Abstract
Posttranslational modifications such as glycosylation can play a fundamental role in signaling pathways that transform an ordinary cell into a malignant one. The development of a protocol to detect these changes in the preliminary stages of disease can lead to a sensitive and specific diagnostic for the early detection of malignancies such as ovarian cancer in which differential glycan patterns are linked to etiology and progression. Small variations in instrument parameters and sample preparation techniques are known to have significant influence on the outcome of an experiment. For an experiment to be effective and reproducible, these parameters must be optimized for the analyte(s) under study. We present a detailed examination of sample preparation and matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry (MALDI-FT-ICR-MS) analysis of O-linked glycans globally cleaved from mucin glycoproteins. Experiments with stable isotope-labeled biomolecules allowed for the establishment of appropriate acquisition times and excitation voltages for MALDI-FT-ICR-MS of oligosaccharides. Quadrupole ion guide optimization studies with mucin glycans identified conditions for the comprehensive analysis of the entire mass range of O-linked carbohydrates in this glycoprotein. Separately optimized experimental parameters were integrated in a method that allowed for the effective study of O-linked glycans.
Keywords: Fourier transform ion cyclotron mass spectrometry (FT-ICR-MS), matrix-assisted laser desorption/ionization (MALDI), mucin, O-linked glycans, protein glycosylations
INTRODUCTION
The arsenal of tools that the world of omics can offer has led to a better understanding of the etiology of diseases such as ovarian cancer1 at the gene, protein, metabolite and carbohydrate levels. Gene expression studies have demonstrated the relationship between tumor behavior and differential expression of gene products, also known as proteins. Investigations with proteomics and glycomics have painted a large and populated hunting ground for biomarker discovery, as clearly shown by the diversity and sheer number of proteins and carbohydrates to be found in the human body. Recent studies2–6 have implicated carbohydrates in the progression of various types of malignancies, including ovarian cancer, and clearly demonstrated that focus on carbohydrate patterns of glycosylated proteins can better direct biomarker discovery so as to improve the chance of success. In this respect, glycan profiling in key biospecimens, including blood,7,8 urine,9,10 cerebrospinal fluid (CSF)11 and peritoneal lavage,12 have demonstrated differential carbohydrate patterns between diseased and control subjects.
Carbohydrates from glycoproteins are important in several fundamental processes within the body such as cell–cell communication, cell adhesion and migration, as well as protein folding and secretion.13 In many diseases such as certain types of cancer2,3,6 and autoimmune disease,14,15 some modification to the glycosylation pattern is observed. In recent studies, changes in the O-glycosolation pattern have been observed in biospecimens from breast cancer16–20, as well as several gynecological malignancies such as ovarian21,22 and cervical23 cancers.
In areas of biomarker discovery where changes between control and disease states are often very subtle, particularly in the early stages of disease progression, the design of a robust methodology is a necessity. Lebrilla and co-workers20,21 have extended and applied well-established reductive β-elimination chemistry for globally cleaving O-linked glycans from glycoproteins24 to the study of glycan pattern changes in cancer. In this approach, protein information is necessarily lost as the primary focus is differential glycan pattern recognition, a less complicated system than the study of the entire serum glycoproteome. The work presented here explores this approach and investigates its effectiveness in glycan profiling of a mucin glycoprotein from porcine stomach,25 along with optimization of sample preparation and MS instrument parameters.
Mucins are highly O-glycosylated proteins that are present in many areas of the body. Mucin glycosylation is based on a simple set of core structures in which N-acetylgalactosamine (GalNAC) is linked to the side chains of serine and threonine. Packer and colleagues26 explored glycosylations in porcine stomach mucin by liquid chromatography/ mass spectrometry(LC/MS) and the use of an in-line flow system to study reducing O-linked oligosaccharides, as well as reductive β-elimination to investigate the reduced glycans. Data on monosaccharide composition by the two approaches was provided.26 Similarly, glycopeptide digests for this type of glycoprotein were investigated by Hotta and colleagues to obtain information on carbohydrate composition.27 Reductive and nonreductive release of O-linked glycans from porcine stomach mucin were compared by Lawson and co-workers through liquid secondary ion mass spectrometry (LSIMS) and direct thin-layer chromatography (TLC)/LSIMS analyses.28 Other reports have explored sulfated oligosaccharides in this glycoprotein through chemical release and alditol formation,29–31 as well as NMR approaches to investigate glycan-protein linkages.32,33
We applied matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry (MALDI-FT-ICR-MS) to study O-linked carbohydrates34 globally cleaved from porcine stomach mucin by reductive β-elimination. MALDI-FT-ICR-MS has generally not been the method of choice for biomolecule quantification due to background from matrix clusters at low m/z, signal suppression between analytes in a sample and reduced sensitivity due to the presence of cation adduct clusters.34–37 We aimed to circumvent many of these problems by adequate sample pretreatment to remove potential contaminants, and proper selection of MALDI matrix so as to improve detection sensitivity and the overall quality of mass spectrometric data. With optimized MALDI sample preparation protocols previously developed in this laboratory,34 we specifically evaluated instrument parameterswith stable isotope-labeled peptides and disaccharides, as well as mucin glycans to achieve an appropriate configuration for quantitative analysis of O-linked carbohydrates. Oligosaccharide identification through accurate mass measurements is given, as well as details of optimization of experimental parameters.
EXPERIMENTAL
Materials or reagents
Mucin type III from porcine stomach, 2,5-dihydroxybenzoic acid (2,5-DHB), trifluoroacetic acid (TFA), hydrochloric acid (HCl), sodium hydroxide (NaOH), sodium borohydride (NaBH4), sucrose, N-acetylglucosamine (GlcNAC), N-acetylneuraminic acid (NeuAc), lacto-N-tetraose (LNT), lacto-N-fucopentose (LNF), lacto-N-difucohexose (LND), phenyl isocyanate (PIC) and angiotensin I (sequence Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu) were obtained from Sigma-Aldrich (St Louis, MO). [UL-13C6fru] Sucrose (Suc*) was purchasedfromOmicron Biochemicals (South Bend, IN). 13C-labeled PIC came from Isotec, Inc. (Miamisburg, OH). Graphitized carbon solid-phase extraction (SPE) cartridges (300 mg bed size, 100 m2/g surface area, 38–125 µm particle size range) were obtained from Alltech (Deerfield, IL). High-performance liquid chromatography (HPLC)-grade Burdick & Jackson water and acetonitrile (ACN) were acquired from VWR International (Suwanee, GA).
Optimization of MALDI-FT-ICR-MS parameters with standard biomolecules
MS data were obtained using a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer interfaced with a ProMALDI ion source (Varian, Lake Forest, CA) in positive-ion mode. Applied Biosystems (Foster City, CA) targets (Part # 433375) were used for sample spotting. A Nd:YAG frequency tripled-laser (355 nm) promoted desorption/ ionization of analyte. The Varian Omega 2XP data station was used for all processing and signal generation. The standard broadband pulse sequence was employed. The instrument has a 9.4 T horizontal bore superconducting magnet with a 128 mm bore (Cryomagnetics Inc., Oakridge, TN).
Stable isotope-labeled peptides
Labeled peptides were quantified by liquid chromatography ultraviolet (LC-UV) analysis (Shimadzu, Columbia, MD), combined in equimolar mixtures and analyzed by MALDI-FT-ICR-MS. Angiotensin I was reacted with PIC and 13C-labeled *PIC through a procedure described elsewhere.38 The matrix of choice was 150 mg/ml 2,5-DHB in 1 : 1 ACN : 0.1% TFA. Mass spectra were acquired using chirp and SWIFT39 excitation waveforms with excitation voltages ranging from 20 to 120 V in 10 V increments, and 90–390 V in 30 V increments, respectively. Ion abundance ratios from MS and UV experiments compared favorably (data not shown).
Stable isotope-labeled disaccharides
Optimized parameters from angiotensin experiments were selected to study and compare a concentration series of actively dried mixtures of sucrose (Suc) and [UL-13C6fru] sucrose (Suc*). In this series, [Suc*] was held constant as the internal standard while [Suc] was varied. A similar excitation voltage range to angiotensin studies (150–420 V in 30 V increments) was applied. The MALDI matrix of choice was 100 mg/ml of 2,5-DHB in 1 : 1 ACN : 50 mm NaCl.34
Analysis of O-glycan patterns from mucin
Release of oligosaccharides from mucin by β-elimination
Oligosaccharides were cleaved from porcine stomach mucin with alkaline borohydride solution (1 m NaOH and 0.1 m NaBH4) by a previously described procedure.21,40 The basic solution was added to starting glycoprotein amounts ranging from 3 to 0.001 mg to determine the limit of detection. The mixture was then incubated in a heat block (VWR Scientific) at a constant 42 °C for 12 h to enable glycan release. Following the reaction, a 1 m solution of HCl was added dropwise to quench excess base. Oligosaccharides released from β-elimination were purified through SPE with a preconditioned graphitized carbon cartridge. Glycans of increasing mass were eluted with 10 and 20% ACN. Standard carbohydrates were employed to optimize the purification procedure.
Mass spectrometric analysis
Samples were spotted on a MALDI target with 2,5-DHB (100 mg/ml in 1 : 1 ACN : 50 mm NaCl) as the matrix. The analyte and matrix were combined in a 1 : 1 ratio and 0.8 µl of the resulting mixture was applied on the target. Active drying (AD) and passive drying (PD) methods for sample spotting were explored.34 Briefly, a cold stream of air was applied to the analyte/matrix droplet in AD experiments, whereas PD involved allowing the droplet to air-dry under ambient conditions. For the determination of MS instrument parameters appropriate to study the entire mass range of released mucin O-glycans, MS data was acquired with the MALDI-FT-ICR quadrupole ion guide optimized at m/z 300–1000.
RESULTS AND DISCUSSION
Prior to the investigation of glycoprotein carbohydrates by MALDI-FT-ICR-MS, instrument parameters were explored with standard small biomolecules. Further optimization of the quadrupole ion guide was then carried out with the target analyte – glycans released from porcine stomach mucin. MALDI sample spotting methods were also studied for mucin glycans. Analysis of mucin carbohydrates from decreasing mass of glycoprotein starting material provided information on limits of detection (LOD) for the approach.
Optimization of MALDI-FT-ICR-MS parameters with standard biomolecules
In any MALDI-FT-ICR experiment, high mass measurement accuracies are possible through the optimization of excitation voltage. Excitation promotes phase coherence of the ions and enables the expansion of the orbit of the ion cloud so as to induce a signal. The radius of the cloud following excitation is called the postexcite radius.38 The role of postexcite radius has been described elsewhere by Hawkridge et al41
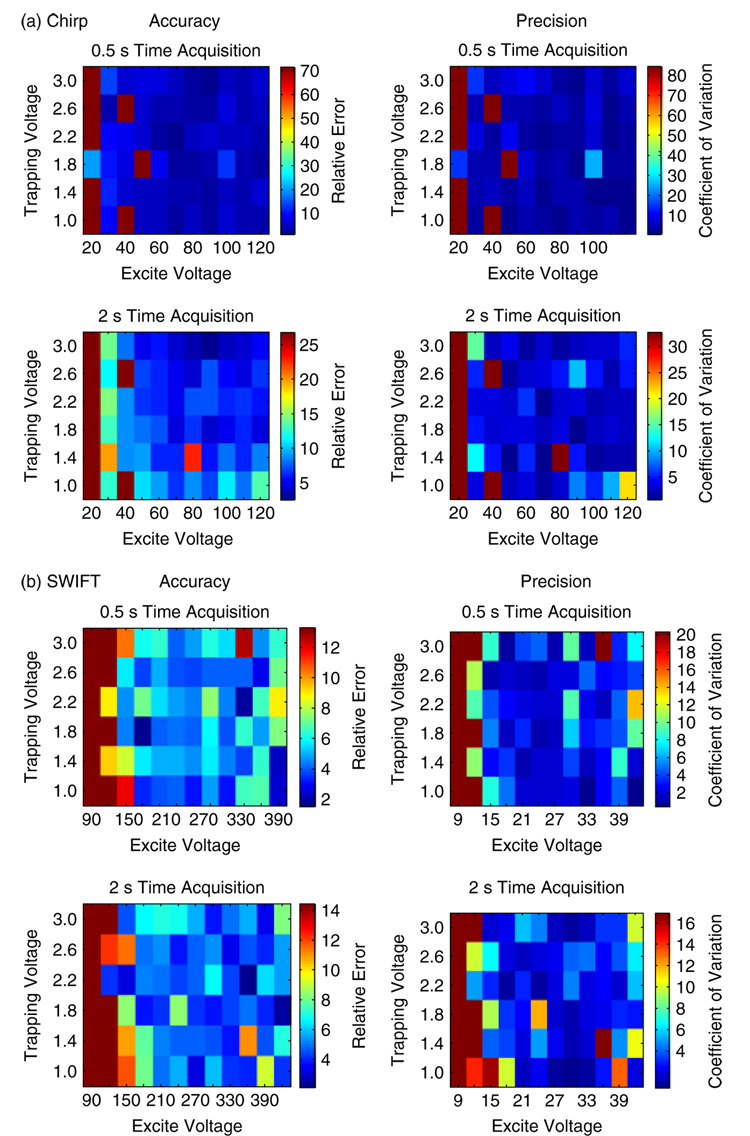
Preliminary experimentswith PIC-labeled and unlabeled angiotensin indicated that MS analyses with SWIFT39 were generally more accurate and precise compared to chirp, based upon percent relative error and coefficient of variation. This is clearly demonstrated in the heat maps described in Fig. 1. Furthermore, a trapping voltage of 1.8 V, an excitation voltage range of 150–420 V, and a 0.5 s time acquisition produced the best results.
Figure 1.
Comparison of relative error and coefficient of variation for (a) chirp and (b) SWIFT excitation waveforms. Due to a smaller relative error and coefficient of variation, SWIFT was determined to be more precise and accurate in MALDI-FT-ICR-MS of small biomolecules.
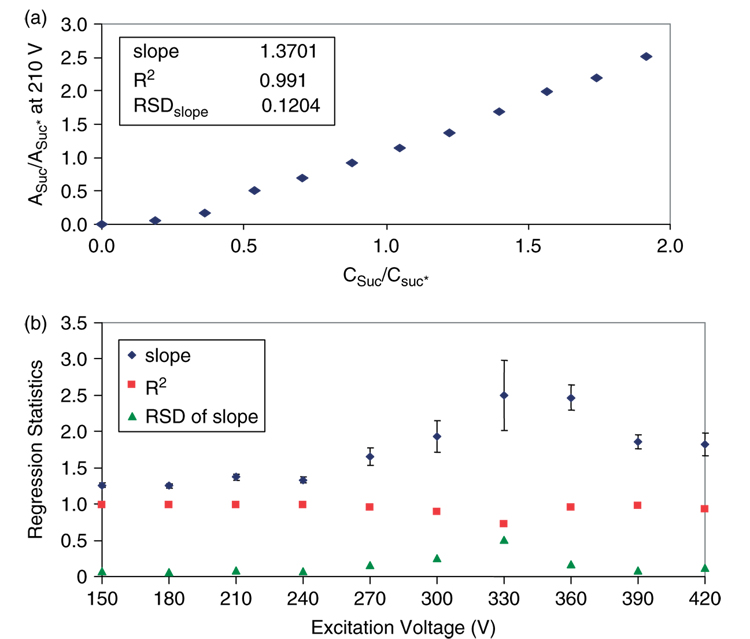
Additional MS investigations with Suc* and Suc were performed within the framework of instrument parameters obtained from angiotensin experiments. MS data were acquired on a concentration series of Suc and Suc*, in which [Suc*] was held constant and [Suc] was varied. Abundance ratios of Suc and Suc* were plotted against their concentration ratios for each excitation voltage studied (Fig. 2). Regression statistics of slope, R2 and relative standard deviation (RSD) of the slope for these plots were then graphed against excitation voltage (Fig. 2). The optimum excitation voltage should provide a slope close to 1, an R2 value close to 1 and a low RSD of the slope. The data in Fig. 2 indicates that the ideal excitation voltage lies in the range of 150–240 V.
Figure 2.
(a) Representative plot of abundance ratios (Asuc/Asuc*) versus concentration ratios (Csuc/Csuc*) of labeled to unlabeled sucrose (excitation voltage of 210 V). (b) Regression statistics from graphs like (a) were plotted against the excitation voltage range studied to determine the optimum excitation voltage range for quantifying carbohydrates with MALDI-FT-ICR-MS.
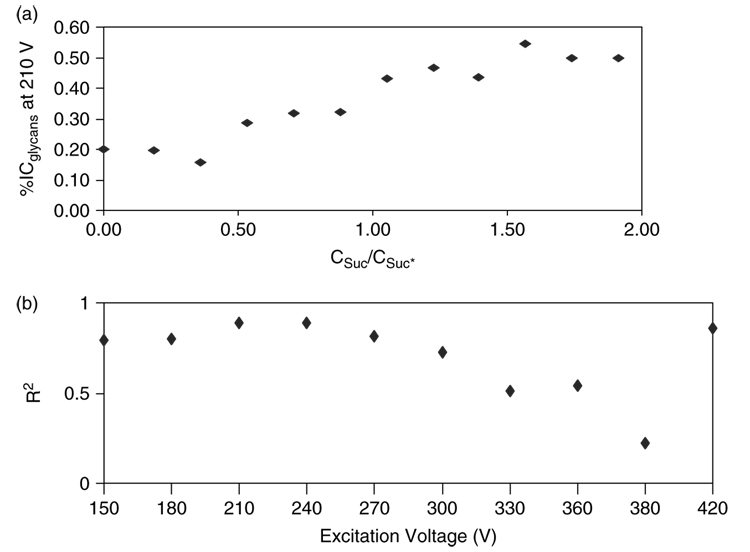
In order to determine the optimum excitation voltage in the above narrowed range, the percent ion composition (%IC) for both labeled and unlabeled disaccharides at each excitation voltage was plotted against their concentration ratios (Fig. 3). R2 values from these plots were then graphed against the excitation voltage range explored (Fig. 3). The most suitable excitation voltage, 210 V, produced an R2 close to 1. These parameters were subsequently applied in the MALDI-FT-ICR-MS of O-linked glycans released from porcine stomach mucin.
Figure 3.
Determination of the optimum excitation voltage for the analysis of oligosaccharides by MALDI-FT-ICR-MS. (a) Plot of percent (%) ion composition of sugar analytes against concentration ratios (Csuc/Csuc*) for labeled and unlabeled sucrose at 210 V excitation voltage. (b) The R2 values determined from plots like (a) over a range of excitation voltages were graphed against the excitation voltage range studied.
Reductive β-elimination
Influence of the amount of β-elimination reagents on MALDI-FT-ICR-MS of released glycans was explored with mucin from porcine stomach. A mixture of standard carbohydrates composed of N-acetylglucosamine (GlcNAC, MW 221.21), N-acetylneuraminic acid (NeuAc, MW 309.27), sucrose (Suc, MW 342), lacto-N-tetraose (LNT, MW 707), lacto-N-fucopentose (LNF,MW853) and lacto-N-difucohexose (LND, MW 999) was used to optimize the SPE of oligosaccharides in glycan analysis. SPE with graphitized carbon cartridges has been shown to be useful in the removal of salts, as well as NaOH and NaBH4 used in β-elimination reactions to release glycans from glycoconjugates.42 A reversed-phase mechanismis involved. Information on the optimumvolume of water for the desalting step of SPE analysis, as well as the distribution of glycan molecular weights in each elution (10 or 20% ACN; higher proportions of ACN were not necessary) was obtained. Troubleshooting for β-elimination and SPE elution of glycans is detailed in Table 1. In general, successful elution of glycans by SPE resulted in the formation of analyte/DHB matrix sample spots on a MALDI target with a thin, microcrystalline interior and a large, macrocrystal periphery. These were produced only with proper removal of excess alkaline borohydride from sample solutions by thorough SPE cleanup. Optimumglycan signals were obtained from the microcrystalline interior of these sample spots.34
Table 1.
Troubleshooting guide for SPE of glycoprotein carbohydrates released by reductive β-elimination
| MALDI analyte/matrix spots | Cause | Comments | ||
|---|---|---|---|---|
| • | Clear, translucent sample spots with no crystal definition | Excess base (NaBH4 + NaOH) from β-elimination reaction | SPE cartridge requires more thorough desalting with H2O prior to elution of glycans | |
| • | Poor MALDI-FT-ICR-MS data | |||
| • | Dark sample spots with no crystal definition | Excess base (NaBH4 + NaOH) from β-elimination reaction and HCl from base neutralization | SPE requires more thorough desalting with H2O prior to elution of glycans | |
| • | Poor MALDI-FT-ICR-MS data | |||
| • | Sample spots appear normal with good crystal definition | Excess H2O desalting of SPE cartridge prior to elution, or too fast a washing rate | • | Slow desalting rate (~1 ml/min) |
| • | Poor MALDI-FT-ICR-MS data | • | Fine-tune volume of desalting wash to sample size | |
Note: In the presence of excess HCl, spots appeared normal with good crystal definition and no apparent effects on MALDI-FT-ICR-MS data.
Small and large carbohydrates were generally eluted in the 10 and 20% ACN fractions, respectively. The oligosaccharides elute in this pattern because increasing mass correlates to increasing hydrophobicity and thereby, increasing attraction to the graphitized carbon surface. Hence, larger glycans require a more hydrophobic solvent for elution to overcome the attraction to graphitized carbon.
MALDI-FT-ICR-MS of mucin glycans
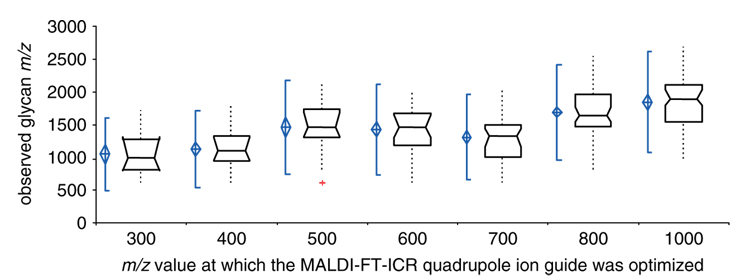
The MALDI-FT-ICR quadrupole ion guide was optimized from m/z 300 to 1000 to obtain a comprehensive picture of mucin O-glycan patterns. Figure 4 depicts a box and whisker plot of carbohydrate m/z ranges observed when the ion guide was optimized at increasing m/z values. Results indicated that the majority of mucin glycan peaks were observed with the ion guide optimized at m/z values of ~400 (small carbohydrates in the 10% ACN fraction of mucin glycans) and ~900 (large carbohydrates in the 20% ACN fraction of mucin glycans).
Figure 4.
Box and whisker plot of m/z range of glycan peaks detected with the quadrupole ion guide optimized at increasing, specific m/z values. Acquiring MS data with optimization at m/z 400 and 900 (data point not shown, but similar to optimization at m/z 1000) enabled the examination of most of the O-glycan peaks from mucin.
In MALDI-FT-ICR-MS, ions are accumulated in the hexapole and passed into the ICR cell through the quadrupole ion guide. Primarily due to the time of flight effect, larger ions arrive at the ICR cell at a slower pace than do the smaller ions.43 Therefore, based on the time during which the gate to the ICR cell is held open, a different mass range of glycan peaks is observed. When the quadrupole ion guide is optimized at a lower m/z, the gate is opened much earlier, permitting only smaller ions into the cell. If the gate is opened after the elapse of a longer time period, ions with a larger mass are selected for analysis and the smaller ions are excluded. As shown in Fig. 4, when this gate is opened at later times, there is an increase in the average molecular weight of ions observed in the mass spectra and a shift in the range of molecular weights toward higher masses. Successive MS analysis of mucin glycans with the quadrupole ion guide optimized at a low (400) and high (900) m/z values allows for the observation of a larger and more diverse range of oligosaccharides from the glycoprotein.
Previous studies with monosaccharides have suggested that AD of sample spots is the preferred method for carbohydrate analysis by MALDI-FT-ICR-MS.34 These results were corroborated with Suc and Suc* mixtures used in experiments for instrument parameter optimizations (data not shown). Our data further demonstrated that AD was preferred over PD in a more complex biological system of cleaved mucin O-glycans. In AD, the constant motion of air over the sample allows for mixing of the analyte and matrix, which in turn creates a more uniform composition and a less defined crystal structure throughout the spot. This allows for higher glycan ion abundances in MS analyses.
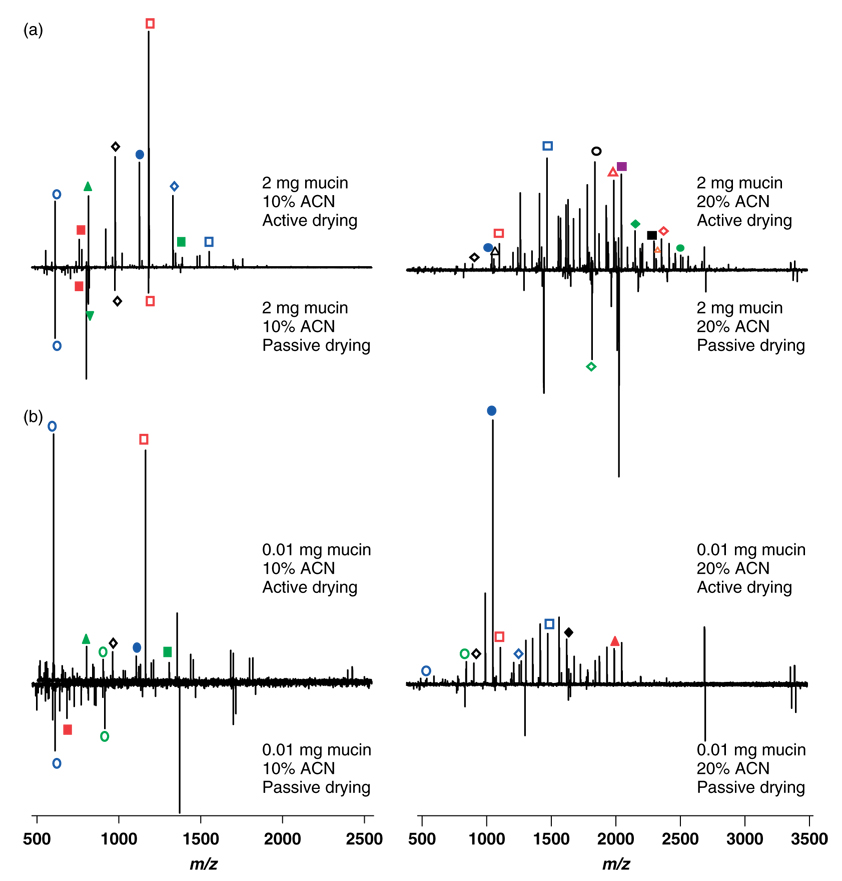
Starting glycoprotein amounts ranging from 3 to 0.001 mg were subjected to reductive β-elimination to release O-glycans, which were subsequently analyzed by AD and PD of MALDI sample spots with optimized MALDI-FT-ICR-MS parameters. Lower LOD (0.01 mg starting mucin) as well as improved glycan diversity and ion abundances were observed with AD (Fig. 5). The 10% ACN fraction of carbohydrates was well represented with the quadrupole ion guide optimized at m/z 400, whereas comprehensive glycan distribution in the 20% ACN fraction was observed with ion guide optimization at m/z 900. Peak identities (accurate mass of Na+ adducted alditol form of glycans) and composition were determined with the OsCal program44 (Table 2 and Table 3). Many other glycans were detected and identified, as shown by peak diversity in the mass spectra, but only a representative number are provided here. Some overlap in glycans was observed in the two fractions.
Figure 5.
Mass spectra of glycans from (a) 2 mg and (b) 0.01 mg (LOD) mucin starting glycoprotein mass. Active drying (AD) and passive drying (PD) sample spotting techniques were compared. Mass spectra were acquired from carbohydrates from the 10 and 20% ACN fractions with the quadrupole ion guide optimized at m/z of 400 and 900, respectively.
Table 2.
Representative sodium-adducted oligosaccharide peaks (alditol form) with possible compositions observed in mucin from the 10% ACN fraction
| Expt. m/z [M +Na]+ | Theo. m/z [M +Na]+ | Composition in [M + Na]+a | Symbolb |
|---|---|---|---|
| 611.224 | 611.227 | 1 Hex, 2 HexNAc | |
| 773.274 | 773.280 | 2 Hex, 2 HexNAc | |
| 814.289 | 814.306 | 1 Hex, 3 HexNAc | |
| 976.339 | 976.359 | 2 Hex, 3 HexNAc | |
| 1122.399 | 1122.417 | 2 Hex, 1 Fuc, 3 HexNAc | |
| 1179.411 | 1179.439 | 2 Hex, 4 HexNAc | |
| 1544.500 | 1544.571 | 3 Hex, 5 HexNAc | |
| 1690.592 | 1690.629 | 3 Hex, 1 Fuc, 5 HexNAc | |
| 1893.669 | 1893.708 | 3 Hex, 1 Fuc, 6 HexNAc | |
| 2112.732 | 2112.783 | 4 Hex, 7 HexNAc | |
| 2404.813 | 2404.898 | 4 Hex, 2 Fuc, 7 HexNAc | |
| 2550.895 | 2550.957 | 4 Hex, 3 Fuc, 7 HexNAc |
Table 3.
Representative sodium-adducted oligosaccharide peaks (alditol form) with possible compositions observed in mucin from the 20% ACN fraction
| Expt. m/z [M + Na]+ | Theo. m/z [M + Na]+ | Composition in [M + Na]+a | Symbolb |
|---|---|---|---|
| 919.345 | 919.338 | 2 Hex, 1 Fuc, 2 HexNAc | |
| 1138.418 | 1138.412 | 3 Hex, 3 HexNAc | |
| 1325.506 | 1325.497 | 2 Hex, 1 Fuc, 4 HexNAc | |
| 1341.503 | 1341.492 | 3 Hex, 4 HexNAc | |
| 1909.728 | 1909.703 | 4 Hex, 6 HexNAc | |
| 2055.790 | 2055.761 | 4 Hex, 1 Fuc, 6 HexNAc | |
| 2217.827 | 2217.814 | 5 Hex, 1 Fuc, 6 HexNAc | |
| 2363.903 | 2363.872 | 5 Hex, 2 Fuc, 6 HexNAc | |
| 2420.905 | 2420.893 | 5 Hex, 1 Fuc, 7 HexNAc |
CONCLUSIONS
In diseases such as ovarian cancer, a limited supply of patient samples is available for study. Therefore, an effective method of analysis must be designed prior to manipulation of valuable biospecimens. In the experiments detailed here, each step of the process for carbohydrate analysis is first optimized with relatively simple systems and subsequently applied to more complex biological specimens. Previous investigations with standard monosaccharides, such as Glc-NAc and NeuAc, revealed significant improvements in carbohydrate ion abundance with AD over PD of carbohydrates spots for MALDI-FT-ICR-MS. These differences were explained by glycan preferences for certain crystal polymorphs of matrix.34,46 Experiments with angiotensin and sucrose indicated that small modifications in mass spectrometric instrument parameters can have favorable effects on the quality of data. The SWIFT waveform, along with a trapping voltage of 1.8 V and an excitation voltage of 210 V yielded precise and accurate quantification for carbohydrates. Optimized MALDI-FT-ICR-MS protocols were applied to study released glycans from a mucin standard glycoprotein. The release of glycans through reductive β-elimination and subsequent analysis was optimized for mucin to obtain information on LOD for the protocol. Results indicated that the majority of glycans investigated in mucin were separated in elutions of increasing organic content by increasing mass of carbohydrate. The entire mass range of released glycans was observed by MALDI-FT-ICR-MS when the quadrupole ion guide was optimized at m/z values of 400 (small oligosaccharides eluted with 10% ACN) and 900 (large glycans eluted with 20% ACN).
While accurate mass is an established method for identification of analytes studied by MALDI-FT-ICR-MS, tandem MS information can provide valuable insight into analyte structure. Indeed, recent experiments in our laboratory with DESI-MS have provided information on glycan composition and configuration.47 In the future, studies with O-glycans can be applied toward the investigation of differential O-glycosylation patterns in glycoproteins from biological specimens in the search for disease biomarkers.
Acknowledgements
We appreciate assistance from D. Keith Williams, Jr (W.M. Keck FT-ICR Laboratory, North Carolina State University, Raleigh, NC) in the analysis of data from MALDI-FT-ICR-MS quadrupole ion guide optimization experiments. Diana Saggese acknowledges North Carolina State University for her undergraduate research award stipend. We gratefully appreciate financial support received from the North Carolina Biotechnology Center, LEAP Technologies, Inc., National Cancer Institute, National Institutes of Health (R33 CA105295), the W.M. Keck Foundation and North Carolina State University.
REFERENCES
- 1.Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. Journal of Proteome Research. 2007;6:2936. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 2.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochimica et Biophysica Acta. 1999;1473:21. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 3.Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Aranganathan S, Senthil K, Nalini N. A case control study of glycoprotein status in ovarian carcinoma. Clinical Biochemistry. 2005;38:535. doi: 10.1016/j.clinbiochem.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Casey RC, Oegema TR, Jr, Skubitz KM, Pambuccian SE, Grindle SM, Skubitz AP. Cell membrane glycosylation mediates the adhesion, migration, and invasion of ovarian carcinomacells. Clinical and Experimental Metastasis. 2003;20:143. doi: 10.1023/a:1022670501667. [DOI] [PubMed] [Google Scholar]
- 6.Dwek MV, Brooks SA. Harnessing changes in cellular glycosylation in new cancer treatment strategies. Current Cancer Drug Targets. 2004;4:425. doi: 10.2174/1568009043332899. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. Journal of Chromatography. A. 2004;1053:79. [PubMed] [Google Scholar]
- 8.Madera M, Mechref Y, Klouckova I, Novotny MV. High-sensitivity profiling of glycoproteins from human blood serum through multiple-lectin affinity chromatography and liquid chromatography/tandem mass spectrometry. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2007;845:121. doi: 10.1016/j.jchromb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 9.Guo JM, Zhang XY, Chen HL, Wang GM, Zhang YK. Structural alterations of sugar chains in urine fibronectin from bladder cancer patients and its enzymatic mechanism. Journal of Cancer Research and Clinical Oncology. 2001;127:512. doi: 10.1007/s004320100245. [DOI] [PubMed] [Google Scholar]
- 10.Valmu L, Alfthan H, Hotakainen K, Birken S, Stenman UH. Site-specific glycan analysis of human chorionic gonadotropin beta-subunit from malignancies and pregnancy by liquid chromatography-electrospray mass spectrometry. Glycobiology. 2006;16:1207. doi: 10.1093/glycob/cwl034. [DOI] [PubMed] [Google Scholar]
- 11.Rudd PM, Mattu TS, Masure S, Bratt T, Van den Steen PE, Wormald MR, Kuster B, Harvey DJ, Borregaard N, Van Damme J, Dwek RA, Opdenakker G. Glycosylation of natural human neutrophil gelatinase B and neutrophil gelatinase Bassociated lipocalin. Biochemistry. 1999;38:13 937. doi: 10.1021/bi991162e. [DOI] [PubMed] [Google Scholar]
- 12.Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Molecular and Cellular Proteomics. 2005;4:409. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Fraser-Reid BO, Tatsuta K, Thiem J, editors. Glycoscience. Berlin: Springer-Verlag, Berlin Heidelberg; 2001. [Google Scholar]
- 14.Zhang XL. Roles of glycans and glycopeptides in immune system and immune-related diseases. Current Medicinal Chemistry. 2006;13:1141. doi: 10.2174/092986706776360897. [DOI] [PubMed] [Google Scholar]
- 15.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd KO, Burchell J, Kudryashov V, Yin BW, Taylor-Papadimitriou J. Comparison of O-linked carbohydrate chains in MUC-1 mucin from normal breast epithelial cell lines and breast carcinoma cell lines. Demonstration of simpler and fewer glycan chains in tumor cells. The Journal of Biological Chemistry. 1996;271:33 325. doi: 10.1074/jbc.271.52.33325. [DOI] [PubMed] [Google Scholar]
- 17.Colpitts TL, Billing P, Granados E, Hayden M, Hodges S, Roberts L, Russell J, Friedman P, Stroupe S. Identification and immunohistochemical characterization of a mucin-like glycoprotein expressed in early stage breast carcinoma. Tumour Biology. 2002;23:263. doi: 10.1159/000068566. [DOI] [PubMed] [Google Scholar]
- 18.Baldus SE, Wienand JR, Werner JP, Landsberg S, Drebber U, Hanisch FG, Dienes HP. Expression of MUC1, MUC2 and oligosaccharide epitopes in breast cancer: prognostic significance of a sialylated MUC1 epitope. International Journal of Oncology. 2005;27:1289. [PubMed] [Google Scholar]
- 19.Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. The Journal of Histochemistry and Cytochemistry. 2006;54:317. doi: 10.1369/jhc.5A6783.2005. [DOI] [PubMed] [Google Scholar]
- 20.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S, Lebrilla CB, Miyamoto S. A serum glycomics approach to breast cancer biomarkers. Molecular and Cellular Proteomics. 2007;6:43. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. Journal of Proteome Research. 2006;5:1626. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 22.Nagata A, Hirota N, Sakai T, Fujimoto M, Komoda T. Molecular nature and possible presence of a membranous glycan-phosphatidylinositol anchor of CA125 antigen. Tumour Biology. 1991;12:279. doi: 10.1159/000217716. [DOI] [PubMed] [Google Scholar]
- 23.Hsu YT, Hou Q, Zhou H, Mohanty S, Litvinchuk A. Differential immunoscreening identifies a glycosylation variant of the epidermal growth factor receptor in ME-180 cervical carcinoma cells. Hybridoma (Larchmt) 2005;24:225. doi: 10.1089/hyb.2005.24.225. [DOI] [PubMed] [Google Scholar]
- 24.Carlson DM. Oligosaccharides isolated from pig submaxillary mucin. The Journal of Biological Chemistry. 1966;241:2984. [PubMed] [Google Scholar]
- 25.Nordman H, Davies JR, Carlstedt I. Mucus glycoproteins from pig gastric mucosa: different mucins are produced by the surface epithelium and the glands. The Biochemical Journal. 1998;331:687. doi: 10.1042/bj3310687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson NG, Packer NH. Analysis of O-linked reducing oligosaccharides released by an in-line flow system. Analytical Biochemistry. 2002;305:173. doi: 10.1006/abio.2002.5657. [DOI] [PubMed] [Google Scholar]
- 27.Iwase H, Ishii-Karakasa I, Hotta K. Isolation and partial characterization of serine- and threonine-rich porcine gastric mucus glycopeptides. Comparative Biochemistry and Physiology. 1992;102B:929. doi: 10.1016/0305-0491(92)90104-y. [DOI] [PubMed] [Google Scholar]
- 28.Chai W, Feizi T, Yuen CT, Lawson AM. Nonreductive release of O-linked oligosaccharides from mucin glycoproteins for structure/function assignments as neoglycolipids: application in the detection of novel ligands for E-selectin. Glycobiology. 1997;7:861. doi: 10.1093/glycob/7.6.861. [DOI] [PubMed] [Google Scholar]
- 29.Thomsson KA, Karlsson H, Hansson GC. Sequencing of sulfated oligosaccharides from mucins by liquid chromatography and electrospray ionization tandem mass spectrometry. Analytical Chemistry. 2000;72:4543. doi: 10.1021/ac000631b. [DOI] [PubMed] [Google Scholar]
- 30.Thomsson KA, Karlsson NG, Hansson GC. Liquid chromatography-electrospray mass spectrometry as a tool for the analysis of sulfated oligosaccharides from mucin glycoproteins. Journal of Chromatography. A. 1999;854:131. doi: 10.1016/s0021-9673(99)00625-1. [DOI] [PubMed] [Google Scholar]
- 31.Slomiany BL, Meyer K. Isolation and structural studies of sulfated glycoproteins of hog gastric mucosa. The Journal of Biological Chemistry. 1972;247:5062. [PubMed] [Google Scholar]
- 32.Van Halbeek H, Dorland L, Veldink GA, Vliegenthart JF, Garegg PJ, Norberg T, Lindberg B. A500-MHz proton-magnetic-resonance study of several fragments of the carbohydrate-protein linkage region commonly occurring in proteoglycans. European Journal of Biochemistry. 1982;127:1. doi: 10.1111/j.1432-1033.1982.tb06830.x. [DOI] [PubMed] [Google Scholar]
- 33.Zenteno E, Vazquez L, Chavez R, Cordoba F, Wieruszeski JM, Montreuil J, Debray H. Specificity of the isolectins from the plant cactus Machaerocereus eruca for oligosaccharides from porcine stomach mucin. Glycoconjugate Journal. 1995;12:699. doi: 10.1007/BF00731267. [DOI] [PubMed] [Google Scholar]
- 34.Williams TI, Saggese DA, Wilcox RJ, Martin JD, Muddiman DC. Effect of matrix crystal structure on ion abundance of carbohydrates by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21:807. doi: 10.1002/rcm.2904. [DOI] [PubMed] [Google Scholar]
- 35.Pan CS, Xu SY, Zhou HJ, Fu Y, Ye ML, Zou HF. Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Analytical and Bioanalytical Chemistry. 2007;387:193. doi: 10.1007/s00216-006-0905-4. [DOI] [PubMed] [Google Scholar]
- 36.Harvey DJ. Matrix-assisted laser desorption ionisation mass spectrometry of oligosaccharides and glycoconjugates. Journal of Chromatography. A. 1996;720:429. doi: 10.1016/0021-9673(95)00307-x. [DOI] [PubMed] [Google Scholar]
- 37.Gusev AI, Wilkinson WR, Proctor A, Hercules DM. Improvement of signal reproducibility and matrix/comatrix effects in Maldi analysis. Analytical Chemistry. 1995;67:1034. [Google Scholar]
- 38.Frahm JL, Velez CM, Muddiman DC. Understanding the influence of post-excite radius and axial confinement on quantitative proteomic measurements using Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21:1196. doi: 10.1002/rcm.2957. [DOI] [PubMed] [Google Scholar]
- 39.Chen L, Wang TC, Ricca TL, Marshall AG. Phase-modulated stored waveform inverse Fourier transform excitation for trapped ion mass spectrometry. Analytical Chemistry. 1987;59:449. doi: 10.1021/ac00130a016. [DOI] [PubMed] [Google Scholar]
- 40.Iyer RN, Carlson DM. Alkaline borohydride degradation of blood group H substance. Archives of Biochemistry and Biophysics. 1971;142:101. doi: 10.1016/0003-9861(71)90263-3. [DOI] [PubMed] [Google Scholar]
- 41.Hawkridge AM, Nepomuceno AI, Lovik SL, Mason CJ, Muddiman DC. Effect of post-excitation radius on ion abundance, mass measurement accuracy, and isotopic distributions in Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 2005;19:915. doi: 10.1002/rcm.1871. [DOI] [PubMed] [Google Scholar]
- 42.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconjugate Journal. 1998;15:737. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 43.Sze TPE, Chan TWD. Time-of-flight effects in matrix assisted laser desorption/ionization Fourier transform mass spectrometry. Rapid Communications in Mass Spectrometry. 1999;13:398. [Google Scholar]
- 44.An HJ, Tillinghast JS, Woodruff DL, Rocke DM, Lebrilla CB. A new computer program (GlycoX) to determine simultaneously the glycosylation sites and oligosaccharide heterogeneity of glycoproteins. Journal of Proteome Research. 2006;5:2800. doi: 10.1021/pr0602949. [DOI] [PubMed] [Google Scholar]
- 45.Murray RK. Glycoproteins: crucial molecules for health. GlycoScience and Nutrition. 2003;4:1. [Google Scholar]
- 46.Cohen DE, Benedict JB, Morlan B, Chiu DT, Kahr B. Dyeing Polymorphs: the MALDI Host 2,5-Dihydroxybenzoic Acid. Crystal Growth and Design. 2007;7:492. [Google Scholar]
- 47.Bereman MS, Williams TI, Muddiman DC. Carbohydrate analysis by desorption electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Analytical Chemistry. 2007;79:8812. doi: 10.1021/ac0713858. [DOI] [PubMed] [Google Scholar]