Abstract
Proteins that directly regulate TNFR signaling play critical roles in regulating cellular activation and survival. A20 Binding and Inhibitor of NFkB (ABIN-1) is a novel protein that is thought to inhibit NFkB signaling (1, 2). Here we show that mice deficient for ABIN-1 die during embryogenesis with fetal liver apoptosis, anemia and hypoplasia. ABIN-1 deficient cells are hypersensitive to TNF-induced PCD, and TNF deficiency rescues ABIN-1 deficient embryos. ABIN-1 inhibits caspase 8 recruitment to FADD in TNF-induced signaling complexes, preventing caspase 8 cleavage and PCD. Moreover, ABIN-1 directly binds polyubiquitin chains and this ubiquitin sensing activity is required for ABIN-1’s anti-apoptotic activity. These studies provide new insights into how ubiquitination and ubiquitin sensing proteins regulate cellular and organismal survival.
To investigate ABIN-1’s potential roles in regulating TNF signals, we targeted the gene that encodes ABIN-1, tnip1, in embryonic stem cells to generate mice deficient for ABIN-1 (Suppl. Fig. 1A–1C). Deletion of exons 12 through 15 of tnip1 eliminated the full length (approximately 80 kD) ABIN-1 protein (Suppl. Fig. 1D). While ABIN-1+/− mice appear normal, very few live born ABIN-1−/− mice were obtained from intercrossed ABIN-1+/− parents (Table 1). ABIN-1−/− embryos were found in Mendelian ratios at post coital days E12.5 through E18.5, but were only rarely obtained as live born mice (Table 1).
Table 1.
ABIN-1 is required for regulating TNF signals in utero.
| ABIN-1 | genotype | |||||
|---|---|---|---|---|---|---|
| TNF genotype | Age | +/+ | +/− | −/− | −/− (%) | Total # |
| E15.5 pc | 12 | 32 | 17 | (27.9%) | 61 | |
| E16.5 pc | 6 | 29 | 11 | (23.9%) | 46 | |
| TNF +/+ | E18.5 pc | 11 | 16 | 11 | (28.9%) | 38 |
| newborn | 12 | 28 | 1 | (2.4%) | 41 | |
| 3–4 week | 37 | 92 | 4 | (3.0%) | 133 | |
| TNF +/− | live born | 19 | 28 | 4 | (7.8%) | 51 |
| TNF +/− | live born | 27 | 61 | 22 | (20%) | 110 |
Numbers of embryos or live born pups of the indicated genotypes at various stages of development obtained from ABIN-1+/− heterozygote intercrosses are shown. Total embryos/pups genotyped at each stage of development for each TNF genotype is indicated at right (total). The percentage of embryos that were homozygous deficient (ABIN-1−/−) among specific TNF genotypes and at specific stages of development are indicated in parentheses.
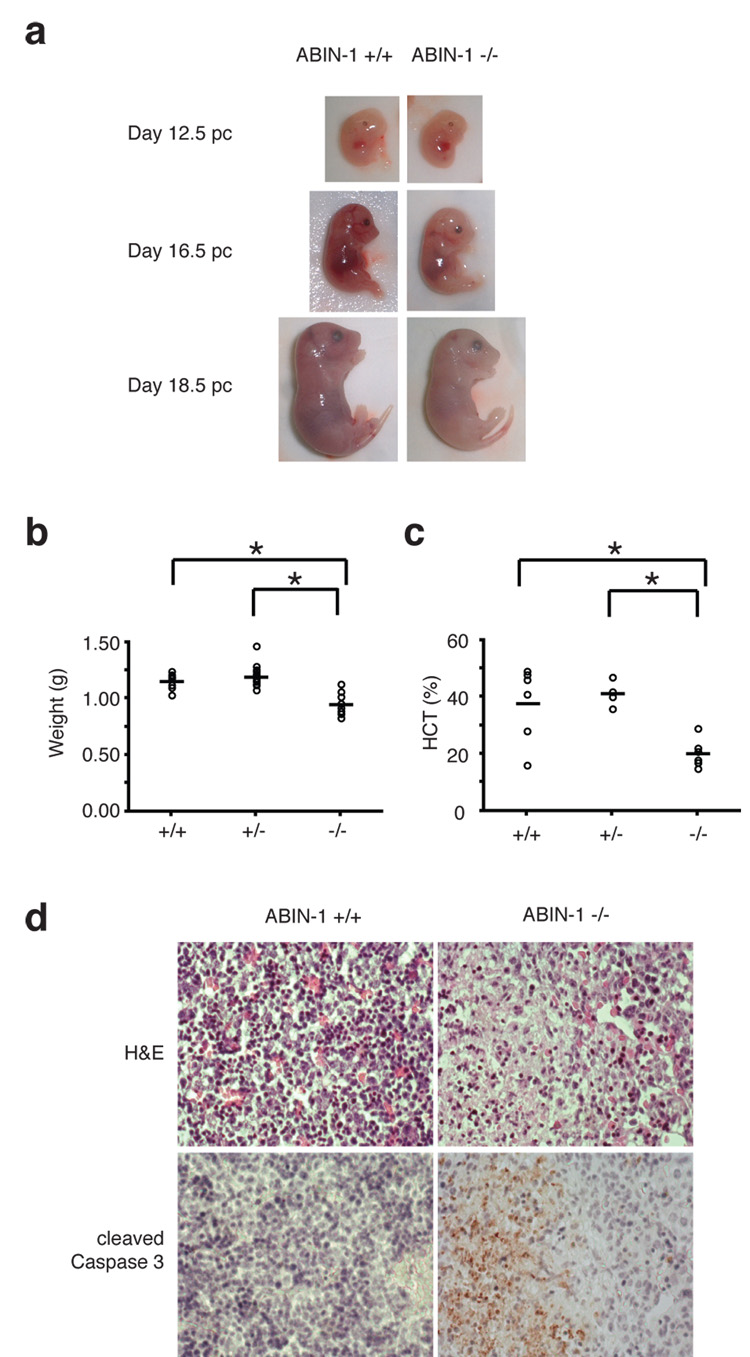
To better understand how ABIN-1 sustains embryonic development, we noted that ABIN-1−/− embryos were smaller and paler than ABIN-1+/+ littermates at E16.5 and E18.5 (Fig. 1A). Quantitation of fetal weights and hematocrits of these embryos revealed that ABIN−/− embryos were significantly smaller and anemic compared to control embryos (Fig. 1B, 1C). Histological analyses of multiple tissues at various stages of development revealed that fetal livers of E16.5 ABIN-1−/− embryos contained multiple areas of hypocellularity (Fig. 1D). Immunohistochemical analyses of caspase 3 cleavage indicated that these areas contained large number of apoptotic cells that were not detected in control embryos (Fig. 1D). Thus, ABIN-1 is critical for preventing fetal liver apoptosis and sustaining fetal hematopoiesis.
Figure 1. ABIN-1 is required for embryonic development.
(A) Gross appearance of ABIN-1+/+ and ABIN-1−/− embryos. (B) Hypoplasia of E18.5 ABIN-1−/− embryos. Weights of individual embryos are shown as circles; horizontal bars indicate mean weights for each genotype. ABIN-1−/− embryos weigh less than ABIN-1+/+ and ABIN-1+/− control embryos (p < 0.01 between ABIN-1+/+ and ABIN-1−/− embryos; and p < 0.01 between ABIN-1+/− and ABIN-1−/− embryos, n=35). (C) Anemia of E18.5 ABIN1−/− embryos. ABIN-1−/− embryos are markedly anemic (p < 0.02 between ABIN-1+/+ and ABIN-1−/− embryos; p < 0.01 between ABIN-1+/− and ABIN-1−/− embryos, n=18). (D) Hematoxilin and eosin (H&E) histology (upper panels) and cleaved caspase 3 immunohistochemistry (lower panels) of sequential sections from ABIN-1+/+ and ABIN-1−/− fetal livers. Apoptotic patches were found in 5 of 7 ABIN-1−/− fetal livers analyzed and in none of 10 ABIN-1+/− and ABIN+/+ fetal livers analyzed (p < 0.01).
The failure to protect embryonic fetal livers from TNF-induced PCD prevents successful development of mice (3–6). We thus interbred ABIN-1+/− mice with TNF−/− mice to determine whether ABIN-1 supports embryonic development by regulating TNF-induced signals. Genotyping of live born mice from ABIN-1+/− TNF+/− intercrosses revealed that ABIN-1−/− animals were obtained in roughly Mendelian ratios when animals were homozygous TNF−/− mice (Table 1). Therefore, ABIN-1 sustains embryogenesis by regulating TNF signals, possibly by protecting cells against TNF-induced PCD.
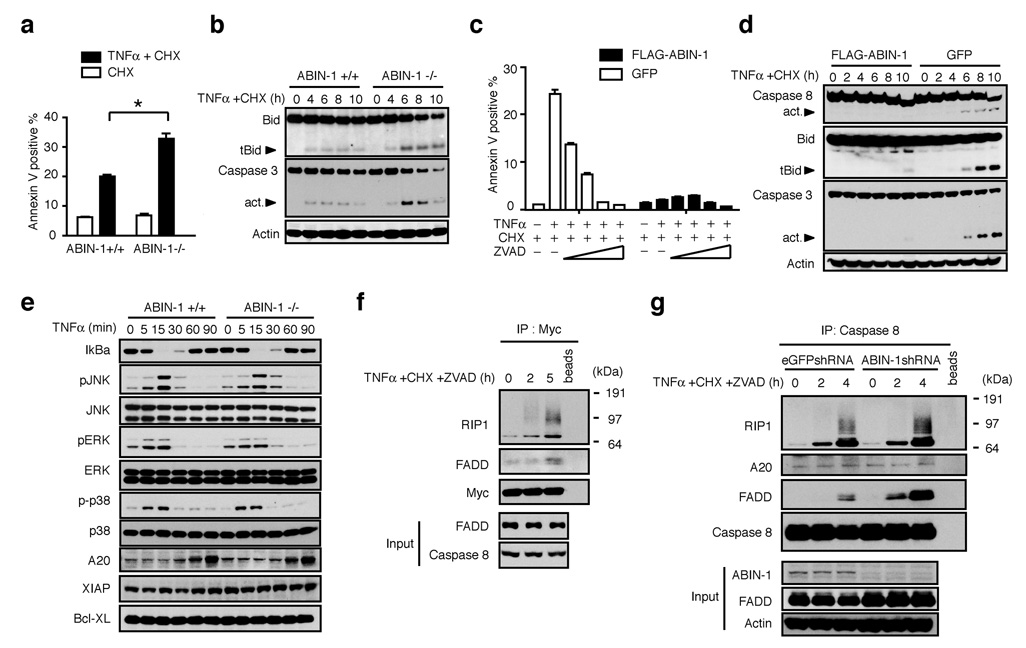
To determine whether ABIN-1 directly protects cells from TNF-induced PCD, we tested the susceptibility of ABIN-1 deficient Jurkat T cells, HepG2 hepatoma cells and HT1080 fibrosarcoma cells as well as embryonic fibroblasts (MEFs) to undergo PCD. ABIN-1 deficient cells and ABIN-1−/− MEFS were more sensitive to TNF-induced PCD in the presence or absence of CHX (Fig. 2A, Suppl. Figs. 2A, 2B, 2C). As ABIN-1 is expressed in unstimulated cells, we performed subsequent experiments in the presence of CHX to distinguish ABIN-1’s anti-apoptotic functions from other translation dependent anti-apoptotic mechanisms. ABIN-1 deficient cells exhibited more caspase 8, caspase 3 and Bid cleavage when compared to control cells after treatment with TNF plus CHX (Fig. 2B and Suppl. Fig. 2D). PCD in ABIN−/− cells was blocked by the caspase inhibitor ZVAD (Fig. 2C), confirming that increased PCD in these cells was caspase mediated. Finally, GFP-FLAG-ABIN-1 protected ABIN-1−/− cells from TNF-induced PCD and prevented caspase 8, Bid and caspase 3 cleavage (Fig. 2C, 2D). These results are consistent with a prior suggestion that ABIN-1 can protect hepatocytes from TNF-induced PCD (8). Our experiments suggest that ABIN-1 directly protects multiple cell types against TNF-induced PCD.
Figure 2. ABIN-1 is required for protecting cells from TNF-induced PCD.
(A) TNF-induced PCD of ABIN-1+/+ and ABIN-1−/− MEF (p < 0.01 between ABIN-1+/+ and ABIN-1−/− cells; means and standard deviations indicated, n=3). (B) TNF-induced caspase-3 and BID expression in ABIN-1+/+ (left) and ABIN-1−/− (right) MEFs. Cleaved, active forms of Bid (tBid) and caspase 3 indicated by arrows. Actin expression shown below as protein loading control. (C) TNF-induced PCD of ABIN-1−/− 3T3s expressing GFP and FLAG-ABIN-1 (black columns on right) or GFP alone (white columns on left), after treatment with the indicated agents. Means and standard deviations indicated, n=3. (D) TNF-induced caspase-8, caspase 3, and BID expression in ABIN-1−/− 3T3s expressing GFP and FLAG-ABIN-1 (left) or GFP alone (right). Cleaved, active forms of caspase 8, caspase 3 and Bid (tBid) indicated by arrows. Actin expression shown below as control. (E) TNF-induced expression of MAP kinase signaling proteins and A20 in ABIN-1−/− and control MEFs. (F) TNF-induced recruitment of RIP1 and FADD to Myc (ABIN-1). Expression of Myc-ABIN-1 in IPs, and expression of FADD and caspase 8 in whole cell lysates (“input”) shown below as controls. (G) TNF-induced recruitment of RIP1, FADD and A20 to caspase 8 in ABIN-1 deficient and control HT1080 cells. Expression of caspase 8 in IPs and expression of ABIN-1, FADD and actin in whole cell lysates (“input”) shown below as controls. All data are representative of 3–5 independent experiments.
Cells that exhibit deficient NFkB signaling are hypersensitive to TNF-induced PCD (4–6). NFkB signaling was not decreased in ABIN-1−/− cells compared to control cells after TNF treatment (Fig. 2E, Suppl Figs. 3A, 3B, 3C). Levels of NFkB dependent survival proteins such as Bcl-xL, XIAP and A20 were expressed at normal levels in ABIN-1−/− cells (Fig. 2E). Prolonged JNK signaling has also been associated with exaggerated PCD after TNF treatment (9–11). However, ABIN-1−/− MEFs and ABIN-1+/+ MEFS exhibited similar kinetics of phospho-JNK activation after TNF treatment, as well as normal p38 and ERK signaling (Fig. 2E). These experiments suggest that ABIN-1 directly protects cells against TNF-induced PCD, and this function is not secondary to aberrant MAP kinase signaling.
Prior studies suggested that ABIN-1 binds A20, a potent restrictor of TNF-induced NFkB signaling, and that ABIN-1 inhibits NFkB signaling (1, 12). ABIN-1−/− cells exhibited slightly greater p-IkBa to IkBa ratios and IKKb kinase activity than control cells, while A20−/− cells displayed significantly prolonged NFkB signaling (Suppl. Figs. 3A, 3B). ABIN-1−/− cells expressed similar levels of NFkB specific DNA binding activity, similar levels of NFkB dependent mRNAs and proteins (Suppl. Figs. 3C, 3D, 3E). By contrast, A20−/− cells expressed significantly prolonged NFkB DNA binding activity and markedly higher levels of most NFkB target genes and proteins compared with either ABIN-1−/− or control cells (Suppl. Figs. 3C, 3D, and 3E). Hence, ABIN-1 deficiency reveals a subtle role for ABIN-1 in restricting proximate NFkB signaling, which is associated with minimal affects on NFkB dependent gene transcription. Meanwhile, A20 deficiency leads to markedly prolonged NFkB signaling and increased NFkB dependent gene transcription.
After TNFR engagement, death inducing signaling complexes (DISCs) including TRADD, FADD, and caspase 8 may trigger caspase 8 cleavage and PCD (13, 14). ABIN-1 is expressed in resting cells, and ABIN-1 regulates TNF-induced PCD in the absence of protein synthesis. We thus hypothesized that ABIN-1 might directly regulate TNF-induced DISCs. We found that ABIN-1 indeed interacts with RIP1 and FADD under conditions that induce PCD (Fig. 2F). To determine whether ABIN-1 directly regulates association of DISC proteins, we stimulated ABIN-1 deficient and control cells with TNF, CHX and ZVAD, immunoprecipitated endogenous caspase 8, and examined the association of DISC proteins. Endogenous RIP-1 was recruited normally to caspase 8 under these conditions (Fig. 2G). By contrast, the amount of endogenous FADD associated with caspase 8 was significantly greater in ABIN-1 deficient cells, indicating that ABIN-1 is essential for inhibiting caspase 8’s interaction with FADD (Fig. 2G).
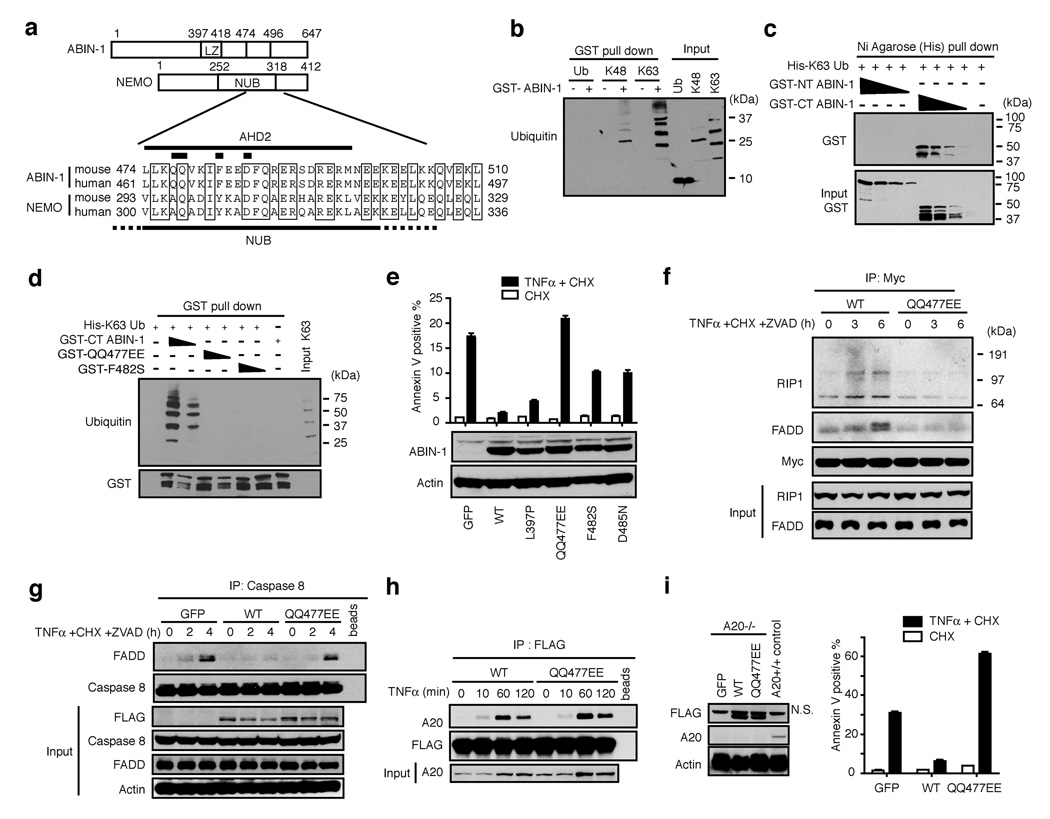
To further investigate how ABIN-1 may regulate TNF-induced PCD, we noted that a peptide sequence overlapping the ABIN homology domain 2 (AHD2) of ABIN-1 is homologous to a larger, recently defined domain within IKKγ/NEMO that binds polyubiquitin chains and ubiquitylated RIP, called the NEMO Ubiquitin Binding (NUB) domain (Fig. 3A) (15–17). In addition, ABIN-1 has a leucine zipper upstream of this domain (Fig. 3A). Hypothesizing that ABIN-1, like IKKγ, may be a ubiquitin binding protein, we tested the ability of recombinant ABIN-1 proteins to bind His tagged ubiquitin chains. These studies showed that GST-ABIN-1 binds to His tagged polyubiquitin chains in a GST pull down assay (Fig. 3B). GST-ABIN-1 binds both K48 linked as well as K63 linked polyubiquitin chains, and displays a preference for chains that are at least three ubiquitin moieties in length (Fig. 3B). GST-ABIN-1 did not bind ubiquitin monomers (Fig. 3B). These findings are consistent with a recent study demonstrating binding of ABIN-1 to ubiquitin chains (18).
Figure 3. ABIN-1 is a ubiquitin sensor that uses a NUB domain to bind to DISC signaling complexes and protect cells from TNF-induced PCD.
(A) Sequence alignment of AHD2 (and neighboring sequences) of ABIN-1 with the NEMO ubiquitin binding (NUB) domain of NEMO/IKKγ. Thick bars mark locations of residues mutated in QQ477EE, F482S, and D485N ABIN-1 mutants. (B) Ubiquitin binding to GST-ABIN-1 in GST pull down assay. Ubiquitin bound to GST-ABIN-1 shown in left six lanes; input of ubiquitin chains is shown as a control in right three lanes. (C) GST-ABIN-1 proteins bound to K63-linked His-polyubiquitin chains in Nickel (Ni) agarose pull down assay. GST-ABIN-1 proteins bound to His-ubiquitin chains shown in upper panel; inputs of GST-ABIN-1 proteins shown in bottom panel as controls. (D) 6xHis-K63 linked ubiquitin chains bound to GST-C-terminal (wild type and mutant) ABIN-1 proteins. Input levels of GST-ABIN-1 proteins shown below as controls. (E) TNF-induced PCD of ABIN-1−/− 3T3s virally complemented with the indicated FLAG-ABIN-1 proteins or GFP alone. FLAG-ABIN-1 expression levels shown below as control. (F) Recruitment of RIP1 or FADD to wild type (WT) or mutant Myc-ABIN-1 proteins. Myc-ABIN-1 expression in anti-Myc immunoprecipitates and RIP1 and FADD expression in whole cell lysates (“input”) shown below as controls. (G) Recruitment of FADD to caspase 8 in presence of wild type, or QQ477EE mutant FLAG-ABIN-1 proteins, or no ABIN-1 proteins (“GFP”). Expression of caspase 8 in IPs and expression of FLAG-ABIN-1, caspase 8, FADD and actin in whole cell lysates (“input”) shown below as controls. (H) Recruitment of A20 to wild type or mutant (“QQ477EE”) FLAG-ABIN-1 proteins. Expression of FLAG-ABIN-1 in IPs and A20 in whole cell lysates (“input”) shown as controls. (I) Anti-apoptotic activity of ABIN-1 in A20−/− 3T3s expressing no ABIN-1 (“GFP”), WT ABIN-1 (“WT”), or mutant ABIN-1 (“QQ477EE”). Means and standard deviations indicated, n=3. Expression of FLAG-ABIN-1, A20, and actin proteins shown as controls at left (N.S. = non-specific band directly above FLAG-ABIN-1 band). All data are representative of 3–5 independent experiments.
To further establish and localize ABIN-1’s ubiquitin binding domain, C-terminal and N-terminal truncation mutants of ABIN-1 were tested for ubiquitin binding. C-terminal GST-ABIN-1, which contains AHD2, binds K63 linked poly-ubiquitin chains while an N-terminal GST-ABIN-1 protein lacking this domain does not (Fig. 3C). Several residues within IKKγ’s NUB domain that are required for ubiquitin binding are conserved in ABIN-1. C-terminal ABIN-1 proteins bearing either QQ (glutamine) to EE (glutamic acid) substitutions at positions 477/478 (“QQ477EE”), or a F (phenylalanine) to S (serine) mutation at position 482 (“F482S”) failed to bind ubiquitin chains (Fig. 3D). Thus, ABIN-1 is a ubiquitin sensing protein that utilizes a NUB-like domain to bind polyubiquitin chains.
To determine whether ABIN-1’s ubiquitin binding activity mediates ABIN-1’s anti-apoptotic function, we introduced wild-type or mutant forms of FLAG tagged ABIN-1 into ABIN-1−/− 3T3s using a GFP expressing virus, purified productively infected cells by FACS sorting, and tested the cells for susceptibility to TNF-induced PCD. These experiments revealed that wild-type ABIN-1 and a leucine zipper mutant L397P protected ABIN-1−/− cells from TNF-induced PCD (Fig. 3E). By contrast, ubiquitin binding domain mutants of ABIN-1 (QQ477EE, F482S, and D485N) failed to fully protect ABIN-1−/− cells from TNF-induced PCD (Fig. 3E). To determine whether ABIN-1’s ubiquitin binding activity mediates ABIN-1’s capacity for regulating DISC formation, we tested whether a ubiquitin binding deficient form of ABIN-1 binds to this complex and whether it regulates the association of DISC proteins. Wild type, but not QQ477EE mutant, ABIN-1 was recruited to RIP1 and FADD (Fig. 3F). Moreover, wild type, but not QQ477EE mutant, ABIN-1 blocked the recruitment of endogenous FADD to caspase 8 (Fig. 3G). Therefore, ABIN-1 requires its ubiquitin binding domain to interact with DISC proteins, to inhibit FADD – caspase 8 binding, and to protect cells from TNF-induced PCD.
ABIN-1 binds to the ubiquitin editing enzyme A20, which also protects cells against TNF-induced PCD, so we investigated whether ABIN-1 might block TNF-induced PCD in an A20 dependent fashion (1, 12). A20 protein was induced by TNF in both ABIN-1+/+ and ABIN-1−/− MEFs, so ABIN-1 is not required for A20 expression (Fig. 2D). A20 association with caspase 8 does not depend upon ABIN-1, arguing against a critical role for ABIN-1 in recruiting A20 to the DISC (Fig. 2G). A20 was recruited normally to both wild type and QQ477EE forms of ABIN-1, indicating that ABIN-1 does not require ubiquitin binding to bind A20 (Fig. 3H). Finally, wild type, but not QQ477EE mutant, ABIN-1 blocks TNF-induced PCD in A20−/− cells, showing that ABIN-1 does not require A20 to perform its anti-apoptotic function (Fig. 3I).
In summary, we have demonstrated that ABIN-1 is a novel ubiquitin sensing protein that inhibits FADD-caspase 8 association in the DISC, protects cells against TNF-induced PCD, and sustains embryonic development. The recent unveiling of non-degradative ubiquitin modifications has led to the discovery of ubiquitin sensors that propagate NFkB signals (16, 17 19–21). Much less is known about ubiquitin sensors and PCD signaling (22, 23). Our current studies provide the first evidence that ubiquitin sensors also restrict signal transduction pathways, and that they regulate PCD signaling. These findings highlight the central roles of ubiquitin sensors in regulating physiological cell survival decisions in vivo.
Methods Summary
Generation of ABIN-1 (tnip1) deficient mice
The generation of a gene targeting construct, targeted ES cells, and tnip1 deficient mice are described in online methods and supplemental figure legend 1.
Lentiviral shRNA knock-down of ABIN in human cell lines
Lentiviruses bearing shRNA sequences specific to ABIN were purchased from Open Biosystems and used to infect Jurkat, HepG2 or HT1080 cells.
Cell-free ubiquitin binding assays
Poly-histidine tagged ubiquitin and polyubiquitin chains were purchased from Boston Biochem. Recombinant GST-ABIN-1 proteins were purified from BL21 bacteria. Ubiquitin binding assays were performed using either glutathione agarose beads or nickel agarose beads for precipitating ABIN-1 - ubiquitin complexes.
Lentiviral transduction of ABIN-1 expression constructs
Lentiviral expression vectors expressing GFP and FLAG-tagged ABIN were used to infect cells. In some experiments, infected GFP positive cells were purified by cell sorting prior to use in cell signaling and cell death assays.
Methods Online
Generation of ABIN-1 (tnip1) deficient mice
Recombineering was used to generate a gene targeting construct from a bacterial artificial chromosome (BAC) containing the tnip1 gene (C57Bl/6J inbred strain). C57Bl/6J inbred PRX-B6T ES cells were transfected with this construct, and successfully targeted ES cells were identified by Southern blot analysis. Blastocyst injections of targeted ES cells were performed by the UCSF Transgenic Core. Mice bearing this targeted allele in the germline were interbred with E2a-Cre transgenic mice to delete intervening sequences including exons 12–15 and generate the null allele. These mice were then bred to B6 mice to eliminate the E2a-Cre transgene and generate ABIN+/− mice.
Programmed cell death assays
ABIN-1 competent and deficient cells were treated with TNF (10ng/ml) plus cycloheximide (10mg/ml) and assayed for programmed cell death by flow cytometric quantitation of Annexin V reactive cells using an LSRII flow cytometer and Flo-Jo software. Cell death signaling in these cells was assessed by preparing whole cell lysates in lysis buffer (50 mM Hepes, 120 mM NaCl, 1mM EDTA, 0.1%NP-40) and immunoblotting for expression and cleavage of cell death proteins using commercial antibodies (caspase 3 [Cell Signaling], caspase 8, [BD Pharmingen, Alexis Biochemicals], BID [Santa Cruz]). In some assays, Z-VAD-FMK was used [Alexis Biochemicals].
Cell signaling assays
Activation of cell signaling cascades was assessed by stimulating cells with the indicated ligands (e.g., TNF), preparing whole cell lysates in lysis buffer (above), and immunoblotting for the expression of various proteins using commercial antibodies (phospho-IkBa, IkBa, phospho-MAPK, MAPK, phospho-JNK, JNK, phospho-p38, p-38, XIAP [Cell Signaling], Bcl-x [BD Pharmingen], ABIN-1 [Zymed], actin [Calbiochem]). A20 protein was detected using either a commercial anti-human A20 antibody [eBioscience], or a polyclonal anti-murine A20 antibody (24). Recruitment of co-associated signaling proteins was assayed by lysing cells in lysis buffer (10mM Tris, 140 mM NaCl, 0.2% NP-40, 10% glycerol and protease inhibitors), immunoprecipitating FLAG-tagged ABIN-1 protein with anti-FLAG beads (Sigma), myc-tagged ABIN-1 with anti-c-myc antibody [Santa Cruz Biotechnology], or caspase 8 with anti-caspase 8 [Santa Cruz Biotechnology] and immunoblotting co-associated FADD, caspase 8, A20, or RIP using commercial antibodies (RIP [BD Transduction Labs, Santa Cruz Biotechnology], FADD [Assay Designs]). IKK kinase assays were performed by immunoprecipitating lysates with anti-IKKγ antibody (Santa Cruz Biotechnology), incubating with recombinant GST-IkBa, and measuring phospho-IkBa by immunoblotting. Control for immunoprecipitation efficiency was performed with anti-IKKg antibody (Becton Dickinson). To perform NFkB EMSAs, 10 ug of protein from MEF nuclear lysates were incubated with a NFkB consensus oligonucleotide (Santa Cruz Biotechnology), separated on 6% acrylamide gels, and developed with a chemiluminescent detection kit (Pierce).
Real Time-PCR assays of mRNA expression
RT-PCR analyses were performed using TaqMan Gene Expression kit (Applied Biosystems), according to manufacturer’s instructions.
Lentiviral shRNA knock-down of ABIN-1 in human cell lines
Lentiviral constructs bearing shRNA sequences specific to ABIN-1 were purchased from Open Biosystems. Constructs were co-transfected with expression constructs for helper virus R89.1, VSV-G, and tat into 293T cells, and replication deficient virus from the resulting supernatants was used to infect Jurkat, HepG2, and HT1080 cells. The degree of ABIN-1 protein reduction was assessed by immunoblotting of whole cell lysates. Two of five shRNA sequences (TRCN8681 [“shRNA 81”] and TRCN8685 [“shRNA 85”]) that caused the greatest reduction of ABIN-1 protein expression were selected for cell signaling and cell death studies. In some experiments, these two shRNA containing viruses were combined in a single infection. Control cells included cells infected with lentiviruses devoid of shRNA sequences, or expressing eGFP specific sequences.
Cell-free ubiquitin binding assays
Poly-histidine tagged ubiquitin and polyubiquitin chains were purchased from Boston Biochem. Wild type and mutant ABIN-1 cDNAs were generated by site-directed mutatgenesis (Quick Change Kit, Stratagene), after which they were subcloned into bacterial expression vectors. Recombinant GST-ABIN-1 proteins were purified from bacteria using glutathione agarose beads. Ubiquitin-binding assays with GST-ABIN-1 proteins were performed either by (i) incubating glutathione agarose beads bound to GST-ABIN-1 proteins with free poly-histidine-ubiquitin for 10 minutes at room temperature in ubiquitin binding buffer (25 mM HEPES, 150 mM KCl, 2 mM MgCl2, 0.5% Triton X-100, 1 mM EGTA, and 1 mg/ml BSA), followed by serial washes with binding buffer and precipitation of glutathione agarose beads by centrifugation, or (ii) by incubating free GST-ABIN-1 proteins with nickel agarose beads bound to poly-histidine-ubiquitin in ubiquitin binding buffer supplemented with 5mM imidazole, followed by serial washes and centrifugation of nickel agarose beads. In both cases, bead bound ABIN-1 – ubiquitin complexes were analysed by immunoblotting for GST (ABIN-1 protein) and/or ubiquitin using commercial antibodies (GST [Cell Signaling], ubiquitin [Santa Cruz Biotechnology]).
Expression of ABIN-1 proteins
Lentiviral expression vectors expressing GFP and FLAG-tagged ABIN constructs were generated from a lentiviral pWPI construct. These constructs were co-transfected along with helper constructs into 293T cells to generate infectious lentivirus, which was then used to infect various cell types. In some assays, productively infected cells were purified by flow cytometry for GFP expressing cells prior to use in cell signaling and cell death assays. In some experiments, transient expression of myc-ABIN-1 was performed in 293T cells.
Supplementary Material
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
This work was supported by the NIH (A.M.), UCSF Liver Center Pathology and Flow Cytometry Facilities, UCSF Transgenic and Targeted Mutagenesis Core Facility, a fellowship from the Crohn’s and Colitis Foundation of America (S.O.), a pre-doctoral NSF fellowship (J.A.C.), and the Rainin Foundation.
References
- 1.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN-1. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opipari AW, Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- 3.Doi TS, Marino MW, Takahashi T, Yoshida T, Sakakura T, Old LJ, Obata Y. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci U S A. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 5.Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 6.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 7.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 8.Wullaert A, Wielockx B, Van Huffel S, Bogaert V, De Geest B, Papeleu P, Schotte P, El Bakkouri K, Heyninck K, Libert C, Beyaert R. Adenoviral gene transfer of ABIN-1 protects mice from TNF/galactosamine-induced acute liver failure and lethality. Hepatology. 2005;42:381–389. doi: 10.1002/hep.20785. [DOI] [PubMed] [Google Scholar]
- 9.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R, Franzoso G. Induction of gadd45beta by NF-kappaB downregulates propoptotic JNK signalling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 10.Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M, Lin A. Inhibition of JNK activation through NF-kappaB target genes. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 14.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwender M, Adam D, Mentlein R, Kabelitz D, Schütze S. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 16.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 18.Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Löhr F, Wu CJ, Ashwell JD, Dötsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 19.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 21.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertz IE, Dixit VM. Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 2008;19:313–324. doi: 10.1016/j.cytogfr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Lee JC, Peter ME. Regulation of apoptosis by ubiquitination. Immunol Rev. 2003;193:39–47. doi: 10.1034/j.1600-065x.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 24.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.