Abstract
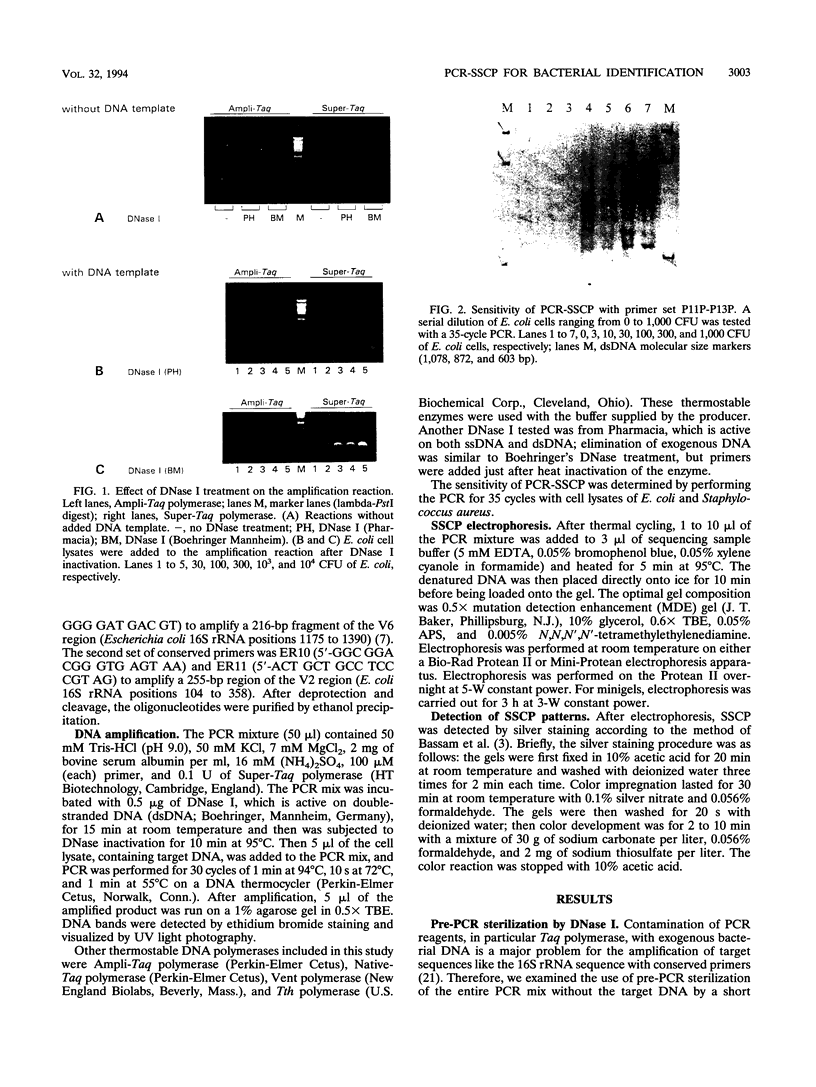
A new molecular biological approach for the identification of bacteria is described. This approach employs PCR of bacterial cell lysates with conserved primers located in the 16S rRNA sequence flanking a variable region, and analysis of the amplified product was based on the principle of single-strand conformation polymorphism (SSCP). The PCR product was denatured and separated on a nondenaturing polyacrylamide gel. SSCP patterns were detected by silver staining the nucleic acids. The mobility of the single-stranded DNA is sequence dependent and could be used to identify the unknown bacteria. Feasibility of the technique was demonstrated for a broad panel of gram-negative and gram-positive bacteria. We tested over 100 strains of bacteria representing 15 genera and 40 species. With the use of only two primer sets, P11P-P13P and ER10-ER11, we were capable to discriminate the tested species at the genus and species levels. Species-specific patterns were obtained for, e.g., Clostridium spp., Listeria spp., Pseudomonas spp., and Enterobacter spp. PCR-SSCP is a sensitive technique; e.g., the sensitivity obtained for Escherichia coli cells was 30 CFU. This technique is a simple and rapid method for the detection and identification of a wide spectrum of bacteria by whole-cell-based PCR amplification with the use of conserved primers and identification by nondenaturing gel electrophoresis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainsworth P. J., Surh L. C., Coulter-Mackie M. B. Diagnostic single strand conformational polymorphism, (SSCP): a simplified non-radioisotopic method as applied to a Tay-Sachs B1 variant. Nucleic Acids Res. 1991 Jan 25;19(2):405–406. doi: 10.1093/nar/19.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry T., Powell R., Gannon F. A general method to generate DNA probes for microorganisms. Biotechnology (N Y) 1990 Mar;8(3):233–236. doi: 10.1038/nbt0390-233. [DOI] [PubMed] [Google Scholar]
- Bassam B. J., Caetano-Anollés G., Gresshoff P. M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991 Jul;196(1):80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- Bej A. K., Mahbubani M. H., Miller R., DiCesare J. L., Haff L., Atlas R. M. Multiplex PCR amplification and immobilized capture probes for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990 Oct;4(5):353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- Bickler S. W., Heinrich M. C., Bagby G. C. Magnesium-dependent thermostability of DNase I. Biotechniques. 1992 Jul;13(1):64–66. [PubMed] [Google Scholar]
- Chen K., Neimark H., Rumore P., Steinman C. R. Broad range DNA probes for detecting and amplifying eubacterial nucleic acids. FEMS Microbiol Lett. 1989 Jan 1;48(1):19–24. doi: 10.1016/0378-1097(89)90139-0. [DOI] [PubMed] [Google Scholar]
- Collins M. D., Wallbanks S., Lane D. J., Shah J., Nietupski R., Smida J., Dorsch M., Stackebrandt E. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991 Apr;41(2):240–246. doi: 10.1099/00207713-41-2-240. [DOI] [PubMed] [Google Scholar]
- Fluit A. C., Torensma R., Visser M. J., Aarsman C. J., Poppelier M. J., Keller B. H., Klapwijk P., Verhoef J. Detection of Listeria monocytogenes in cheese with the magnetic immuno-polymerase chain reaction assay. Appl Environ Microbiol. 1993 May;59(5):1289–1293. doi: 10.1128/aem.59.5.1289-1293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisen K., Loeffelholz M., Purohit A., Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994 Feb;32(2):335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. PCR-SSCP: a method for detection of mutations. Genet Anal Tech Appl. 1992 Jun;9(3):73–79. doi: 10.1016/1050-3862(92)90001-l. [DOI] [PubMed] [Google Scholar]
- Jensen M. A., Webster J. A., Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993 Apr;59(4):945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino R., Yazyu H., Kishimoto Y., Sekiya T., Hayashi K. F-SSCP: fluorescence-based polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) analysis. PCR Methods Appl. 1992 Aug;2(1):10–13. doi: 10.1101/gr.2.1.10. [DOI] [PubMed] [Google Scholar]
- Meier A., Persing D. H., Finken M., Böttger E. C. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J Clin Microbiol. 1993 Mar;31(3):646–652. doi: 10.1128/jcm.31.3.646-652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohabeer A. J., Hiti A. L., Martin W. J. Non-radioactive single strand conformation polymorphism (SSCP) using the Pharmacia 'PhastSystem'. Nucleic Acids Res. 1991 Jun 11;19(11):3154–3154. doi: 10.1093/nar/19.11.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., de Waal E. C., Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993 Mar;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Rand K. H., Houck H. Taq polymerase contains bacterial DNA of unknown origin. Mol Cell Probes. 1990 Dec;4(6):445–450. doi: 10.1016/0890-8508(90)90003-i. [DOI] [PubMed] [Google Scholar]
- Rys P. N., Persing D. H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J Clin Microbiol. 1993 Sep;31(9):2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L., Lévesque R. C., Lalonde M. Identification of endomycorrhizal fungi colonizing roots by fluorescent single-strand conformation polymorphism-polymerase chain reaction. Appl Environ Microbiol. 1993 Dec;59(12):4211–4215. doi: 10.1128/aem.59.12.4211-4215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torensma R., Visser M. J., Aarsman C. J., Poppelier M. J., Fluit A. C., Verhoef J. Monoclonal antibodies that react with live Listeria spp. Appl Environ Microbiol. 1993 Aug;59(8):2713–2716. doi: 10.1128/aem.59.8.2713-2716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. H., Blitchington R. B., Greene R. C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990 Sep;28(9):1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfhagen M. J., Fluit A. C., Torensma R., Jansze M., Kuypers A. F., Verhage E. A., Verhoef J. Comparison of typing methods for Clostridium difficile isolates. J Clin Microbiol. 1993 Aug;31(8):2208–2211. doi: 10.1128/jcm.31.8.2208-2211.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamballerie X., Zandotti C., Vignoli C., Bollet C., de Micco P. A one-step microbial DNA extraction method using "Chelex 100" suitable for gene amplification. Res Microbiol. 1992 Oct;143(8):785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]








