Abstract
Diatoms are ecologically important algae that acquired their plastids by secondary endosymbiosis, resulting in a more complex cell structure and an altered distribution of metabolic pathways when compared with organisms with primary plastids. Diatom plastids are surrounded by 4 membranes; the outermost membrane is continuous with the endoplasmic reticulum. Genome analyses suggest that nucleotide biosynthesis is, in contrast to higher plants, not located in the plastid, but in the cytosol. As a consequence, nucleotides have to be imported into the organelle. However, the mechanism of nucleotide entry into the complex plastid is unknown. We identified a high number of putative nucleotide transporters (NTTs) in the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum and characterized the first 2 isoforms (NTT1 and NTT2). GFP-based localization studies revealed that both investigated NTTs are targeted to the plastid membranes, and that NTT1 most likely enters the innermost plastid envelope via the stroma. Heterologously expressed NTT1 acts as a proton-dependent adenine nucleotide importer, whereas NTT2 facilitates the counter exchange of (deoxy-)nucleoside triphosphates. Therefore, these transporters functionally resemble NTTs from obligate intracellular bacteria with an impaired nucleotide metabolism rather than ATP/ADP exchanging NTTs from primary plastids. We suggest that diatoms harbor a specifically-adapted nucleotide transport system and that NTTs are the key players in nucleotide supply to the complex plastid.
Keywords: chloroplast, complex plastids, nucleotide synthesis, nucleotide transport
Diatoms, unicellular algae of high environmental relevance, dominate the phytoplankton and are responsible for up to 40% of primary productivity in the ocean (1, 2). Particularly, the 2 distantly-related model diatoms, the centric species Thalassiosira pseudonana and the pennate diatom Phaeodactylum tricornutum, became highly investigated objects of research. Increasing molecular and physiological data provided insights into the metabolic situation of diatoms and revealed interesting differences to other algae and plants (1–4). Plastids of plants, glaucophytes, and red and green algae arose from a single, ancient primary endosymbiosis between a cyanobacterium and a heterotrophic eukaryote, and they are surrounded by 2 envelope membranes (5, 6). Diatoms, however, gained their plastids by secondary endosymbiosis between a eukaryotic host and a red alga. This important evolutionary process was responsible for generating an enormous amount of algal biodiversity (5, 6). Secondary plastids possess up to 4 envelope membranes, thus protein import and solute trafficking apparently are more complex than in primary plastids (6).
Nucleotides are probably the most important molecules in living cells. They are building blocks of DNA and RNA and crucial components of metabolic processes, and ATP serves as the major energy currency. In plants all enzymatic reactions of the purine nucleotide de novo synthesis and 5 of 6 catalytic steps of the pyrimidine nucleotide generation occur in the plastid (7). Newly-synthesized purines leave the organelle and enter cytosolic metabolism via the adenine nucleotide uniporter. This carrier is a member of the functionally diverse mitochondrial carrier family (MCF) (8). It is located in the inner plastid envelope and catalyzes the export of AMP, ADP, or ATP (9). Until now it was unclear how pyrimidine nucleotides leave the plastid (7). When photosynthetic ATP production is missing, energy has to be supplied to the organelle. The required energy provision is mediated by plastidial nucleotide transporters (NTTs), which catalyze a highly-specific ATP import in counter exchange with ADP (10, 11). These plastidial ATP/ADP transporters are structurally and phylogenetically different from MCF-type carriers but related to functionally heterogeneous NTTs from obligate intracellular bacteria (11–13).
Compared with nucleotide metabolism and transport in plants our knowledge about the corresponding processes in diatoms is rudimentary. In T. pseudonana, a cytosolic localization of the pyrimidine de novo synthesis similar to that of heterotrophs was suggested (3). Furthermore, the plastidial oxidative pentose phosphate pathway, which provides ribose-5-phosphate for nucleotide synthesis during dark phases in plant plastids, is not present in diatom plastids (4, 14). Here, we identified that in a centric and a pennate diatom pyrimidine and purine nucleotides are generated in the cytosol. Therefore, diatom plastids rely on nucleotide uptake by a specialized import system in their membranes that interconnects cytosolic nucleotide formation and nucleotide consumption in the organelle. Because net nucleotide importers are not required in primary plastids and have not been described in secondary plastids, the prediction of possible candidate nucleotide importers is complicated. Because diatom plastids are surrounded by 4 membranes, whereas primary plastids possess only 2, it seems justified to assume that diatoms have extra transporters in the additional membranes. Interestingly, we identified a comparatively high number of putative NTTs in the genomes of diatoms. We performed a comprehensive characterization of the first diatom NTTs and demonstrate that these carriers might play an important role in purine and pyrimidine nucleotide supply rather than in energy provision to the plastid.
Results and Discussion
Diatom Plastids Depend on Nucleotide Import from the Cytosol.
To enlighten the cellular localization of nucleotide biosynthesis in diatoms we conducted an in silico analysis. In diatoms, nuclear-encoded proteins with plastidial destination are synthesized as precursor proteins with an N-terminal bipartite presequence consisting of a signal peptide and a transit peptide domain. Moreover, a conserved phenylalanine (or in rare cases tryptophan, tyrosine or leucine) in the sequence motif (ASAFAP) at the cleavage site of the signal peptide was shown to be essential for successful stroma targeting (15). The latter characteristics help to identify plastidial precursor proteins (15). The fact that all putative enzymes of the pyrimidine de novo synthesis lack typical plastid targeting informations argues for an extra-plastidial localization of this pathway in T. pseudonana (3) and P. tricornutum (Table S1). Moreover, also purine biosynthesis does not take place in the complex plastid (Table S1). Most reactions of the nucleotide biosynthesis are catalyzed in the cytosol of diatoms and only very few intermediary steps might occur in mitochondria (Table S1).
The extra-plastidial localization of the nucleotide biosynthesis in diatoms implies that nucleotides have to be imported into the plastids to maintain organellar metabolism, such as DNA and RNA synthesis. In search for possible candidate nucleotide importers, we identified an unexpected high number of genes encoding putative NTTs, 8 in T. pseudonana (TpNTT1–8) and 6 in P. tricornutum (PtNTT1–6) (Tables S2–S4). This finding was surprising in so far as red algae and the higher plant Arabidopsis thaliana possess only 1 and 2 (ATP/ADP exchanging) NTTs, respectively (11). However, some intracellular bacteria use up to 5 different NTTs for energy import and net uptake of various nucleotides (12, 13, 16). The occurrence of numerous NTT sequences in diatoms could indicate that NTTs also reside in the extra membranes of the complex plastid and/or that they fulfill specialized, additional functions, such as net nucleotide provision to the organelle.
We performed an amino acid alignment of the 14 candidate NTTs and included ATP/ADP transporters from primary plastids and bacterial NTTs to get an idea about the possible function of the diatom carriers. This analysis revealed that diatom NTT1 sequences resemble ATP/ADP transporters, particularly the one from the red alga Galdieria sulfuraria (TpNTT1 61%, PtNTT1 63% amino acid sequence similarity) (Table S3) (11). Accordingly, diatom NTT1 proteins most likely facilitate ATP/ADP exchange to provide energy to the plastid. PtNTT2 and TpNTT2 resemble ATP/ADP transporters (40–49% amino acid similarity) and NTTs from chlamydiae and rickettsiae with different substrate spectra and transport modes (≤45% amino acid similarity) (Table S3). We did not identify a sequence homologous to TpNTT3 in P. tricornutum. TpNTT3 possesses up to 43% amino acid similarity to nondiatom NTTs (Table S3). The other diatom NTTs exhibit lower sequence similarities (≤ 30%) to characterized NTTs. Here, we focus on the diatom carriers NTT1 and NTT2, because they are highly related to bacterial and plastidial NTTs.
Diatom NTTs Exhibit a Broader Substrate Spectrum than ATP/ADP Transporters from Primary Plastids.
Nucleotide import measurements on Escherichia coli cells synthesizing the recombinant carriers are a suitable and frequently-used method to determine the substrate spectra and transport modes of NTTs (10–13, 16–19). Uptake studies with α32P-labeled nucleotides revealed that the recombinant diatom transporters NTT1 and NTT2 mediated a time-dependent nucleotide import (Fig. S1). TpNTT1 and PtNTT1 imported adenine nucleotides, with preference for ADP, followed by AMP and ATP, whereas dATP was transported with lowest rates (Fig. 1A). Therefore, the substrate spectra of the diatom NTT1 homologues differ remarkably from those of plastidial or chlamydial ATP/ADP transporters, which accept solely ATP and ADP as substrates (10, 11, 13). Interestingly, the diatom NTT2 isoforms exhibited an even broader substrate spectrum as they were able to import all nucleoside and deoxynucleoside triphosphates. Ribonucleotides were generally imported with higher rates than the corresponding deoxy forms and CTP seems to be the favored substrate (Fig. 1B). The substrate spectra of diatom NTT1 and NTT2 transporters (Fig. 1) were confirmed by competition analyses with nonlabeled nucleotides and structurally related compounds (Table S5). Apart from the above-mentioned substrates other tested molecules did not markedly affect nucleotide uptake mediated by the diatom NTTs.
Fig. 1.
Uptake of α32P-labeled nucleotides into E. coli cells expressing diatom NTTs. Net import of [α32P] nucleotides was calculated by subtraction of the corresponding control values (import into noninduced E. coli cells). The highest net value for each imported nucleotide was set to 100%, and the remaining uptake rates were calculated as percentage of this maximal transport. Data are the mean of at least 3 independent experiments. Standard errors are given. (A) Adenine nucleotide import by TpNTT1 (dark gray bars) and PtNTT1 (light gray bars). (B) Nucleotide and deoxynucleotide import by TpNTT2 (dark gray bars) and PtNTT2 (light gray bars).
Apparent affinities and maximal velocities of the NTT1 and NTT2 proteins were determined by application of rising exterior nucleotide concentrations (Table 1). It is necessary to mention that the Vmax but not the apparent KM values are influenced by the amount of functional recombinant transporters in the E. coli membrane. Hence, this system allows the evaluation of all biochemical parameters of 1 transporter and the comparison of KM values of different transporters. TpNTT1 exhibited moderate affinities for all tested substrates ranging from 21 μM for ADP to 68 μM for dATP and highest transport velocities for ATP and ADP. PtNTT1 showed high to medium affinities (9–39 μM) and Vmax values of ≈5–10 nmol·mg protein−1·h−1 for ADP, ATP, and dATP. AMP was transported with lower affinity but higher maximal velocity when compared with the other substrates of PtNTT1 (Table 1). The KM values of the diatom NTT1 proteins for ATP and ADP are in the same range as described for plastidial NTTs (13). dATP was imported with ≈5–20 times higher affinities than described for the ATP/ADP transporter from Caedibacter caryophilus that also accepts dATP as substrate (17). TpNTT2 exhibited higher affinities for CTP, ATP, GTP, and dCTP and higher maximal velocities for CTP, dATP, and GTP when compared with the remaining nucleotides (Table 1). PtNTT2 showed high affinities for dCTP and CTP followed by GTP, dGTP, and UTP and low affinities for ATP, dATP, and TTP (Table 1). Ribonucleotides were transported by PtNTT2 with higher Vmax than the corresponding dNTPs. The determined affinities of diatom NTT2 isoforms are similar to, or even higher than, that of bacterial NTTs involved in nucleotide exploitation (12). Because of the capacity to transport not only nucleoside triphosphates but also the corresponding deoxy forms diatom NTT2 carriers are the first NTTs known to accept such a broad range of substrates.
Table 1.
KM values and Vmax values of nucleotide transport mediated by recombinant diatom NTTs
| Substrate | Transporter |
|||
|---|---|---|---|---|
| TpNTT1 | TpNTT2 | PtNTT1 | PtNTT2 | |
| ATP | 57.9 (105) | 48.9 (4) | 38.6 (9) | 197.2 (322) |
| GTP | – | 58.9 (9) | – | 78.4 (444) |
| CTP | – | 33.1 (20) | – | 49.0 (621) |
| UTP | – | 89.9 (6) | – | 85.6 (262) |
| ADP | 21.4 (109) | – | 8.8 (5) | – |
| AMP | 52.6 (51) | – | 168.8 (24) | – |
| dATP | 68.0 (38) | 665.4 (13) | 31.7 (5) | 270.6 (130) |
| dGTP | – | 74.3 (3) | – | 82.2 (79) |
| dCTP | – | 59.8 (7) | – | 31.2 (196) |
| TTP | – | 250.7 (5) | – | 428.1 (103) |
Nucleotide uptake in the presence of rising concentrations of substrate (5–1,000 μM) was allowed for time spans in the linear phase of corresponding transport at 50 μM. KM values are given in μM. Vmax values (in parentheses) are given in nmol·mg protein−1·h−1. Data are the mean of at least 3 independent experiments. Standard errors were <10%.
Transport Modes of NTT1 and NTT2.
To clarify whether diatom NTTs act in a counter exchange mode we investigated the capacity of nonlabeled import substrates to induce export of radioactively-labeled nucleotides (previously loaded into E. coli cells) in a back-exchange approach. Additionally, the molecular nature of exported substrates was identified by thin-layer chromatography (12). E. coli cells expressing PtNTT1 released low amounts of nucleotides independent of the presence or absence of import substrates (≈1.5% of total label). However, PtNTT2 exported significant amounts (>11% of total label) of triphosphorylated nucleotides but solely when import substrates were added (Fig. S2) and thus catalyzes the counter exchange of nucleotides.
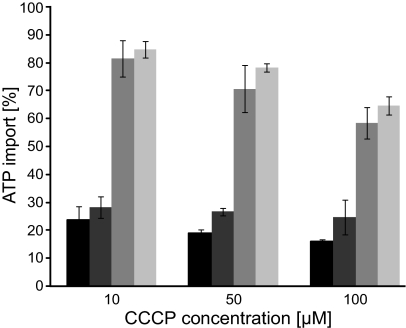
In a second approach we investigated the influence of the proton gradient on diatom NTT-mediated transport (18). Nucleotide import in the presence of the protonophore m-chlorophenylhydrazone (CCCP) was calculated compared with the noneffected transport (control without CCCP, set as 100%). Already very low CCCP concentrations (10 μM) substantially reduced ATP import of the NTT1 proteins (Fig. 2). The high inhibitory effect of CCCP indicates protons as the driving force required for NTT1-mediated transport. This result was surprising because all eukaryotic NTTs known so far are ATP/ADP antiporters and proton-driven nucleotide import was assumed to be an exclusive property of bacterial NTTs with lower amino acid similarities to ATP/ADP transporters from chlamydiae or plant plastids (12, 18). In contrast to NTT1, transport activities of NTT2 proteins were not profoundly affected by the presence of CCCP (Fig. 2). Our data indicate that NTT2 proteins most likely facilitate the counter exchange of NTPs or dNTPs, whereas NTT1 proteins mediate H+-adenine nucleotide symport (Figs. 1 and 2 and Fig. S2).
Fig. 2.
Analysis of the proton dependency of diatom NTTs. Effect of the protonophore CCCP on [α32P] ATP uptake by TpNTT1 (black bars), PtNTT1 (dark gray bars), TpNTT2 (gray bars), and PtNTT2 (light gray bars). Rates of nucleotide uptake are given as percentage of the control rates (nonaffected transport = 100%).
Cellular Localization and Physiological Role of NTT1 and NTT2.
It is commonly accepted that NTT proteins from plant and algae are located in the inner plastid envelope where they mediate energy provision to the organelle (10, 11). Because (i) the biochemical properties of the selected transporters differed from that of all plastidial NTTs described so far and (ii) NTT proteins were also detected in the plasma membrane and mitochondrial relicts of microsporidia (19) it is important to analyze the cellular localization of the diatom NTTs.
The NTT precursor proteins from plant and red algae and the diatom NTTs possess an N-terminal extension when compared with bacterial transporters (Fig. S3). The extensions of diatom NTT1 and NTT2 (and also of 3 other diatom NTTs) exhibit a bipartite presequence structure with a signal peptide followed by a transit peptide (Fig. S4A and Table S4). Furthermore, the NTT1 presequences contain a sequence with a phenylalanine at the signal peptide cleavage site that resembles the “ASAFAP” motif of stroma proteins (Table S4). The ability of the bipartite NTT1 and NTT2 leader sequences to target the transporters to the complex plastid was explored by GFP-fusion constructs (Fig. S4A) expressed in P. tricornutum. In a similar approach, we analyzed the cellular localization of the corresponding (full-length) transporters.
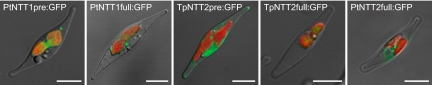
NTT1 proteins (full-length precursors) fused to GFP are located in a plastid membrane and the bipartite NTT1-presequences directed the reporter protein to the plastid, most likely into the stroma (Fig. 3 and Fig. S4B). The latter demonstrates that the bipartite leaders of PtNTT1 and TpNTT1 are sufficient for correct plastid targeting and suggests an insertion of NTT1 in the innermost plastid membrane via the stroma. It is necessary to mention that the catalytic activity of the NTT1 isoforms depends on a proton gradient across the corresponding membrane (Fig. 2). To our knowledge it has not been proven explicitly that a proton gradient exists at the innermost membrane of diatom plastids. However, transport of different solutes into the stroma of primary plastids is driven by a proton gradient generated by a P-type H+-ATPase in the inner envelope (20, 21).
Fig. 3.
Analysis of the cellular localization of diatom NTTs. Cellular targeting of GFP fused to the NTT presequences or the full-length NTTs. Selected merged images (GFP fluorescence in green, chlorophyll autofluorescence in red, and Nomarski differential interference contrast in white) are displayed. See Fig. S4 for additional information. (Scale bars: 5 μm.)
In contrast to the leaders of the NTT1 proteins the selected prepeptides of TpNTT2 and PtNTT2 did not target GFP into the plastid (Fig. 3 and Fig. S4). Although the presequence of TpNTT2 was not sufficient for plastid targeting, the full-length fusion protein might be located in a plastid membrane (Fig. 3). PtNTT2 (full-length precursor) fused to GFP labels the silhouette of the plastid [possibly the chloroplast ER (CER)] and the nucleus (possibly the nuclear envelope), and cytoplasmatic structures (possibly ER cisternae) (Fig. 3 and Fig. S4). The latter could represent a localization in the ER membrane system and implicitly suggests a role of PtNTT2 in nucleotide exchange at the CER. However, the identical biochemical characteristics and the high sequence similarities of TpNTT2 and PtNTT2 argue for an identical physiological role and thus for an identical cellular localization. We cannot rule out that the attached GFP hindered efficient translocation of PtNTT2 across the second plastid membrane (counted from the outside) that led to an accumulation of the fusion protein in the CER and the connected ER.
In the following we propose a model explaining the role of NTTs as possible components of a sophisticated nucleotide import system in the complex plastids of diatoms. Nucleotides might cross all 4 membranes solely by the interaction of net nucleotide importers and antiporters (NTTs); however, vesicle trafficking between membranes 2 and 3 might be involved (22). Nevertheless, independent of the mechanism of nucleotide provision, the proposed localization of the net adenine nucleotide importer (NTT1) in the innermost membrane requires the presence of nucleotides in the intermembrane space (Fig. S5). NTT1 catalyzes net uptake of ATP, ADP, or AMP into the stroma. Imported ATP could directly act as a counter exchange substrate of NTT2 and thus drive the import of other metabolically relevant (d)NTPs. Imported AMP and ADP first have to be converted into ATP to become substrates of NTT2 (Fig. S5). This suggested interaction of NTT1 and NTT2 presupposes that both transporters reside in the innermost envelope. We demonstrated that NTT1 is located in the innermost plastid membrane and that TpNTT2 is plastid associated (Fig. 3 and Fig. S4). Therefore, a localization of these NTTs within the same membrane is conceivable. If PtNTT2 resides in the outermost plastid membrane (CER) one of the yet-uncharacterized PtNTTs might facilitate the required pyrimidine nucleotide transport across the innermost envelope.
Our analyses raise the question of how energy is supplied to the diatom plastid. Although highly related to ATP/ADP transporters, the characterized diatom NTTs apparently do not facilitate ATP/ADP exchange. Therefore, alternative strategies for energy generation or import must exist in diatom plastids. Interestingly, in addition to the cytosolic glycolysis diatoms possess a similar set of enzymes in the plastid (4). Plastidial glycolysis might allow ATP generation in the dark. However, it is also possible that one of the uncharacterized NTTs (TpNTT3–8 or PtNTT3–6) acts as an ATP/ADP carrier. Furthermore, outsourcing of energy consuming processes (like nucleotide biosynthesis) might lower the plastid's energy demand. Characterization of the remaining NTTs will help to clarify nucleotide transport of diatoms.
Evolutionary Origin of Diatom NTTs.
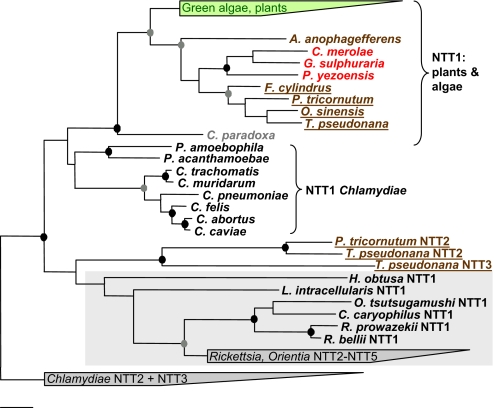
To gain insights into the evolutionary origin of diatom NTT proteins we performed a detailed phylogenetic analysis. The overall topology of the obtained trees is consistent with previous analyses on smaller datasets (13, 23–26). NTT1 proteins of T. pseudonana and P. tricornutum form a monophyletic cluster together with putative NTTs from other diatoms (Fig. 4; for more details see Fig. S6). NTTs from red algae constitute a sister group of diatom NTT1 proteins, suggesting a common origin of diatom NTT1 proteins from the red algal progenitor engulfed during secondary endosymbiosis. Furthermore, all NTT1 proteins from plants and algae formed a monophyletic group, with the transporter from the glaucophyte Cyanophora paradoxa as the deepest branch in this cluster (Fig. 4). This branching pattern is in gross agreement with the phylogeny of these organisms and the evolutionary history of their plastids (1, 27) and is thus evidence for a single primary endosymbiotic event leading to a “protoplastid” and for subsequent coevolution of this organelle with its eukaryotic host. Plastidial NTTs consistently grouped together with those from the Chlamydiae with all treeing algorithms applied in our analysis, albeit with low statistical support. A well-supported monophyletic grouping of plastidial with chlamydial NTTs was observed earlier and, together with recent phylogenomic analyses, suggests that the primary photosynthetic eukaryote most likely acquired its NTT gene from a donor most closely related to extant chlamydiae (13, 23, 24, 28–30).
Fig. 4.
Phylogenetic relationships of diatom NTTs with known bacterial and plastidial NTTs. An amino acid-based phylogenetic tree calculated by using the TREE-PUZZLE algorithm is shown. Black circles indicate nodes, which are supported by TREE-PUZZLE support, maximum parsimony, and ProML bootstrap values >80% (1,000 resamplings). Gray circles indicate nodes that are supported by 2 of the 3 above-mentioned treeing methods with support values >80%. TREE-PUZZLE support and bootstrap values <80% are not shown. Bar indicates 10% estimated evolutionary distance. Green algae and plant NTTs are marked in green, red algae are in red, heterokonts are in brown, diatoms are underlined, and C. paradoxa is in light gray. The rickettsial NTT group including NTT1 from L. intracellularis is highlighted by a gray box. A more detailed version of this figure and protein IDs are given in Fig. S6.
All chlamydial and plastidial NTT1 proteins characterized biochemically so far function as ATP/ADP antiporters (13). The H+-adenine nucleotide symporters from diatoms are thus the only exception in this clade, revealing a hitherto unseen functional diversity within these closely-related transport proteins. The change of transport mode in diatom NTT1 proteins most likely took place during establishment of the secondary endosymbiosis, possibly facilitated this process and obviously is associated with the extra-plastidial nucleotide biosynthesis.
Compared with the diatom NTT1 proteins, the situation is more complex with the diatom NTT2 proteins. These (deoxy-)NTTs form a deep branch together with the NTT3 protein of T. pseudonana, which groups with functionally diverse NTTs from rickettsiae, intracellular symbionts of protozoa, and the deltaproteobacterial Lawsonia intracellularis (25) (Fig. 4). Two scenarios could explain this branching pattern. First, diatom NTT2 proteins and TpNTT3 might have originated from gene duplications of the red algal NTT1 proteins that occurred during establishment of the secondary endosymbiosis. Reconstruction of this relationship could be blurred by the different functional constraints that apply to these transporters, which led to a comparatively low degree of sequence conservation and because of long branch attraction place diatom NTT2/TpNTT3 proteins next to the rickettsiae. The alternative scenario would be that diatoms gained the genes encoding NTT2/TpNTT3 proteins by a lateral gene transfer from a rickettsial donor, possibly through an infection by these intracellular bacterial parasites. A similar scenario has recently been proposed to explain the origin of menA genes for menaquinone biosynthesis in diatoms (31). At present it is, however, impossible to decide which of these alternative explanations is more likely. On the one hand, the low statistical support for the grouping of diatom NTT2/TpNTT3 proteins with those of the rickettsiae might be evidence for the first scenario. However, analyses of the P. tricornutum and T. pseudonana genomes revealed that lateral gene transfers from bacteria to diatoms are more common than previously thought (32).
Our study revealed that diatoms possess plastidial NTT proteins with biochemical properties similar to those of some bacterial NTTs but previously not observed for any eukaryotic NTT. The identified characteristics of the diatom NTTs allow net nucleotide provision to the plastid and therefore, metabolically connect plastidial nucleotide demand with the extra-plastidial nucleotide synthesis. This essential metabolic role is similar to that of chlamydial and rickettsial NTTs that provide host-derived nucleotides to metabolically-impaired intracellular bacteria.
In the future, it will be interesting to analyze whether a cytosolic localization of the nucleotide de novo synthesis and net nucleotide supply to the plastid is an exclusive feature of diatoms, a general principle of all heterokonts, all organisms with red algal derived plastids, or several protists that evolved by secondary endosymbiosis.
Materials and Methods
Culture of Algae.
P. tricornutum Bohlin (strain 646; University of Texas Culture Collection, Austin) and T. pseudonana Hasle and Heimdal (strain CCMP 1335; Provasoli-Guillard National Center for Culture of Marine Phytoplankton, West Boothbay Harbor, ME) were cultivated in L1 medium or f/2 medium using Tropic Marin salt (Dr. Biener, GmbH, Wartenberg, Germany) at a final concentration of 50% compared with natural seawater (33). Both diatom strains were grown under alternating light conditions (12-h light/12-h dark) in 1,000-mL aerated glass tubes. For nuclear transformation P. tricornutum was grown in liquid culture or at solid media containing 1.2% Bacto Agar (Difco) under continuous illumination at 35 μmol·photons·m−2·s−1.
Identification and Sequence Analysis of Selected Proteins from Diatoms.
The JGI T. pseudonana v1.0–3.0 (3) and the P. tricornutum v1.0–v2.0 (32) databases (http://genome.jgi-psf.org) were screened for sequences of interest by using the BLAST algorithm (34). Comparative sequence analysis of NTT proteins was performed with ClustalX (35). For predictions of presequences, of a cleavage site or chloroplast transit peptide-like domains the programs SignalP, ChloroP, and TargetP (all at www.cbs.dtu.dk/services) were applied. For a detailed description see ref. 36. Protein IDs from the diatom sequences and the corresponding databases are included in Tables S1 and S2. Protein accession numbers are given in Fig. S6.
Phylogenetic Analyses of NTT Proteins.
A database containing NTT sequences available from the public databases European Molecular Biology Laboratory/GenBank/ DNA Data Base in Japan was established by using the ARB software package (37). Amino acid sequences were aligned with MAFFT (38) and imported into ARB. Phylogenetic trees were constructed with the PHYLIP distance matrix (Fitch), maximum parsimony, ProML (using the JTT amino acid replacement model) (39), and TREE-PUZZLE (using the VT model of amino acid substitution) (40), methods implemented in ARB. Bootstrap analysis was performed with 1,000 resamplings. A filter considering only those alignment positions that were conserved in at least 10% of all sequences (resulting in a total number of 460 alignment columns) was used for all treeing calculations.
Preparation of cDNA and PCR.
Poly(A+) RNA was isolated from T. pseudonana and P. tricornutum by using the RNeasy plant mini kit and the Oligotex kit (Qiagen). RNA isolation and reverse transcription with SuperscriptII (Invitrogen) were performed according to the supplier's instructions. PCR was conducted with oligonucleotides (Eurofins MWG Operon) allowing compatible insertion into the expression vectors (Table S6). Amplification products were gel-purified by using the NucleoSpin Extract II Kit (Macherey & Nagel) and inserted into the vector pBSK (Stratagene) or directly into the expression vectors.
Generation of Constructs for Expression in E. coli or P. tricornutum.
For generation of the E. coli expression constructs the coding sequences of the diatom NTTs were inserted in-frame with the histidine tag into the IPTG-inducible expression vector pET16b (Novagen). The shuttle vector pPha-T1 [GenBank accession no. AF219942 (41)] was used for the transformation of P. tricornutum. The enhanced-GFP gene (BD Bioscience) was inserted into pPha-T1 via EcoRV (fcpA promoter). The full-length sequences and the presequences were inserted via StuI, EcoRI/StuI, or EcoRI/BamHI into the generated GFP-vectors (pPha-T1-StuI-GFP or pPha-T1-BamHI-GFP). Correctness of constructs was verified by restriction analyses (Fermentas) and sequencing (Nano+ Bio Center, TU Kaiserslautern, or Gesellschaft für Analyse-Technik und Consulting Biotech, Konstanz).
Heterologous Expression in E. coli and Import Measurements.
Heterologous synthesis of the diatom NTTs in the E. coli strain BLR (DE3) and transport measurements were conducted as described (12, 16). To analyze transport properties of the recombinant diatom NTTs either induced or noninduced (control) E. coli cells harboring the corresponding plasmids were incubated in phosphate buffer (KPi) containing 50 μM or the indicated concentrations of α32P-labeled substrates. Uptake was allowed at 30 °C for the indicated time spans and terminated by removal of external substrate using vacuum filtration and washing (12). Radioactivity in the samples was quantified in a scintillation counter (Tricarb 2500; Canberra-Packard).
Back-Exchange Analysis and Thin-Layer Chromatography.
E. coli cells synthesizing PtNTT1 or PtNTT2 were incubated in KPi containing 50 μM [α32P] ATP. Import (loading of the cells) was allowed for 5 min, cells were harvested by centrifugation and washed 2 times, and internal label was quantified. For induction of the back-exchange, cells were resuspended in KPi with 500 μM of the given nonlabeled nucleotides. Back-exchange was terminated after 2 min by rapid centrifugation. Radioactively labeled (exported) compounds in the supernatant were analyzed by thin-layer chromatography (12) and quantified in a scintillation counter.
Nuclear Transformation and Microscopy.
Nuclear transformation of P. tricornutum was performed as described (33). Cellular localization of GFP fusion proteins was analyzed with a confocal laser scanning microscope LSM 510 META (Zeiss) using a Plan-Apochromat 63 × 1.4 oil-immersion Nomarski differential interference contrast objective (Zeiss).
Supplementary Material
Acknowledgments.
We thank D. Ballert for help with the transformation and cultivation of P. tricornutum. This work was supported by the University of Konstanz, Deutsche Forschungsgemeinschaft Grants Project KR 1661/3-4 (to P.G.K.) and Project SPP 1131 “Life Inside Cells” NE 418/8-2 (to H.E.N.), and Austrian Science Fund Grants P19252-B17 and Y277-B03 (to S.S.-E. and M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808862106/DCSupplemental.
References
- 1.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 2.Roberts K, Granum E, Leegood RC, Raven JA. Carbon acquisition by diatoms. Photosynth Res. 2007;93:79–88. doi: 10.1007/s11120-007-9172-2. [DOI] [PubMed] [Google Scholar]
- 3.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 4.Kroth PG, et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE. 2008;3:e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delwiche CF. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Philos Trans R Soc London Ser B. 2003;358:109–133. doi: 10.1098/rstb.2002.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
- 8.Walker JE, Runswick MJ. The mitochondrial transport protein superfamily. J Bionenerg Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- 9.Leroch M, et al. Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J Biol Chem. 2005;280:17992–18000. doi: 10.1074/jbc.M412462200. [DOI] [PubMed] [Google Scholar]
- 10.Winkler HH, Neuhaus HE. Nonmitochondrial ATP transport. Trends Biochem Sci. 1999;24:64–68. doi: 10.1016/s0968-0004(98)01334-6. [DOI] [PubMed] [Google Scholar]
- 11.Linka N, et al. Phylogenetic relationship of nonmitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene. 2003;306:27–35. doi: 10.1016/s0378-1119(03)00429-3. [DOI] [PubMed] [Google Scholar]
- 12.Haferkamp I, et al. Tapping the nucleotide pool of the host: Novel nucleotide carrier proteins of Protochlamydia amoebophila. Mol Microbiol. 2006;60:1534–1545. doi: 10.1111/j.1365-2958.2006.05193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz-Esser S, et al. ATP/ADP translocases: A common feature of obligate intracellular amoebal symbionts related to chlamydia and rickettsia. J Bacteriol. 2004;186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michels A, Wedel N, Kroth P. Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol. 2005;137:911–920. doi: 10.1104/pp.104.055285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber A, et al. Protein targeting into complex diatom plastids: Functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- 16.Haferkamp I, et al. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature. 2004;432:622–625. doi: 10.1038/nature03131. [DOI] [PubMed] [Google Scholar]
- 17.Daugherty RM, et al. The nucleotide transporter of Caedibacter caryophilus exhibits an extended substrate spectrum compared to the analogous ATP/ADP translocase of Rickettsia prowazekii. J Bacteriol. 2004;186:3262–3265. doi: 10.1128/JB.186.10.3262-3265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjaden J, et al. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for the transport of energy. J Bacteriol. 1999;181:1196–1202. doi: 10.1128/jb.181.4.1196-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsaousis AD, et al. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 20.Shingles R, North M, McCarty RE. Ferrous ion transport across chloroplast inner envelope membranes. Plant Physiol. 2002;128:1022–1030. doi: 10.1104/pp.010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shingles R, Roh MH, McCarty RE. Nitrite transport in chloroplast inner envelope vesicles. I. Direct measurement of proton-linked transport. Plant Physiol. 1996;112:1375–1381. doi: 10.1104/pp.112.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs SP. The route of entry of cytoplasmically synthesized proteins into chloroplasts of algae possessing chloroplast ER. J Cell Sci. 1979;35:253–266. doi: 10.1242/jcs.35.1.253. [DOI] [PubMed] [Google Scholar]
- 23.Tyra HM, Linka M, Weber AP, Bhattacharya D. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 2007;8:R212. doi: 10.1186/gb-2007-8-10-r212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greub G, Raoult D. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl Environ Microbiol. 2003;69:5530–5535. doi: 10.1128/AEM.69.9.5530-5535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz-Esser S, et al. Lawsonia intracellularis encodes a functional rickettsia-like ATP/ADP translocase for host exploitation. J Bacteriol. 2008;190:5746–5752. doi: 10.1128/JB.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf YI, Aravind L, Koonin EV. Rickettsia and Chlamydiae. Evidence of horizontal gene transfer and gene exchange. Trends Genet. 1999;15:173–175. doi: 10.1016/s0168-9525(99)01704-7. [DOI] [PubMed] [Google Scholar]
- 27.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Gogarten JP. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8:R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker B, Hoef-Emden K, Melkonian M. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol Biol. 2008;8:203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moustafa A, Reyes-Prieto A, Bhattacharya D. Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS ONE. 2008;3:e2205. doi: 10.1371/journal.pone.0002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross J, Meurer J, Bhattacharya D. Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol Biol. 2008;8:117. doi: 10.1186/1471-2148-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;13:239–44. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 33.Kroth PG. Genetic transformation: A tool to study protein targeting in diatoms. Methods Mol Biol. 2007;390:257–267. [PubMed] [Google Scholar]
- 34.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson DJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felsenstein J. PHYLIP: Phylogeny Inference Package, Version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- 40.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 41.Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman A, Apt KE. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol. 2000;36:379–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.