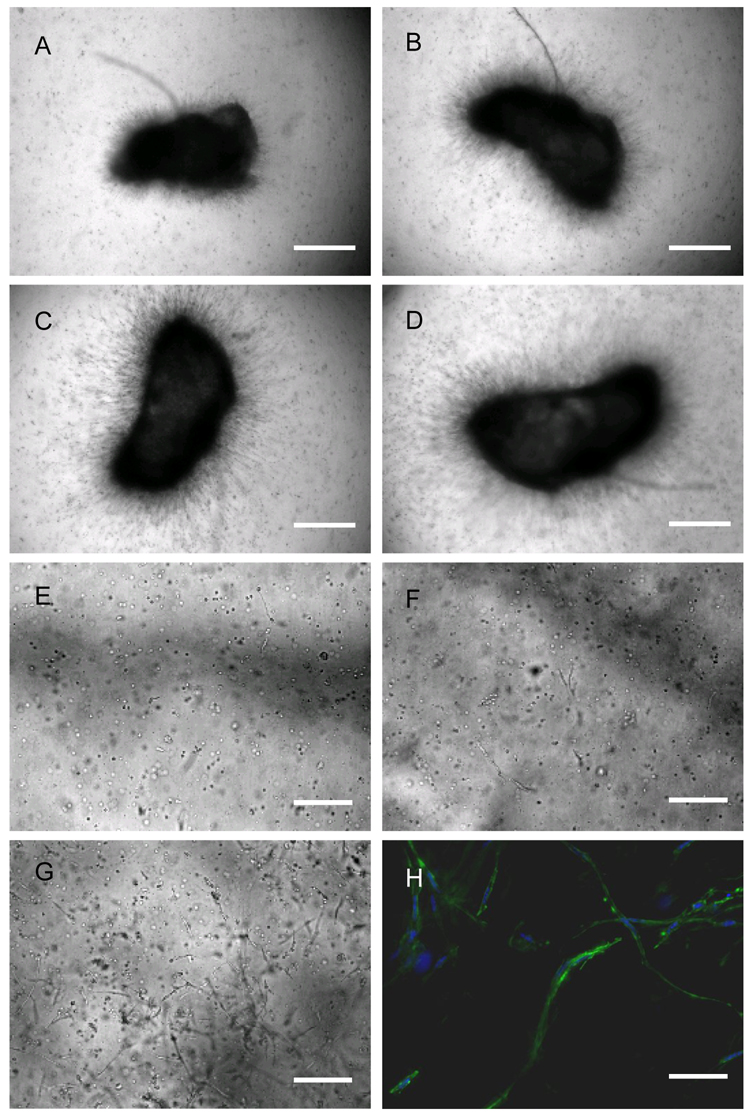
High levels of protease expression are required for cells to survive and migrate in MMP sensitive PEG hydrogels [36]. D1 cells at passages 3, 6 and 10 were found to express high amounts of proteases (Figure 5) and to be able to migrate inside MMP-degradable hydrogels when seeded as clusters or encapsulated homogenously (Figure 6). The migration rate of D1 cells out of fibrin clusters was different depending on the day of incubation, with rates at 0 to 24-hours equaling 7.3±1.8 µm/hour, at 24 to 69-hours equaling 5.0±1.8 µm/hour, at 69 to 122-hours equaling 5.7±2.4 µm/hour and at 122 to 155-hours equaling 11.0±3.2 µm/hour. Actin staining of cells at day 13 revealed spread spindle like cells (Figure 6E) as have been previously observed for cells grown inside enzymatically degradable PEG hydrogels [30, 32, 35].
Figure 5.
Zymogram gel electrophoresis of D1 cell conditioned medium at passage 3 (lane 1), 6 (lane 2) and 10 (lane 3). BSA (lane 4), 10 ng MMP-2 (lane 5) and DMEM with 10% serum (lane 6) were run for comparison. All conditioned medium samples as well as BSA and DMEM were run at a total protein concentration of 27 µg as determined by Bradford assay.
Figure 6.
Migration of D1 cells in MMP-degradable PEG hydrogels. Cells were placed in the gel either as a cluster for 24-hours (A), 69-hours (B), 122-hours (C) and 155-hours (D) or homogeneously for 48-hours (E), 96-hours (F), and 240-hours (G). A representative picture of cells migrating out of a fibrin cluster at 312-hour is shown (H, green stain is actin, blue stain is the nuclei). Objectives used are 10x (A–G) and 20x (H). Scale bar equals 600 µm (A–D), 200 µm (E–G) and 100 µm (H).
Cell viability studies were performed to determine the toxicity of encapsulated DNA/PEI polyplexes to infiltrating cells. Viable cells were visible (LIVE/DEAD assay) for up to 15-days with few cells infiltrating into the scaffold showing red fluorescence, which is indicative of dead cells (Figure 7). There were no significant differences in viability between hydrogels without DNA (Figure 7 A–C) and those loaded with DNA/PEI (Figure 7 D–F). In both types of hydrogels, most dead cells were found in the fibrin cluster, indicating that cell death might be a result of the fibrin clot cell encapsulation procedure or the lack of nutrient flow inside the fibrin clot. Additionally, LIVE/DEAD staining of hydrogels that contained polyplexes but no cells revealed that the polyplexes did stain red, indicating that the ethidium bromide monoazide (the DEAD component in the assay) is able to stain encapsulated polyplexes (Figure 7 G–I). Thus, the red fluorescence observed in the hydrogels which contained cells and polyplexes could be from dead cells as well as the encapsulated polyplexes. For all images the exposure time was 200-ms.
Figure 7.
Cellular viability of cells seeded as clusters inside MMP-degradable PEG hydrogels not containing polyplexes (A–C) or containing DNA/PEI polyplexes (D–F). Cells were stained with the LIVE/DEAD assay, which stains live cells green and dead cells red. Pictures were taken using an inverted microscope (10x) at 4-days (A, D), 8-days (B, E) and 15-days (C, F) and are shown as a single merged image of the red and green pictures. As a control, an acellular hydrogel containing polyplexes was stained (G-green, H-red and I-merged red & green). Scale bar equals 200 µm. All images were acquired using 200-ms exposure times.
Reporter gene selection for assessing gene transfer inside hydrogels
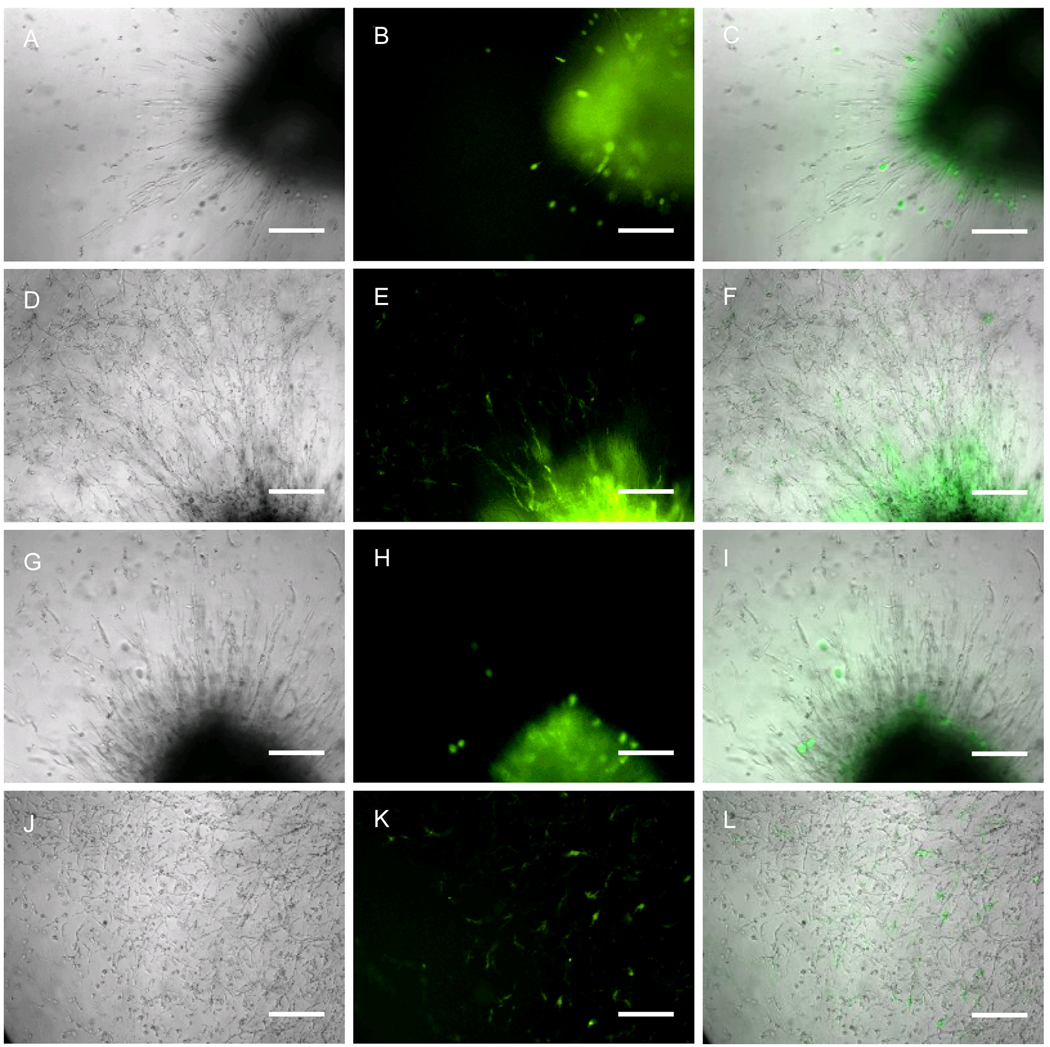
Although gene transfer to cells inside hydrogel scaffolds is typically characterized using GFP reporter plasmids, we found that for D1 cells and enzymatically degradable PEG hydrogels the use of green fluorescence to assess transgene expression repeatedly resulted in false positives. Hydrogels were formed using either GFP or β-galactosidase encoding plasmids to form the DNA/PEI polyplexes to be encapsulated inside the hydrogel scaffolds. Cells were seeded as clusters and green fluorescence was monitored as the cells infiltrated the scaffold using a fluorescence microscope (Figure 8). Although not all pβGAL/PEI polyplex containing hydrogels showed green fluoresce in infiltrating cells, some clearly showed false positives (Figure 8H, K). In all experiments at least two of the pβGAL/PEI polyplex-containing gels contained infiltrating cells that fluoresced green at a 2000-ms exposure time. Thus, although green fluorescent cells were observed in all samples of pEGFP-LUC/PEI polyplex containing hydrogels when using 2000-ms exposure time (Figure 8B,E), this fluorescence could not be unequivocally attributed to transfection, but rather to an artifact of growing the cells inside the hydrogel material. Further, all cell clusters showed intense green fluorescence at day 0 even for gels that contained no DNA, indicating that clustered cells autofluoresce (data not shown). To allow comparisons to be made, all pictures were taken using the same exposure times and magnifications.
Figure 8.
Gene transfer to infiltrating D1 cells into DNA/PEI polyplex loaded hydrogels. D1 cells were seeded inside the MMP-degradable hydrogels as a cluster (fibrin clot) in gels that contained pEGFP-LUC/PEI polyplexes (A–F) or pβGAL (G–L). Pictures were taken with a 10x objective at 5-days (A–C and G–I) and 13-days (D–F and J–L). Pictures shown are the phase picture, the green fluorescence picture and the merge of green and phase. Scale bar equals 200 µm. All images were acquired using 2000-ms exposure times.
Because of the limitations of green fluorescent protein, we investigated the use of the pSEAP reporter plasmid, which expresses secreted alkaline phosphatase. Alkaline phosphatase activity can be quantified using commercially available kits. To ensure secreted alkaline phosphatase protein can freely diffuse out of the PEG hydrogels, D1 cells were transfected with pSEAP for 24-hours prior to either seeding in the hydrogel scaffold homogeneously or plating in the 24-well tissue culture plate. Medium from both the hydrogel seeded cells and the tissue culture plastic plated cells were collected after 24-hours and analyzed for SEAP activity. There was no significant difference found for the total SEAP secreted to the medium from cells seeded in the gel and plated in the 24-well plate (Figure 9). This result indicated that the SEAP protein was able to diffuse out of the PEG gel freely.
Figure 9.
Diffusion of SEAP protein out of MMP-degradable PEG hydrogels. 150,000 D1 cells transfected with pSEAP/PEI were either seeded inside the MMP degradable PEG hydrogel homogenously (Hydrogel) or plated in 24-well tissue culture plate (TCP) for another 24-hours. Secreted alkaline phosphatase protein released into the total medium was quantified.
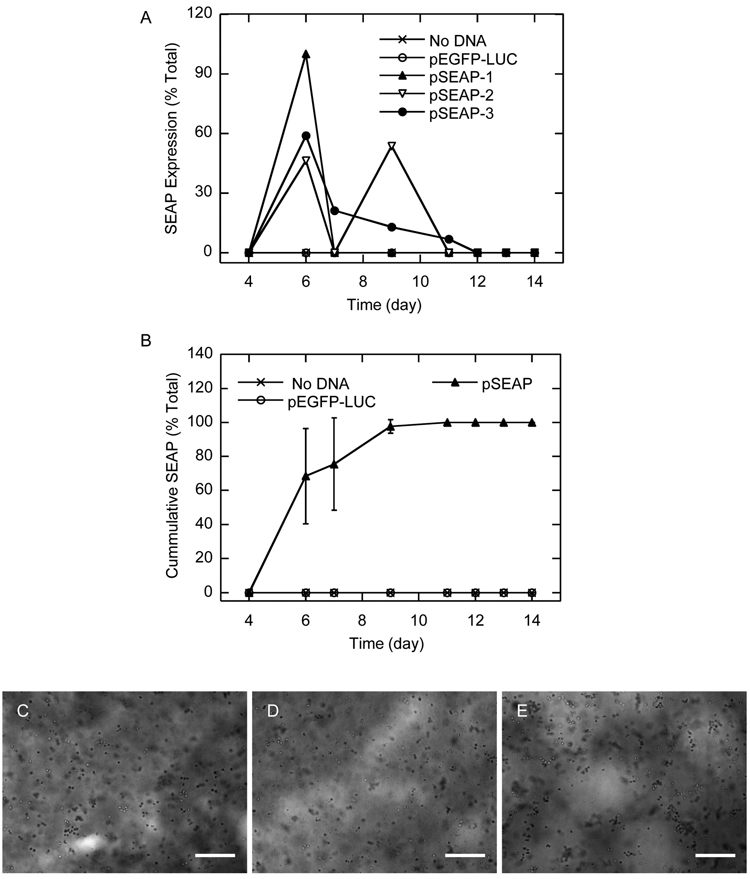
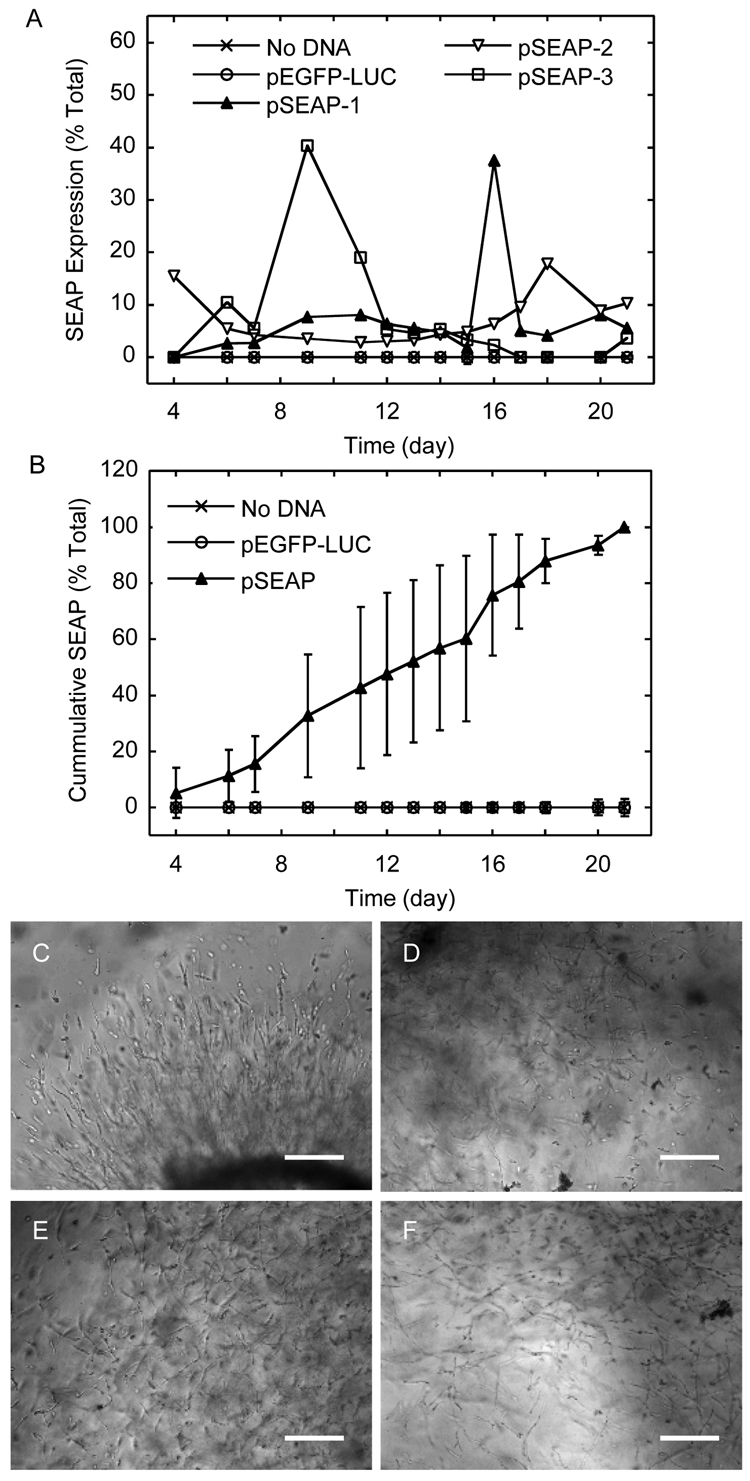
Gene transfer to infiltrating cells
The ability of cells grown inside MMP-degradable hydrogels to internalize and express encapsulated DNA/PEI polyplexes was studied over time. For cells seeded using the homogeneous approach, kinetic SEAP expression showed a peaked at day 6 (Figure 10B) and cumulative SEAP expression leveled off between days 7 and 10 (Figure 10B). In contrast, gene transfer from infiltrating cells showed kinetic SEAP expression with multiple peaks (Figure 11A) and cumulative SEAP expression that continued to rise throughout the 21-day incubation (Figure 11B). As controls, gels with no DNA or DNA encoding for pEGFP-LUC were used. In all the control hydrogels, the SEAP assay reading was below 1000 RLU, which is the reading considered zero SEAP expression based on the reading obtained for untransfected cells (data not shown).
Figure 10.
Kinetic (A) and cumulative (B) SEAP transgene expression from D1 cells plated homogeneously in MMP-degradable PEG hydrogels containing pSEAP/PEI polyplexes. The medium was collected at predetermined time points and assayed for SEAP gene expression. Hydrogels containing no polyplexes or polyplexes with pEGFP-LUC were used as controls and did not show SEAP expression at any time point. Data in A is presented as the average of three gels for the no DNA and pEGFP-LUC controls and three individual gels for pSEAP in order to more clearly see the different kinetics SEAP expression. Phase images were taken with a 10x objective at 2-days (C), 4-days (D), and 10-days (E). Scale bar equals 200 µm.
Figure 11.
Kinetic (A) and cumulative (B) SEAP transgene expression from D1 cells plated as clusters in MMP-degradable PEG hydrogels containing pSEAP/PEI polyplexes. The medium was collected at predetermined time points and assayed for SEAP gene expression. Hydrogels containing no polyplexes or polyplexes with DNA pEGFP-LUC were used as controls and did not show SEAP expression at any time point. Data in A is presented as the average of three gels for the no DNA and pEGFP-LUC controls and three individual gels for pSEAP in order to more clearly see the different kinetics SEAP expression. Phase images were taken with a 10x objective at 4-days (C), 9-days (D), and 13-days (E) and 17-days. Scale bar equals 200 µm.
Discussion
The ability to genetically modify MSCs seeded inside synthetic hydrogel scaffolds would offer an alternative approach to protein delivery to guide their differentiation into functional tissues ex vivo. Additionally, MSC-like progenitor cells are believed to reside in most adult tissues and to be responsible for adult tissue regeneration. Therefore, the design of hydrogel materials that allow for cellular infiltration and deliver genes to infiltrating cells would be ideal to guide regeneration in vivo. In this report, we explored gene transfer to MSCs infiltrating MMP-degradable hydrogels that were loaded with DNA/PEI polyplexes. Cloned mouse bone marrow derived MSCs (D1) were used in this study as a model MSC cell type. D1 cells have similar characteristics to human MSCs and can be passaged up to 25 times under regular tissue culture conditions [37–41]. Cellular migration inside MMP degradable hydrogels has been demonstrated to occur primarily by proteolytic degradation of the scaffold [30]. Further, MMP activity inside MMP-degradable hydrogels has been found to be highest at the cell surfaces with MMP activity dramatically decreasing away from the cell surfaces [29] (Scheme 1). Thus, inside MMP-degradable hydrogels cells can only degrade the gel immediately surrounding them and do not contribute to hydrogel degradation far away from their surface [29]. Gene transfer inside MMP-degradable PEG gels is, therefore, expected to occur as the cells proteolytically migrate through the hydrogel. Polyplexes will be internalized as the cells encounter them during the migration (Scheme 1).
D1 cells were found to express high levels of MMP enzymes (Figure 5) and to be able to grow inside the hydrogel scaffold when seeded as single cells or as cell clusters (Figure 6). Live/Dead cell staining has been widely used to assess the viability of cells seeded inside hydrogel scaffolds [33, 42–46] and was used in this study to assess viability. Most dead cells were observed inside the fibrin clot used to cluster the cells and force cellular infiltration to the scaffold. Thus, we conclude that the DNA/PEI polyplexes at the tested concentrations are not toxic to cells seeded inside the hydrogel scaffold. Gene transfer was studied using a secreted alkaline phosphatase (SEAP) encoding plasmid. The use of a secreted plasmid rather than the more common luciferase and GFP plasmids allowed us to quantify the reporter gene expression over time of the same hydrogel by simply analyzing the cell culture media, which is ideal when characterizing gene transfer over long time periods. SEAP protein (~64 kDa) was found to efficiently diffuse out of the PEG gel (Figure 9). The SEAP expression pattern was found to be a function of the cell seeding method, clustered or homogeneous. D1 cells seeded as a cluster of cells showed multiple surges in SEAP expression throughout the 21-day incubation. Because gene transfer in these gels depended on the ability of the cells to infiltrate the scaffold, they did not result in the exact same transgene expression with time. After the 21-day incubation, all the samples resulted in about 180 pg of SEAP protein produced and showed an increasing cumulative transgene expression profile during the 21-day incubation (Figure 11B). However, the observed surges of SEAP expression (positive slopes in Figure 11A) throughout the 21-day incubation occurred at different days for all the samples. We propose that the multiple surges in SEAP transgene expression occurred as a results of the cells encountering new polyplexes as they migrated into the hydrogel scaffold. Previous reports of fibroblast cell migration inside MMP-degradable hydrogels showed that cells seeded as clusters migrated much further than single cells [29]. The use of hydrogel scaffolds that allow cellular infiltration to deliver DNA may result in long lasting signals in vivo, which are needed for the regeneration of functional tissues.
Cells seeded homogeneously generally showed only one surge in SEAP transgene expression (Figure 10A), with cumulative transgene expression leveling off after 7-days and reaching less than 60 pg in total protein produced (Figure 10B), indicating that the cells were transfected only once during the 21-day incubation. As mentioned above, homogenously suspended cells have limited migration inside the gel compared to clustered cells, suggesting that they are able to internalize only nearby polyplexes and that they do not encounter new polyplexes during the 21-day incubation.
Although reporter plasmids encoding for luciferase and green fluorescent protein are widely utilized for quantifying non-viral gene transfer and GFP has been previously used to quantify gene transfer in hydrogel scaffolds [28, 47], we found that these reporter plasmids are not ideal for characterizing gene transfer over time inside MMP-degradable hydrogel scaffolds. Luciferase plasmids are inconvenient to determine gene transfer in hydrogel scaffolds as a function of time since they require the sacrifice of the hydrogel sample for analysis. Further, the lysis of cells seeded inside hydrogels is not trivial, requiring the use of enzymes to degrade the hydrogel which can affect the stability of the released luciferase protein after lysis (data not shown). GFP reporter plasmid allows for the monitoring of the same hydrogel with time, however, we have found that cells fluoresce green when they are seeded as a cluster (Figure 8B, E, H), when the cells are rounded (Figure 8H), and, in some cases when the cells are highly spread (Figure 8K). Thus, although we observed green fluorescent cells in gels that contain pEGFP-LUC/PEI polyplexes (Figure 8B, E), we cannot be sure that this fluorescence is due to transgene expression. We hypothesize that this false positives are due intracellular autofluorescence, which has been previously described by others for cells seeded inside PEG hydrogels [29, 48]. Researches have shown that all cultured cells have some level of intracellular autofluoresence, which peaks in the blue and green wavelengths due to the existence of NADH, riboflavins and flavin coenzyme [49, 50]. Intracellular autoflurescence increases as the culture time and numbers of cells expands [49, 50]. For MSCs, the ratio of green over blue intracellular autofluoresence rises as the cell density increases and the stem cells differentiate [51]. Because of the limitations of green fluorescent protein and luciferase reporter plasmids we looked for a reporter plasmid that resulted in a secreted protein. Although soluble growth factors would be well suited for this application, their quantification typically requires the use of expensive assays (i.e. ELISA). We found that reporter plasmids encoding for secreted alkaline phosphatase to result in an effective approach to assay gene expression from hydrogel scaffolds over time.
The physical characteristics of DNA/PEI loaded PEG hydrogels were characterized for polyplex quality, polyplex release, and hydrogel mechanical properties before and after polyplex encapsulation. Confocal microscopy images of fluorescently labeled DNA showed that the polyplexes did not aggregate inside the PEG hydrogel significantly (Figure 1). In vitro transfections showed that 65% of the polyplexes remained active post gel encapsulation (Figure 4), indicating that a certain percentage of the encapsulated complexes were deactivated through the covalent modification of the polyplexes with the gel precursors or polyplex mechanical deformation during gel formation. The vinyl sulfone functional group has been reported to react with amine functional groups in PEI [52] and the PEGylation of PEI has been observed to reduce the transfection efficiency of polyplexes in vitro. Thus, it is possible that DNA/PEI polyplexes were becoming PEGylated during hydrogel formation and this PEGylation contributed to the reduced activity of the encapsulated polyplexes. Further since the mesh size in this type of a hydrogel is in the order of 25 nm [30] and the complex size is 150 nm (data not shown), the polyplexes will be larger than the average mesh size of the hydrogel, which might result in the mechanical deformation of the polyplexes during gel formation. Polyplex released from the hydrogel scaffold in PBS and D1 cell conditioned media was close to zero after an initial bust release of 15% (Figure 3), which suggested that the initial release was due to surface associated polyplexes. Release of the polyplexes was not expected since, as mentioned above, the hydrogel mesh size is significantly smaller than the average diameter of the polyplexes. The mechanical properties of the hydrogel were unchanged by the addition of up to 0.30 µg DNA/µL gel. However, the addition of 0.5 µg DNA/µL gel resulted in a statistical increase in the storage modulus of the hydrogel (Figure 2A). The change in mechanical strength could be explained by the increase of the overall mass of the hydrogel per volume or by covalent modification, where the polyplex acts as a crosslinker. Thus, although we found conditions in which encapsulated DNA/PEI polyplexes inside MMP-degradable hydrogels were active and did not change the mechanical properties of the hydrogel, different strategies must still be devised to increase the DNA loading capacity inside hydrogel scaffolds.
Conclusion
DNA/PEI polyplexes were encapsulated uniformly into MMP-degradable PEG hydrogel without changing the hydrogel mechanical properties significantly. Polyplexes have limited diffusion inside the PEG hydrogel and 65% of them remained active post encapsulation. The encapsulated polyplexes were found to be non-toxic to the cells suspended homogenously inside the gel or infiltrating the gel. Gene transfer to both homogenously seeded cells and clustered cells were observed, but the transgene expression patterns were different. Infiltrating cells (originally seeded as clusters) showed gene expression that rise several times through the incubation indicating that the cells were able to be transfected several times as they migrated through the scaffold. Clustered cells resulted in cumulative transgene expression that increased through the 21-day incubation, while homogeneously seeded cells resulted in cumulative transgene expression that plateau after 7-days of culture.
Acknowledgements
The authors would like to thank Professor Pirouz Kavehpour (UCLA) and Kevin Lu (UCLA) for their assistance with the test of gel mechanical properties; Quinn Ng (UCLA) and Talar Tokatlian (UCLA) for the helpful discussions and the Institute for Cell Mimetic Space Exploration (CMISE) at UCLA for the fluorescent microscope. We like to thank the National Institute of Health for funding the research (R21EB007730(TS)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Suh H. Tissue restoration, tissue engineering and regenerative medicine. Yonsei Medical Journal. 2000;41:681–684. doi: 10.3349/ymj.2000.41.6.681. [DOI] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Ando S, Putnam D, Pack DW, Langer R. PLGA microspheres containing plasmid DNA: preservation of supercoiled DNA via cryopreparation and carbohydrate stabilization. J Pharm Sci. 1999;88:126–130. doi: 10.1021/js9801687. [DOI] [PubMed] [Google Scholar]
- 5.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 6.Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. Stability of peptide-condensed plasmid DNA formulations. J Pharm Sci. 1998;87:678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 7.Berry M, Gonzalez AM, Clarke W, Greenlees L, Barrett L, Tsang W, et al. Sustained effects of gene-activated matrices after CNS injury. Mol Cell Neurosci. 2001;17:706–716. doi: 10.1006/mcne.2001.0975. [DOI] [PubMed] [Google Scholar]
- 8.Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid-collagen hydrogels via a substrate-mediated approach. Biomaterials. 2005;26:1575–1584. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trentin D, Hall H, Wechsler S, Hubbell JA. Peptide-matrix-mediated gene transfer of an oxygen-insensitive hypoxia-inducible factor-1 alpha variant for local induction of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2506–2511. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature Reviews Genetics. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 11.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 14.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Ma N, Gao SJ, Yu H, Leong KW. Transgene expression in the brain stem effected by intramuscular injection of polyethylenimine/DNA complexes. Molecular Therapy. 2001;3:658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 16.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Therapy. 2003;10:1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 17.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. Intranasal gene delivery with a polyethylenimine-PEG conjugate. Journal of Controlled Release. 2002;81:379–388. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 18.Rudolph C, Schillinger U, Plank C, Gessner A, Nicklaus P, Muller RH, et al. Nonviral gene delivery to the lung with copolymer-protected and transferrin-modified polyethylenimine. Biochimica Et Biophysica Acta-General Subjects. 2002;1573:75–83. doi: 10.1016/s0304-4165(02)00334-3. [DOI] [PubMed] [Google Scholar]
- 19.Gautam A, Densmore CL, Golunski E, Xu B, Waldrep JC. Transgene expression in mouse airway epithelium by aerosol gene therapy with PEI-DNA complexes. Molecular Therapy. 2001;3:551–556. doi: 10.1006/mthe.2001.0300. [DOI] [PubMed] [Google Scholar]
- 20.Segura T, Schmokel H, Hubbell JA. RNA interference targeting hypoxia inducible factor 1alpha reduces post-operative adhesions in rats. J Surg Res. 2007;141:162–170. doi: 10.1016/j.jss.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 21.Iwai M, Harada Y, Tanaka S, Muramatsu A, Mori T, Kashima K, et al. Polyethylenimine-mediated suicide gene transfer induces a therapeutic effect for hepatocellular carcinoma in vivo by using an Epstein-Barr virus-based plasmid vector. Biochemical and Biophysical Research Communications. 2002;291:48–54. doi: 10.1006/bbrc.2002.6383. [DOI] [PubMed] [Google Scholar]
- 22.Aoki K, Furuhata S, Hatanaka K, Maeda M, Remy JS, Behr JP, et al. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Therapy. 2001;8:508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 23.Coll JL, Chollet P, Brambilla E, Desplanques D, Behr JP, Favrot M. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10:1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 24.Chun KW, Lee JB, Kim SH, Park TG. Controlled release of plasmid DNA from photo-cross-linked pluronic hydrogels. Biomaterials. 2005;26:3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Quick DJ, Anseth KS. DNA delivery from photocrosslinked PEG hydrogels: encapsulation efficiency, release profiles, and DNA quality. Journal of Controlled Release. 2004;96:341–351. doi: 10.1016/j.jconrel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Megeed Z, Haider M, Li DQ, O'Malley BW, Cappello J, Ghandehari H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. Journal of Controlled Release. 2004;94:433–445. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. Journal of Controlled Release. 2005;102:263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. Journal of Controlled Release. 2007;120:233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Moon JJ, Miller JS, West JL. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials. 2007;28:3163–3170. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: A novel model system for proteolytically mediated cell migration. Biophysical Journal. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutolf MP, Weber FE, Schmoekel HG, Schense JC, Kohler T, Muller R, et al. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 33.Adeloew C, Segura T, Hubbell JA, Frey P. The effect of enzymatically degradable poly(ethylene glycol) hydrogels on smooth muscle cell phenotype. Biomaterials. 2008;29:314–326. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4:713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 35.Ehrbar M, Rizzi SC, Schoenmakers RG, San Miguel B, Hubbell JA, Weber FE, et al. Biomolecular hydrogels formed and degraded via site-specific enzymatic reactions. Biomacromolecules. 2007;8:3000–3007. doi: 10.1021/bm070228f. [DOI] [PubMed] [Google Scholar]
- 36.Rizzi SC, Ehrbar M, Halstenberg S, Raeber GP, Schmoekel HG, Hagenmuller H, et al. Recombinant protein-co-PEG networks as cell-adhesive and proteolytically degradable hydrogel matrixes. Part II: Biofunctional characteristics. Biomacromolecules. 2006;7:3019–3029. doi: 10.1021/bm060504a. [DOI] [PubMed] [Google Scholar]
- 37.Cui Q, Wang GJ, Balian G. Steroid-induced adipogenesis in a pluripotential cell line from bone marrow. Journal of Bone and Joint Surgery-American Volume. 1997;79A:1054–1063. doi: 10.2106/00004623-199707000-00012. [DOI] [PubMed] [Google Scholar]
- 38.Cui QJ, Wang YS, Saleh KJ, Wang GJ, Balian G. Alcohol-induced adipogenesis in a cloned bone-marrow stem cell. Journal of Bone and Joint Surgery-American Volume. 2006;88A:148–154. doi: 10.2106/JBJS.F.00534. [DOI] [PubMed] [Google Scholar]
- 39.Shen FH, Visger JM, Balian G, Hurwitz SR, Diduch DR. Systemically administered mesenchymal stromal cells transduced with insulin-like growth factor-I localize to a fracture site and potentiate healing. Journal of Orthopaedic Trauma. 2002;16:651–659. doi: 10.1097/00005131-200210000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Hsiong SX, Carampin P, Kong HJ, Lee KY, Mooney DJ. Differentiation stage alters matrix control of stem cells. Journal of Biomedical Materials Research Part A. 2008;85A:145–156. doi: 10.1002/jbm.a.31521. [DOI] [PubMed] [Google Scholar]
- 41.Hsiong SX, Huebsch N, Fischbach C, Kong HJ, Mooney DJ. Integrin-adhesion ligand bond formation of preosteoblasts and stem cells in three-dimensional RGD presenting matrices. Biomacromolecules. 2008;9:1843–1851. doi: 10.1021/bm8000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 43.Nuttelman CR, Tripodi MC, Anseth KS. In vitro osteogenic differentiation of human mesenchymal stem cells photoencapsulated in PEG hydrogels. Journal of Biomedical Materials Research Part A. 2004;68A:773–782. doi: 10.1002/jbm.a.20112. [DOI] [PubMed] [Google Scholar]
- 44.Wang DA, Williams CG, Li QA, Sharma B, Elisseeff JH. Synthesis and characterization of a novel degradable phosphate-containing hydrogel. Biomaterials. 2003;24:3969–3980. doi: 10.1016/s0142-9612(03)00280-1. [DOI] [PubMed] [Google Scholar]
- 45.Lee HJ, Lee JS, Chansakul T, Yu C, Elisseeff JH, Yu SM. Collagen mimetic peptide-conjugated photopolymerizable PEG hydrogel. Biomaterials. 2006;27:5268–5276. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biology. 2005;24:208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Quick DJ, Anseth KS. Gene delivery in tissue engineering: A photopolymer platform to coencapsulate cells and plasmid DNA. Pharmaceutical Research. 2003;20:1730–1737. doi: 10.1023/b:pham.0000003368.66471.6a. [DOI] [PubMed] [Google Scholar]
- 48.Lee SH, Miller JS, Moon JJ, West JL. Proteolytically degradable hydrogels with a fluorogenic substrate for studies of cellular proteolytic activity and migration. Biotechnology Progress. 2005;21:1736–1741. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 49.Aubin JE. Autofluorescence of Viable Cultured Mammalian-Cells. Journal of Histochemistry & Cytochemistry. 1979;27:36–43. doi: 10.1177/27.1.220325. [DOI] [PubMed] [Google Scholar]
- 50.Benson RC, Meyer RA, Zaruba ME, Mckhann GM. Cellular Autofluorescence - Is It Due to Flavins. Journal of Histochemistry & Cytochemistry. 1979;27:44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 51.Reyes JMG, Fermanian S, Yang F, Zhou SY, Herretes S, Murphy DB, et al. Metabolic changes in mesenchymal stem cells in osteogenic medium measured by autofluorescence spectroscopy. Stem Cells. 2006;24:1213–1217. doi: 10.1634/stemcells.2004-0324. [DOI] [PubMed] [Google Scholar]
- 52.Sagara K, Kim SW. A new synthesis of galactose-poly(ethylene glycol)-polyethylenimine for gene delivery to hepatocytes. Journal of Controlled Release. 2002;79:271–281. doi: 10.1016/s0168-3659(01)00555-7. [DOI] [PubMed] [Google Scholar]