Abstract
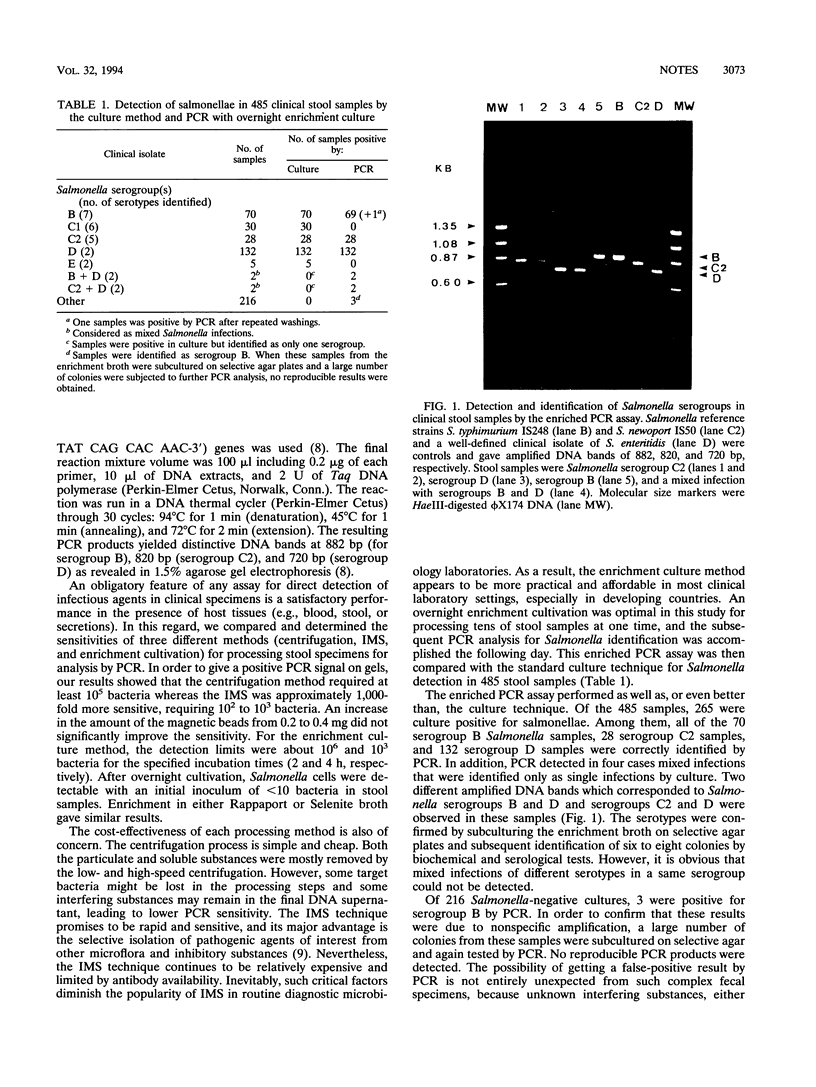
Three different stool sample-processing methods (centrifugation, immunomagnetic separation, and selective enrichment cultivation) for the identification of Salmonella serogroups by PCR were studied. The corresponding sensitivities in an ethidium bromide stained-agarose gel were 10(5), 10(3), and 10 bacteria, respectively. The PCR assay with overnight enrichment performed as well as, or even better than, the conventional culture technique. Of 485 clinical stool samples, PCR correctly identified all 230 culture-positive samples as well as mixed Salmonella infections in four cases.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gumerlock P. H., Tang Y. J., Meyers F. J., Silva J., Jr Use of the polymerase chain reaction for the specific and direct detection of Clostridium difficile in human feces. Rev Infect Dis. 1991 Nov-Dec;13(6):1053–1060. doi: 10.1093/clinids/13.6.1053. [DOI] [PubMed] [Google Scholar]
- Islam D., Lindberg A. A. Detection of Shigella dysenteriae type 1 and Shigella flexneri in feces by immunomagnetic isolation and polymerase chain reaction. J Clin Microbiol. 1992 Nov;30(11):2801–2806. doi: 10.1128/jcm.30.11.2801-2806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Romana L. K., Reeves P. R. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J Gen Microbiol. 1992 Sep;138(9):1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
- Luk J. M., Kongmuang U., Reeves P. R., Lindberg A. A. Selective amplification of abequose and paratose synthase genes (rfb) by polymerase chain reaction for identification of Salmonella major serogroups (A, B, C2, and D). J Clin Microbiol. 1993 Aug;31(8):2118–2123. doi: 10.1128/jcm.31.8.2118-2123.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk J. M., Lindberg A. A. Rapid and sensitive detection of Salmonella (O:6,7) by immunomagnetic monoclonal antibody-based assays. J Immunol Methods. 1991 Mar 1;137(1):1–8. doi: 10.1016/0022-1759(91)90387-u. [DOI] [PubMed] [Google Scholar]
- Luk J. M., Zhao C. R., Karlsson K. M., Lindberg A. A. Specificity of monoclonal antibodies binding to the polysaccharide antigens (Vi, O9) of Salmonella typhi. FEMS Microbiol Lett. 1992 Oct 1;76(1-2):173–178. doi: 10.1016/0378-1097(92)90382-x. [DOI] [PubMed] [Google Scholar]
- Persing D. H. Polymerase chain reaction: trenches to benches. J Clin Microbiol. 1991 Jul;29(7):1281–1285. doi: 10.1128/jcm.29.7.1281-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Nishibuchi M., Ramamurthy T., Bhattacharya S. K., Pal S. C., Takeda Y. Polymerase chain reaction for detection of the cholera enterotoxin operon of Vibrio cholerae. J Clin Microbiol. 1991 Nov;29(11):2517–2521. doi: 10.1128/jcm.29.11.2517-2521.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Romana L. K., Reeves P. R. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992 Mar;130(3):429–443. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde J., Eiden J., Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990 Jun;28(6):1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]