Summary
The envelope of Gram-negative bacteria consists of inner and outer membranes surrounding the peptidoglycan wall. The outer membrane (OM) is rich in integral membrane proteins (OMPs), which have a characteristic β-barrel domain embedded in the OM. The Omp85 family of proteins, ubiquitous among Gram-negative bacteria and also present in chloroplasts and mitochondria, is required for folding and insertion of OMPs into the outer membrane. Bacterial Omp85 proteins are characterized by a periplasmic domain containing five repeats of polypeptide transport-associated (POTRA) motifs. Here we report the crystal structure of a periplasmic fragment of YaeT (the E. coli Omp85) containing the first four POTRA domains in a new extended conformation consistent with recent solution X-ray scattering data. Analysis of the YaeT structure reveals conformational flexibility around a hinge point between POTRA2 and 3 domains. The structure’s implications for the substrate binding and folding mechanisms are also discussed.
Introduction
The cell envelope of Gram-negative bacteria is composed of inner and outer membranes separated by the peptidoglycan wall. The outer membrane (OM) consists of lipopolysaccharide (LPS), phospholipids and proteins, and constitutes a selective physical barrier impermeable to many antibiotics and resistant to detergents (Nikaido, 2003). At the same time, outer membrane proteins (OMPs) integral to the OM facilitate, among other functions, the influx of necessary nutrients. OMPs have characteristic β-barrel structures spanning the membrane (Schulz, 2003). The correct folding and insertion of β-barrels in the OM is essential for the bacteria, and several key players in these processes have recently come to light (Bos et al., 2007a; Ruiz et al., 2006).
After translocation through the inner membrane (IM) and cleavage of the signal sequence, OMPs traverse the aqueous periplasm with the assistance, at least in some cases, of periplasmic chaperones such as SurA and Skp (Mogensen and Otzen, 2005). The insertion process in the outer membrane appears to be conserved in bacteria as well as in evolutionarily-related mitochondria and chloroplasts, and is mediated by the Omp85 family of OMPs (Voulhoux et al., 2003; Voulhoux and Tommassen, 2004; Wu et al., 2005). All members of the Omp85 family consist of an N-terminal periplasmic domain and a membrane-embedded C-terminal β-barrel domain (Voulhoux and Tommassen, 2004). The periplasmic domain contains a variable number of the so-called polypeptide transport-associated (POTRA) motifs. The number of POTRA domains ranges from one, in the case of the mitochondrial Sam50, to five for bacterial Omp85 proteins (Gentle et al., 2005). The POTRA domains are hypothesized to mediate protein-protein interactions and even to have chaperone-like qualities (Gentle et al., 2005; Sanchez-Pulido et al., 2003; Voulhoux et al., 2003).
In E. coli the Omp85 protein is known as YaeT. Together with four lipoproteins – YfiO, YfgL, NlpB and SmpA, YaeT forms a multiprotein complex required for OMP insertion in the outer membrane (Sklar et al., 2007a; Wu et al., 2005). YaeT and YfiO are essential for cell viability, whereas null mutants of YfgL, NlpB or SmpA are viable, but show OM permeability defects due to impaired OMP assembly (Malinverni et al., 2006; Sklar et al., 2007a; Wu et al., 2005). A large number of OMPs require YaeT for insertion into the outer membrane and they appear to be targeted to YaeT by a species-specific C-terminal signature sequence (Robert et al., 2006). Consistent with a fundamental role in OMP insertion, depletion of YaeT results in a marked increase of misfolded β-barrels in the periplasm and a decrease of correctly inserted β-barrel OMPs (Kim et al., 2007; Voulhoux et al., 2003; Werner and Misra, 2005; Wu et al., 2005).
Recent work by Kim et al., investigated the functional importance of the individual POTRA domains in E. coli YaeT (Kim et al., 2007). Whereas POTRA1 and POTRA2 deletion mutants retain partial function, POTRA3-5 domains are essential for cell viability. Interestingly, only POTRA5 appears to be required for cell viability in Neisseria as deletion of the first four POTRA domains in its Omp85 homolog was tolerated with only a slight reduction of cell viability and some defects in the folding of large OMPs (Bos et al., 2007b). It is notable, however, that all bacterial homologs of Omp85 have five POTRA domains (Gentle et al., 2005). The crystal structures of FhaC (Clantin et al., 2007) and an N-terminal fragment of YaeT containing the first four POTRA domains (Kim et al., 2007) provided the first glimpse of the POTRA architecture. The YaeT structure showed a curved, fishhook-like arrangement of POTRA domains. However, recent small angle X-ray scattering and NMR data suggests that the POTRA domains adopt a more extended conformation in solution (Knowles et al., 2008). Here we present the crystal structure of E. coli YaeT periplasmic domain containing POTRA1-4 in a new extended conformation. This new structure highlights apparent conformational flexibility of the YaeT periplasmic domain and shows critical features of POTRA3, not visible in the previous structure, shedding new light onto the substrate recognition process.
Results
Crystallization and Structure Determination of Four POTRA Domains of YaeT
YaeT is an outer membrane protein comprised of an N-terminal periplasmic domain and a C-terminal β-barrel domain. The first 20 amino acids in E. coli YaeT represent the signal peptide that is cleaved in the mature protein. However, the exact boundary between the POTRA-containing periplasmic domain and the β-barrel domain has not been precisely defined. For crystallization purposes, we systematically searched for a well expressing periplasmic fragment resistant to proteolysis, and initially isolated a fragment containing amino acids 21-410 (YaeT21:410). Crystallization of this fragment was achieved in 0.1 M MES pH 6.5, 1.35M (NH4)2SO4, 6% PEG400, 10% Dioxane. A native data set to 3.3Å resolution was collected at the ALS from these crystals, which belonged to space group P3121. Further refinement of the expression construct identified a shorter fragment containing amino acids 21-359 (YaeT21:359) as a well behaved crystallization target. Seleno-methionine substituted YaeT21:359 readily crystallized in conditions similar to YaeT21:410 (0.1 M MES pH 6.5, 1.55M (NH4)2SO4, 6% PEG400, 10% Dioxane), yielding large diamond shaped crystals isomorphous to those of YaeT21:410. A three-wavelength data set to 3.3Å resolution collected on these crystals was used to determine the YaeT21:359 structure using MAD techniques as described in Experimental Procedures. In spite of the modest resolution, density modification by solvent flipping dramatically improved the electron density map, probably due to the large solvent content of these crystals (approximately 70%). The resulting map was readily interpretable and allowed modeling of most residues in the first four POTRA domains (Supplemental Figure 2A). The model was then refined against the YaeT21:410 native data set to attempt modeling of the additional amino acids present in this construct. However, only residues 346-349 corresponding to the beginning of POTRA5 could be modeled. This was attributed to conformational flexibility due to POTRA5 being incomplete and, therefore, unfolded. The final structure contains residues 23 through 349 comprising all residues in the first four POTRA domains and the first three residues of POTRA5. Data collection and refinement statistics are shown in Table 1, and an example of unbiased electron density is shown in Supplemental Figure 2. This structure of the YaeT periplasmic domain was determined independently and from a different crystal form than the one recently reported by Kim et al (Kim et al., 2007).
Table 1.
Data collection, phasing and refinement statistics
| Native YaeT21:410 | Se-Met YaeT21:359 | |||
|---|---|---|---|---|
| Data Collection Statistics | ||||
| Space Group | P3121 | P3121 | ||
| Cell Parameters: | ||||
| a=b (Å) | 92.51 | 92.36 | ||
| c (Å) | 142.43 | 142.13 | ||
| α=β (°) | 90.00 | 90.00 | ||
| γ (°) | 120.0 | 120.0 | ||
| Peak | Inflection | Remote | ||
|
|
||||
| Wavelength (Å) | 1.00 | 0.9796 | 0.9797 | 0.9643 |
| Resolution (Å)a | 50.0-3.3 (3.42-3.3) | 50.0-3.3 (3.42-3.3) | 50.0-3.3 (3.42-3.3) | 50.0-3.3 (3.42-3.3) |
| Rsymb (%) | 5.6 (42.4) | 9.7 (44.2) | 9.6 (42.7) | 9.7 (44.1) |
| I / σ | 20.6 (2.3) | 14.5 (2.0) | 14.3 (2.1) | 14.0 (2.1) |
| Data Completeness (%) | 99.6 (99.9) | 99.4 (95.9) | 99.4 (95.9) | 99.5 (97.1) |
| Redundancy | 3.5 (3.6) | 4.0 (3.3) | 4.0 (3.3) | 4.0 (3.3) |
| Phasing | ||||
| R.m.s. FH/εc | 0.8 | 0.6 | 0.8 | |
| FOM before DMd | 0.39 (0.18) | |||
| FOM after DM | 0.64 (0.26) | |||
| Refinement Statistics | ||||
| Resolution (Å) | 47.5 – 3.3 (3.45 – 3.3) | |||
| No. reflections | 11004 | |||
| No. atoms | 2514 | |||
| Rwork | 26.7 (36.7) | |||
| Rfreee | 29.5 (42.1) | |||
| Mean B-value (Å2) | 95.5 | |||
| RMS dev. bonds (Å) | 0.009 | |||
| RMS dev. angles (°) | 1.4 | |||
| Ramachandran statistics: | ||||
| Residues in favored region (%) | 67.4 | |||
| Residues in allowed region (%) | 32.6 | |||
Values in parentheses are for the highest-resolution shell.
Rsym=ΣhΣi |(Ii(h)-< I(h)>|/ΣhΣI Ii(h), where Ii(h) is the I-th measurement of reflection h, and <I(h)> is the weighted mean of all measurements of h. Bijvoet measurements were treated independently for the MAD phasing data sets.
R.m.s FH/εc = (1/n Σ FH2)1/2 / (1/n Σ ε2)1/2 where FH is the structure factor amplitude for anomalous scatterers and ε is the lack of closure expression for each wavelength in the MAD dataset.
FOM before DM indicates the Figure Of Merit before Density Modification
Rwork = Σ|Fobs-Fcalc|/ΣFobs Where Fobs = observed structure factor amplitude and Fcalc = structure factor calculated from model. Rfree is computed in the same manner as Rcryst, using the test set of reflections.
Each of the four POTRA domains in the structure has the characteristic POTRA fold comprising two α-helixes packed against a three-strand mixed β-sheet, as previously reported (Kim et al., 2007) (Figure 1A). Despite low sequence conservation, the four POTRA domains are structurally well conserved, with POTRA2, 3 and 4 superimposing on POTRA1 with root-mean square deviations (RMSDs) of 1.35, 1.62 and 1.79 Å respectively (Figure 1B). Some minor conformational differences are visible in the α1 helix and in loop regions, most pronounced in loop4 (L4, Figure 1B). Nevertheless, some specific differences are noteworthy: (1) POTRA1 contains a 3 amino acid deletion in L3 between α2 and β2 (Figure 1B, green); and (2) POTRA3 is unique, with a 10 amino acid insertion in L2 (between α1 and α2), and the presence of a β-bulge in β2 (Figure 1B, yellow). The tandem arrangement of POTRA domains adopts a rather extended, cane-shaped conformation in these crystals (Figure 1C). With approximate dimensions of 100×50Å, the structure is dramatically different from the much more compact arrangement reported by Kim et al (Kim et al., 2007).
Figure 1. Overall structure of YaeT periplasmic domain.
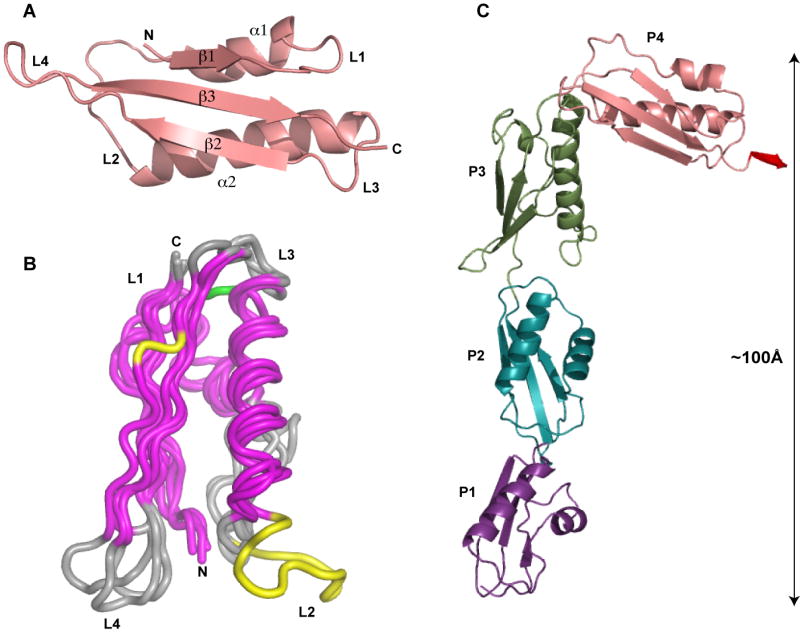
(A) Representative structure of a POTRA domain with secondary structure elements labeled as follows: α - alpha helix, β - beta strand, L – loop, N- amino terminus, C – carboxy terminus. (B) Superposition of the individual POTRA domains. Conserved secondary elements between the POTRA domains are colored in magenta. Loop3 (L3) of POTRA1 containing a 3 amino acid deletion is colored in green; Loop2 (L2) of POTRA3 containing a 10 amino acid insertion, and a β-bulge in β2 of POTRA3 are colored in yellow. (C) Cartoon representation of YaeT21:359. Individual domains are colored as follows: POTRA1 – purple; POTRA2 – cyan; POTRA3 – green; POTRA4 – salmon; the small extension containing residues 345-349 from POTRA5 is colored red.
Conformational Flexibility of the YaeT Periplasmic Domain
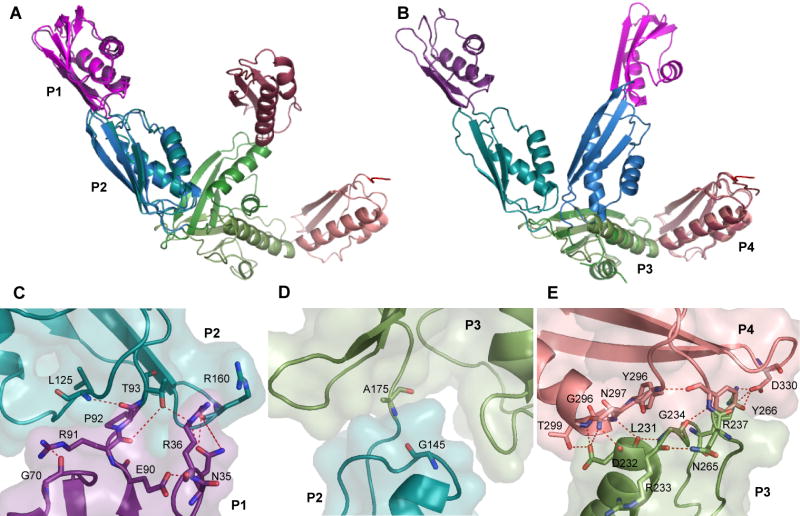
Comparison of the YaeT periplasmic domain structure presented here and the one reported by Kim et al. reveals that, with the exception of POTRA3 (see below), the individual POTRA domains superimpose well between the two structures with RMSDs between 0.6 and 0.8 Å. However, the overall conformation is quite different. A superposition of both structures onto either the first two (Figure 2A) or the last two POTRA domains (Figure 2B) shows that the reason for this difference is in the degree of bending in the connection between POTRA2 and POTRA3. Quantitatively, the angle between POTRA2 and POTRA3 (assessed between the β3 strands in each domain) is approximately 130° in the structure presented here compared to 100° in the structure solved by Kim et al (Kim et al., 2007).
Figure 2. Conformational flexibility of the YaeT periplasmic domain.
(A-B) Superposition of the structure of YaeT21:359 presented here with that of YaeT21-351 determined by Kim et al. (PDB: 2qdf). The two structures are superimposed on POTRA1 and 2 (A) or POTRA3 and 4 (B). The color scheme for YaeT21:359 is the same as in Figure 1. The color scheme for Kim’s structure is as follows: POTRA1 – magenta; POTRA2 – blue; POTRA3 – dark green; POTRA4 – raspberry. (C-E) Interfaces between POTRA domains 1 and 2 (C); 2 and 3 (D) and 3 and 4 (E). Interacting residues are shown as sticks and secondary structure elements are shown as cartoon representation. A semitransparent surface representation is shown to highlight the extent of surface interaction between the domains.
Several inter-domain interactions stabilize the connection between POTRA1 and 2 (Figure 2C and Supplemental Table1) resulting in a relatively large interface and a conformation conserved between the two structures (Figure 2A). Likewise, the interface between POTRA3 and 4 is fairly extensive and stabilized by a number of hydrophobic contacts and hydrogen bonding interactions (Figure 2E and Supplemental Table1) leading to a conformation that superimposes well between the two structures (Figure 2B). In contrast, a small interface with little inter-domain interaction is observed between POTRA2 and 3 (Figure 2D). The linker between these two domains appears to be a hinge point that affords flexibility to the periplasmic domain of YaeT. Interestingly, the bent conformation observed in the Kim et al. structure is stabilized by a crystal packing interaction whereas a different lattice environment results in the extended conformation we report here. This extended conformation is consistent with recent Small Angle X-ray Scattering (SAXS) data obtained for the periplasmic domain of YaeT in solution (Knowles et al., 2008).
Unique Characteristics of POTRA3 in YaeT
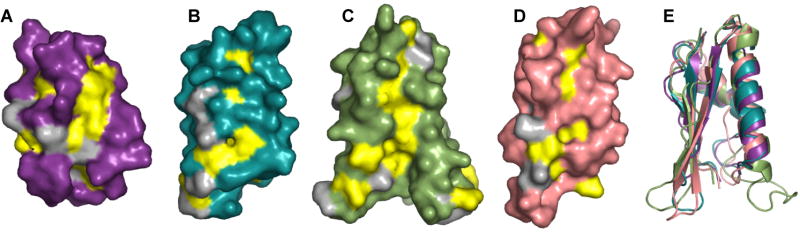
Despite the structural similarity among POTRA domains, POTRA3 contains several unique features. It has been proposed that binding of OMPs to YaeT may occur at the edge of the POTRA’s β-sheet by a process called β-augmentation (Kim et al., 2007). We notice a surface groove located between the β-sheet and the long helix α2 that is deeper and more hydrophobic in POTRA3 than in the other POTRA domains (Figure 3). Despite relatively low sequence conservation in bacterial Omp85 proteins, the hydrophobic character of the residues lining the groove is conserved (supplemental data Figure 1). Furthermore, the groove is approximately 30Å in length, which is comparable to the average height of an OM β-barrel protein (Tamm et al., 2004). This suggests that the groove may represent a binding site for OMPs before they are inserted into the outer membrane.
Figure 3. Conserved hydrophobic patches in the four POTRA domains.
Surface representations of individual POTRA domains in the same orientation: (A) POTRA1, (B) POTRA2, (C) POTRA3, (D) POTRA4. Hydrophobic residues are colored in grey and residues conserved as hydrophobic between gram-negative bacteria (see Supplemental Figure 1) are colored in yellow. Other residues in each POTRA domain are colored as in Figure 1. (E) Cartoon representation showing a superposition of the four POTRA domains oriented as in (A-D).
In their structure determination, Kim and co-workers noted that the C-terminal tail of the crystallized construct was bound to the POTRA3 β-sheet of a neighboring molecule in the crystal lattice (Kim et al., 2007). This tail adopted a β-strand conformation extending the β-sheet by what is called “β-augmentation” (Harrison, 1996) (Figure 4A). They proposed that this “β-augmentation” might represent a form of substrate binding to YaeT (Kim et al., 2007). Despite the different crystallization conditions and completely different crystal packing, we also observe β-augmentation of the POTRA3 β-sheet by the C-terminal tail of a neighboring molecule (Figure 4B). Interestingly, the additional strand is in an antiparallel orientation in our structure rather than the parallel arrangement observed by Kim et al (Figure 4A and B).
Figure 4. Unique Features in POTRA3.

(A) Parallel β-augmentation of the β-sheet from POTRA3 with short β-strand from POTRA5 (red) belonging to the second copy of YaeT in the asymmetric unit as observed in the structure of YaeT21-351 determined by Kim et al. (Kim et al., 2007). (B) Anti-parallel β-augmentation of the β-sheet from POTRA3 with the β-strand of a symmetry-mate containing the last few residues of YaeT21:359 (bright red). (C) Comparison of POTRA3 in the present structure (light green) and that of Kim et al. (dark green and magenta).
POTRA3 also contains a β-bulge consisting of residues I240 and D241 in the β2 strand (Figure 1B, yellow) (Kim et al., 2007). This feature is not present in the other POTRA domains. Mutational analyses have shown that the position of D241 is important for binding of YfgL, one of the lipoproteins in the E. coli YaeT complex (Kim et al., 2007). Therefore, the unique β-bulge in POTRA3 may be important for stabilization of the YaeT complex.
The L2 loop between the α1 and α2 helices is 10 amino acids longer in POTRA3 compared to the other POTRA domains. We were able to unambiguously trace the residues in this region, which was incompletely modeled in the structure previously solved by Kim et al (Kim et al., 2007). Furthermore, superposition of the two structures shows markedly different conformations for the L2 region (Figure 4C). Interestingly, the two structures diverge at a point in the α2 helix previously identified by mutational analysis as important for YaeT function. An insertion of two amino acids between K218 and L219 was the only YaeT mutant that produced a phenotype in the E. coli chemical conditionality tests and displayed defects in OMP assembly (Ruiz et al., 2005; Wu et al., 2005).
In our structure, the α2 helix is 5.5 turns long and the loop is close to the β-sheet in a “closed” conformation (light green in Figure 4C). In contrast, the structure determined by Kim et al, shows that the α2 helix is two turns shorter and the L2 loop is splayed outwards in what may represent an “open” conformation (dark green and magenta in Figure 4C). The loop is also incompletely traced, presumably due to conformational flexibility. We note, however, that this area is involved in crystal contacts in the Kim et al. structure, which might affect its conformation.
Discussion
Outer-membrane proteins (OMPs) are synthesized in the cytoplasm and need to travel through both the inner membrane (IM) and the periplasm to reach their final destination in the outer membrane. The process of IM translocation is well described, and advances have been made in understanding the chaperone-assisted transport of OMPs through the periplasm (Bitto and McKay, 2003; Mogensen and Otzen, 2005; Sklar et al., 2007b; Walton and Sousa, 2004). However, little is known about the mechanisms of OMP targeting, delivery and insertion into the OM. Nevertheless, the Omp85 family of proteins has been identified as a crucial player in the outer membrane insertion process both in bacteria, and in double-membrane eukaryotic organelles such as mitochondria and choloroplasts (Gentle et al., 2004; Gentle et al., 2005; Ruiz et al., 2006; Voulhoux and Tommassen, 2004). In E. coli the Omp85 family is represented by YaeT, which together with the lipoproteins YfgL, NlpB, YfiO and SmpA, forms the YaeT complex (Wu et al., 2005).
All bacterial Omp85s have a membrane embedded C-terminal β-barrel domain and a periplasmic domain containing five POTRA domains (Gentle et al., 2005; Voulhoux and Tommassen, 2004). This periplasmic domain is essential for the in vivo function of YaeT (Bos et al., 2007b; Kim et al., 2007; Stegmeier and Andersen, 2006). It has been suggested, that the POTRA domains may be involved in reception and partial folding of OMPs, perhaps with the assistance of periplasmic folding catalysts, prior to insertion in the OM (Bos and Tommassen, 2004). We have solved the structure of a fragment of YaeT containing the first four POTRA domains. The construct used to refine the structure contains additional C-terminal residues that correspond to a portion of the POTRA5 constituting a C-terminal “tail” that could be unambiguously modeled in the electron density.
The residues of the C-terminal tail adopt a β-strand conformation and interact with the POTRA3 domain of a neighboring molecule in the crystal lattice augmenting its β-sheet. The β-sheet augmentation of POTRA3 is evident not only in our structure, but also in the structure previously solved by Kim et al, who first proposed β-augmentation as the mechanism underlying OMP binding to YaeT (Kim et al., 2007). Interestingly, there seems to be no directionality requirement for this β-augmentation. The additional β-strand binds to the β-sheet of POTRA3 in an antiparallel orientation in our structure, whereas it is in a parallel arrangement in the structure determined by Kim et al. The fact that β-augmentation of POTRA3 is observed in both structures despite the different crystallization conditions and unrelated crystal packing, together with the tolerance for alternative orientations of the β-strand, further support the hypothesis of β-augmentation as a form of substrate binding by YaeT. This mechanism would be relatively insensitive to substrate sequence, consistent with the YaeT role in insertion of a large variety of OMPs. Furthermore, it would promote formation of β-strands in the substrates and may represent a required step in OMP folding into β-barrels.
Running parallel to the β-sheet where β-augmentation is observed, POTRA3 contains a large surface groove lined with hydrophobic residues. The hydrophobic character of this groove is conserved in bacterial Omp85 proteins (see Figure1 in supplemental information). Polypeptides bound by β-augmentation project their side chains across this groove, which may serve to accommodate the non-polar side chains expected in membrane protein substrates. The 30Å size of this groove is well suited to accommodate the β-strands of the substrate OMP barrels as they usually are 27 to 35Å in length (Tamm et al., 2004).
The POTRA3 domain of YaeT appears strikingly similar to SecB—the cytoplasmic chaperone that binds newly synthesized OMP precursors before translocation across the inner membrane (Figure 5). Analogous to YaeT, SecB has a 30Å long hydrophobic groove at the edge of its β-sheet and binds peptides by β-augmentation (Xu et al., 2000). It has been proposed that the hydrophobic groove in SecB is composed of two subsites. Subsite1 located at the top of the groove, contains mostly aromatic residues, whereas Subsite2 is composed of mostly hydrophobic (but not aromatic) conserved residues (Xu et al., 2000). Despite having no sequence homology, similar traits are observed in the YaeT POTRA3 domain (Figure 5). These similarities and the fact that SecB and YaeT share the same family of substrates (OMPs) further support a role for the POTRA3 domain in the mechanism of YaeT recognition of substrate OMPs.
Figure 5. Comparison of the structures of YaeT POTRA3 and SecB.

Cartoon representations of SecB (PDB: 1fx3) (A) and YaeT POTRA3 (B). Aromatic residues lining the top of a hydrophobic groove (Subsite1) are highlighted in brown-red. Hydrophobic (but not aromatic) residues forming extended hydrophobic groove (Subsite2) are shown in light magenta.
The periplasmic domain of YaeT appears to have a hinge point in the linker connecting POTRA2 and 3. A comparison of the structure obtained here with the one previously determined by Kim et al., shows that the structures have two conformationally conserved “arms”. One formed by POTRA domains 1 and 2 and the other formed by POTRA3 and 4. Recent NMR data collected on a YaeT fragment containing POTRA 1 and 2 suggested that the connection between these two domains is also quite flexible (Knowles et al., 2008). However, this suggestion was made based on the lack of observable NOEs between the two domains under the experimental conditions. The conservation in the orientation of these two domains in the crystal structures despite the differences in crystal lattices, together with the extent and conservation of the interactions at the interface between the two domains imply that the connection between POTRA 1 and 2 is relatively rigid.
A flexible linker joins the two arms in the periplasmic domain of YaeT and behaves as a hinge that allows the protein to adopt the distinct conformations observed in the two crystal structures. The extended conformation reported here is consistent with recent SAXS data showing that an extended arrangement of POTRA domains is favored in solution (Knowles et al., 2008). The degree of bending in the periplasmic domain of YaeT may be modulated by the interactions and conformation of the unique L2 loop in POTRA3. This loop is 10 amino acids longer than the L2 loop in the other POTRA domains and mutations adjoining the loop interfere with YaeT function (Wu et al., 2005). Moreover, the different bends in the periplasmic domain of YaeT observed in the structures correlate with dramatic differences in the conformation of the L2 loop in POTRA3. Therefore, binding of substrates or other factors to this loop may promote specific conformations in YaeT during the OMP binding and insertion cycle. Insertion of β-barrels into the membrane is thought to occur via β-hairpins (Tamm et al., 2004). We speculate, that bending of the periplasmic domain of YaeT may assist the creation of β-hairpins in OMPs after individual β-strands are formed on POTRA domains by β-augmentation. Consistent with this hypothesis, peptides can bind as β-strands to POTRA3 as described above, and recent NMR data suggest that peptides derived from OMP β-barrels also bind “at the edges” of the β-sheets in POTRA1 and 2 (Knowles et al., 2008). Along the same lines, the differences in hydrophobic character of the surface groove adjoining the β-sheet in each POTRA domain would help accommodate the diverse sequences present in OMP β-barrels.
Strains of E. coli expressing YaeT mutants missing POTRA 1 or 2 individually can survive the depletion of wild type YaeT. However, these strains grow poorly and display a strong stress response, dramatically increasing the expression of DegP, a chaperone-protease known to degrade OMP aggregates in the periplasm (Kim et al., 2007). It is interesting to note that the length of the periplasmic domain of YaeT, in the extended conformation presented here, and despite missing POTRA5, is comparable to the TolC α-helical domain (Koronakis et al., 2000). This opens the possibility of YaeT spanning the periplasmic space and reaching the outer leaflet of the inner membrane, as proposed for TolC (Koronakis et al., 2004). Consistent with this possibility, the presence of five POTRA domains is conserved in all Gram-negative bacteria (Gentle et al., 2005). On the other hand, only the POTRA5 of Omp85 is essential for viability of Neisseria meningitidis, and deletion of the first four POTRA domains is relatively well tolerated (Bos et al., 2007b). Neisseria is unique among LPS-producing Gram-negative bacteria as it is the only one not requiring LPS or capsular polysaccharide for viability (Bos and Tommassen, 2005). In addition, Omp85 is thought to be homo-oligomeric in Neisseria (Robert et al., 2006) whereas it appears to be monomeric, albeit in complex with lipoproteins, in E. coli (Kim et al., 2007; Wu et al., 2005). This points to possible differences in the OMP biogenesis pathway between Neisseria and other Gram-negative bacteria represented by E. coli.
Experimental Procedures
Cloning of YaeT N-terminal Domain Constructs
The E. coli YaeT N-terminal domain (residues 21-359 or 21-410) was amplified by polymerase chain reaction (PCR) from genomic E. coli DNA with primers introducing NcoI and XmaI restriction sites. The PCR amplification was performed with AccuPrime Pfx DNA polymerase (Invitrogen) according to manufacturer’s instructions. The NcoI/XmaI digested gene was purified and then ligated into the pMS174 vector (an engineered variant of the pET28 vector that generates an N-terminal His-tag fusion that can be efficiently and specifically cleaved with the Tobacco etch virus (TEV) protease). The resulting plasmids, pMS436 (YaeT21:359) or pMS331 (YaeT21:410), were sequenced to confirm that no random mutations had been introduced.
Protein Expression and Purification of YaeT21:410
The plasmid pMS331 (YaeT21:410) was transformed into E.coli Rosetta (DE3) cells (Novagen). Overnight culture from a single colony containing 50 μg/ml kanamycin was used to inoculate 2 × 1L LB medium supplemented with 50 μg/ml kanamycin. Cultures were grown at 37°C to OD600 of 0.6. Expression was induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG, Gold Bio Technology Inc.) and cultures were allowed to grow an additional 3h at 37°C. Cells were harvested by centrifugation, and the cell pellet re-suspended in lysis buffer containing 25 mM Tris-Cl pH 8.0, 1mM EDTA, 300mM NaCl, 10mM imidazole (pH8.0) and Complete EDTA-free protease inhibitor (Roche). Cells were lysed on ice by sonication. Removal of cell debris was achieved by centrifugation at 16,000 rpm for 20 min at 4°C. The supernatant was applied to a Ni-NTA column (Qiagen) pre-equilibrated with lysis buffer. The protein bound to Ni-NTA beads was washed with 1 column volume of lysis buffer, followed by a wash with a buffer containing 20mM imidazole. The protein was eluted with buffer containing 250mM imidazole. Fractions containing the protein were incubated with TEV protease to achieve cleavage of the His tag. After removal of the tag and the TEV protease (which is also 6His tagged) using Ni-NTA beads, the protein was loaded on a size exclusion (HiLoad 26/60 Superdex 200, Amersham Pharmacia Biotech) column pre-equilibrated with 25 mM Tris-Cl pH 8.0, 150 mM NaCl and eluted in the same buffer. The protein eluted as a monomer from the size exclusion column.
Protein Expression and Purification of Se-Methionine labeled YaeT21-359
The plasmid pMS436 (YaeT21:359) was transformed into E.coli Rosetta (DE3) cells (Novagen). A 50 ml culture from single colony containing 50 μg/ml kanamycin was grown overnight at 37°C. Four 10 ml aliquots were taken, spun down to remove LB media and re-suspended in 10 ml M9 minimal media supplemented with 50 μg/ml kanamycin. The re-suspended bacterial pellets were used to start 4 × 1 L cultures in M9 minimal media containing 50 μg/ml kanamycin and grown at 37°C to an OD600 ~ 0.6. At that point methionine synthesis was inhibited by adding 100 mg/l D-lysine, D-phenylalanine, and D-threonine; 50 mg/l D-isoleucine and D-valine. In addition, 60 mg/l D/L selenomethionine (Se-Met, Sigma) was added. Protein expression was induced with 0.4 mM IPTG and cells were grown for additional 2.5 hrs at 37°C. Cells were harvested by centrifugation and the protein was purified as described above for YaeT21:410.
Protein Crystallization and Data Collection
Crystals of the YaeT N-terminal constructs were grown by the hanging drop method of vapor diffusion at 16°C (protein : precipitant 1.5 μl : 1.5 μl). The precipitant was 1.3-1.6M (NH4)2SO4, 3-6% polyethylene glycol 400, 10% Dioxane, 0.1M MES pH 6.5. Prior to X-ray data collection, the crystals were transferred to a cryo-protecting solutions composed of mother liquor without added Dioxane and containing 20% glycerol, and flash cooled in nitrogen stream.
Data collection on Se-Methionine YaeT21-359 and native YaeT21-410 was performed at the Advanced Light Source in Lawrence Berkeley National Laboratory. Data were indexed and integrated with DENZO and scaled with SCALEPACK (Otwinowski and Minor, 1997). X-ray data collection statistics for both sets are shown in Table 1.
Structure Determination and Refinement
Crystals of both YaeT21:410 and YaeT21:359 belong to space group P3121 and are isomorphous with cell dimensions described in Table 1. The Selenium peak wavelength was used to search for heavy atom sites using the program SOLVE (Terwilliger, 2003) as implemented in PHENIX (Adams et al., 2002). Three sites were identified and initial phases calculated from these sites were improved by density modification using RESOLVE/PHENIX. The resulting electron density map was readily interpretable and used to build most of the first four POTRA domains of YaeT using the program O (Jones, 1978). Iterative cycles of refinement in PHENIX followed by manual rebuilding in O were carried out until no further improvement of the Rfree factor was observed. The final model contains residues 21 to 349. Phasing and refinement statistics are summarized in Table1. It is worth noting that residues 350-410 are not modeled, presumably because they are unfolded. However, these residues contribute to the scattering resulting in the relatively high R factors observed in refinement. Atomic coordinates and structure factors have been deposited in the PDB with accession number 3EFC.
Supplementary Material
Acknowledgments
We thank Corie Ralston and Anthony Rozales at the Advanced Light Source for their help during data collection. Structural biology research at the University of Colorado at Boulder is supported in part by the William M Keck Foundation. This work is based on research performed at the ALS, which is funded by the Department of Energy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystal Sect D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bitto E, McKay DB. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Ann Rev Microbiol. 2007a;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007b;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Curr Opin Microbiol. 2004;7:610–616. doi: 10.1016/j.mib.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Bos MP, Tommassen J. Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect Immun. 2005;73:6194–6197. doi: 10.1128/IAI.73.9.6194-6197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Peptide-surface association: the case of PDZ and PTB domains. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- Jones A. A Graphics Model Building and Refinement System for Macromolecules. J Appl Cryst. 1978;11:268–272. [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Ann Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Schulz GE. Transmembrane beta-barrel proteins. Adv Protein Chem. 2003;63:47–70. doi: 10.1016/s0065-3233(03)63003-2. [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007a;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007b;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier JF, Andersen C. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J Biochem (Tokyo) 2006;140:275–283. doi: 10.1093/jb/mvj147. [DOI] [PubMed] [Google Scholar]
- Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzym. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol. 2004;155:129–135. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Xu Z, Knafels JD, Yoshino K. Crystal structure of the bacterial protein export chaperone secB. Nature Struct Biol. 2000;7:1172–1177. doi: 10.1038/82040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.