Abstract
The quantum yield and kinetics of decay of cob(II)alamin formed by pulsed-laser photolysis of adenosylcobalamin (AdoCbl) in coenzyme B12 (AdoCbl)-dependent ethanolamine ammonia-lyase (EAL) from Salmonella typhimurium have been studied on the 10-7 - 10-1 s time scale at 295 K by using transient ultraviolet-visible absorption spectroscopy. The aim is to probe the mechanism of formation and stabilization of the cob(II)alamin-5′-deoxyadenosyl radical pair, which is a key intermediate in EAL catalysis, and the influence of substrate binding on this process. Substrate binding is required for cobalt-carbon bond cleavage in the native system. Photolysis of AdoCbl in EAL leads to a quantum yield at 10-7 s for cob(II)alamin of 0.08 ±0.01, which is 3-fold less than for AdoCbl in aqueous solution (0.23 ±0.01). The protein binding site therefore suppresses photoproduct radical pair formation. Three photoproduct states, Pf, Ps, and Pc, are identified in holo-EAL by the different cob(II)alamin decay kinetics (subscripts denote fast, slow, and constant, respectively). These states have the following first-order decay rate constants and quantum yields: Pf (2.2×103 s-1; 0.02), Ps (4.2×102 s-1; 0.01), and Pc (constant amplitude, no recombination; 0.05). Binding of the substrate analog, (S)-1-amino-2-propanol, to EAL eliminates the Pf state, and lowers the quantum yield of Pc (0.03) relative to Ps (0.01), but does not significantly change the quantum yield or decay rate constant of Ps, relative to holo-EAL. The substrate analog thus influences the quantum yield at 10-7 s by changing the cage escape rate from the geminate cob(II)alamin-5′-deoxyadenosyl radical pair state. However, the predicted substrate analog binding-induced increase in the quantum yield is not observed. It is proposed that the substrate analog does not induce the radical pair stabilizing changes in the protein that are characteristic of true substrates.
Coenzyme B12-dependent enzymes catalyze radical mediated rearrangement reactions in both bacteria and mammals (1-3). The first step in the catalytic cycle is the homolytic cleavage of the cobalt-carbon (Co-C) bond in coenzyme B12 (adenosylcobalamin, AdoCbl; Figure 1), which results in the formation of the cob(II)alamin-5′-deoxyadenosyl radical pair. The C5′ radical center of the 5′-deoxyadenosyl moiety then migrates over 5-6 Å (4, 5) to abstract a hydrogen atom from the substrate, which activates the substrate for rearrangement. Cleavage of the Co-C bond is accelerated by >1011-fold in the enzymes, relative to the cleavage in solution (6-8). A long-standing issue in AdoCbl-dependent enzyme catalysis is the molecular mechanism of the rate acceleration (9, 10), and how substrate binding, which is required for cleavage, is coupled to the reaction (1-3). In order to address the mechanism of formation and stabilization of the cob(II)alamin-5′-deoxyadenosyl radical pair in the protein, we use pulsed-laser photolysis of the Co-C bond to prepare the radical pair population, followed by a UV-visible absorption probe of its time evolution. This method overcomes the kinetic complexity and asynchrony of steady-state kinetic studies, and the evolution of the radical pair can be observed on time scales that are several orders of magnitude shorter than in previous stopped-flow studies of cob(II)alamin formation in the AdoCbl-dependent enzymes, methylmalonyl-CoA mutase (11), glutamate mutase (12), ribonucleotide triphosphate reductase (13), and ethanolamine ammonia-lyase (14). The photolysis measurements are performed by using ethanolamine ammonia-lyase (EAL) [EC 4.3.1.7; cobalamin (vitamin B12)-dependent enzyme superfamily (15, 16)] from Salmonella typhimurium (1, 17). EAL catalyzes the conversion of aminoethanol and 2-aminopropanol to the corresponding aldehydes and ammonia in bacteria (18).
Figure 1.
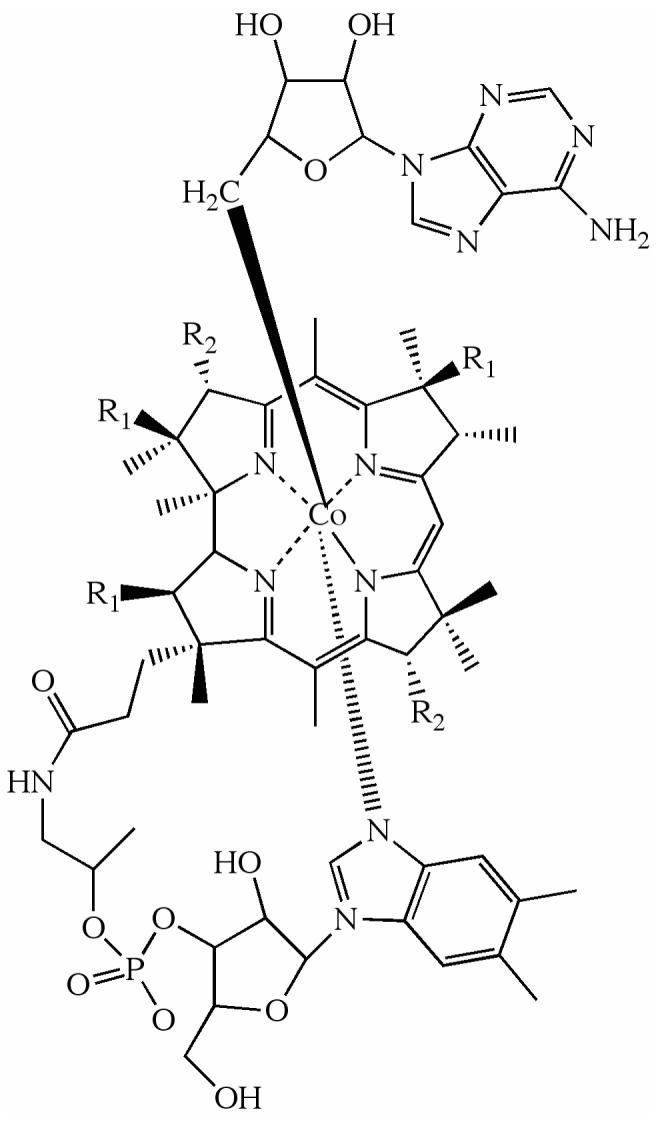
Depiction of the structure of AdoCbl. The β-axial ligand is 5′-deoxyadenosyl. R1 and R2 represent acetamide and propionamide side chains. The dimethylbenzimidazole α-axial ligand of the coenzyme remains coordinated when the coenzyme is bound to EAL (53, 54).
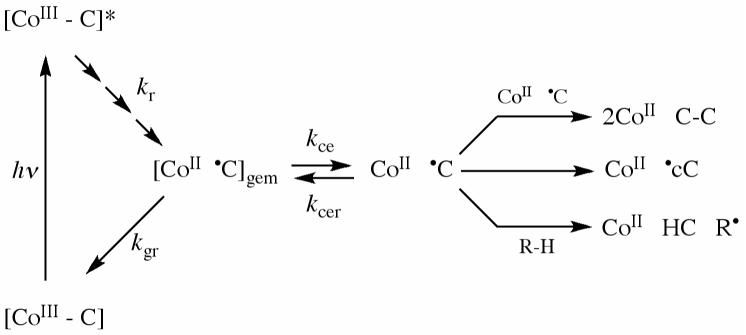
Figure 2 shows a simplified kinetic scheme of the canonical states and steps involved in the photolysis experiment. Following photo-excitation, a fraction of the photoproducts relaxes to the ground state in <10-9 s (19, 20), forming the geminate cob(II)alamin-5′-deoxyadenosyl radical pair. The excited state formation and sequence of early photoproduct intermediates for alkylcobalamins in solution, which are not shown explicitely in Figure 2, have been described by Sension and coworkers (20, 21). The geminate radical pair exists within a “cage” of surrounding solvent molecules, and can recombine promptly (geminate recombination), or diffuse apart (cage escape) and recombine on a slower time scale (22). The radical pair formation and decay are detected optically with relatively high sensitivity by monitoring the UV-visible absorption changes associated with the interconversion of the cobalamin between the CoIII state (visible wavelength maximum, λmax=525 nm in water) and CoII state (λmax=470 nm in water). In studies of AdoCbl, MeCbl, and other alkylCbl in solution, time scales from electronic excited state formation and decay (femtoseconds) to solvent separation of the cob(II)alamin-radical pairs (milliseconds) have been monitored (19, 20, 23-33). Following AdoCbl photolysis in water, the geminate cob(II)alamin-5′deoxyadenosyl radical pair recombination and cage escape occur with rate constants of 1.4×109 s-1 and 0.6×109 s-1, respectively, leading to a fraction (0.29) of radical pairs that escape the solvent cage (20, 31).
Figure 2.
Simplified schematic diagram of the states and pathways of formation following photolysis of AdoCbl in solution. The cobalamin and 5′-deoxyadenosyl moiety are represented by cobalt (Co) and C5′-methylene center (C), as follows: [CoIII-C], intact coenzyme; [CoIII-C]*, excited singlet state; [CoII •C]gem, geminate radical pair; CoII gem •C, cage escape radical pair. The cage escape radical pair can recombine to reform the geminate radical pair, or can undergo radical-radical annihilation (top), internal rearrangement to form cycloadenosyl or other species (middle), or react with solvent (R-H) by hydrogen atom abstraction (bottom), which leads to irreversible cob(II)alamin formation (19, 50). Intermediate excited and relaxed states (20, 26), which are not shown, are represented by the sequence of arrows leading from [CoIII-C]*. Rate constants are defined as follows: kr, excited to ground state relaxation; kgr, geminate recombination; kce, cage escape; kcer, reformation of geminate radical pair from cage escaped radical pair.
Quantitative time-resolved measurements of AdoCbl photolysis and recombination in proteins have thus far only been performed for AdoCbl-dependent glutamate mutase (GluM) on the ultrafast time scale (<9 ns) (34, 35). The results showed that binding of AdoCbl to GluM led to a reduction of quantum yield of cob(II)alamin (at 9 ns) from 0.23 in solution to 0.05 in the protein. This was caused primarily by a decrease in the cage escape rate constant to 5-6 ×107 s-1 (34, 35), relative to the value of 5.7×108 s-1 reported for pure water. The protein reduced the geminate recombination rate by only 30% (23). These results show that AdoCbl is photolyzable in situ and that the protein influences (reduces) the quantum yield. However, these studies were restricted to ultrafast time scales that do not address recombination of the cage escaped radical pair. If the cage escaped 5′-deoxyadenosyl radical diffuses along the native radical pair separation coordinate (36-38) in the active site, then the time scale of recombination may be governed by the activation free energy barriers that control the native radical pair separation process on the micro- to millisecond time scales. The measurements in GluM were also carried out with holoenzyme in the absence of substrate. Therefore, the influence of substrate binding on the photoproduct yields and decay reactions was not addressed.
Here, we report transient optical absorption measurements of the quantum yield and radical pair recombination kinetics following phototolysis of AdoCbl on time scales from 10-7 to 10-1 s in solution and in EAL. The influence of substrate on the quantum yield and radical pair recombination kinetics is assessed by using (S)-1-amino-2-propanol, an inactive substrate analog, which binds to the substrate binding site in EAL, but does not form the cob(II)alamin-substrate radical pair state. This analog has a methyl group at the pro-(S) position of stereospecific hydrogen atom abstraction from the carbinol carbon of the native substrate (39, 40), which blocks the hydrogen atom transfer reaction. A branched kinetic mechanism is proposed to account for the results, in which the observed transients represent metastable cob(II)alamin-5′-deoxyadenosyl radical pair states. We conclude that the substrate analog partially fulfills native function by suppressing non-productive reactions of the radical pair, but is not capable of inducing the full “substrate trigger” of Co-C bond cleavage that is characteristic of the native substrates.
EXPERIMENTAL PROCEDURES
Enzyme Preparation
EAL was purified from the Escherichia coli over expression strain incorporating the cloned S. typhimurium EAL coding sequence (41) essentially as described (42), with the exception that the enzyme was dialyzed against buffer containing 100 mM HEPES (pH 7.5), 10 mM potassium chloride, 5 mM dithiothreitol, 10 mM urea, and 10% glycerol (43). Enzyme activity was determined as described (44) by using the coupled assay with alcohol dehydrogenase/NADH. The specific activity of the purified enzyme with aminoethanol as substrate was 35-45 μmol/min/mg.
Sample Preparation
Adenosylcobalamin and (S)-1-amino-2-propanol were purchased from Sigma-Aldrich Chemical Co. Samples of enzyme (20-100 μM active sites) with cofactor (10-50 μM) bound were prepared in 10 mM potassium phosphate (pH 7.5) and sonicated at 277 K to reduce light scattering. Samples with substrate analog bound were prepared as above with 10 mM of the substrate analog, (S)-1-amino-2-propanol (dissociation constant for interaction with holo-EAL, 39 μM), present. Anaerobic solutions of AdoCbl were prepared by placing small volumes of cofactor stock solution under vacuum, followed by backfilling with argon before addition to anerobic buffer. Argon bubbling for 1 h was utilized to degass the buffer solution. Components were injected into a 3 ml quartz anerobic cuvette with a sealed septum that had been flushed with argon for at least 5 min prior to sample mixing.
Static Absorption Spectra
Static absorption spectra from 300 nm to 650 nm were collected by using a Shimadzu UV-1601 absorption spectrometer with 0.5 nm wavelength accuracy. Spectra were collected before and after transient absorption measurements and again after laser saturation of the sample. A scattering base line of each enzyme sample was taken before the addition of cofactor. All measurements were performed at 295 ±1 K.
Transient Absorption Spectroscopy
Transient absorption of the cob(II)alamin state was monitored at 470 nm (probe) following laser pulse photolysis at 532 nm. Measurements were made with 0.1-10 μs dwell times and corresponding time constants of 0.01-1 μs. All measurements were performed at 295 ±1 K.
Transient Absorption Spectrophotometer
Transient absorption measurements were performed by using a transient spectrophotometer of home design and construction. The transient absorption spectrometer has a sensitivity to change in absorbance (A) of 2×10-3 over 300-650 nm, and a deadtime of ≤20 ns. The second harmonic output (532 nm) of a Nd:YAG laser (SpectraPhysics GCR-10; 10 ns pulse width), with pulse energy adjusted by a glan prism polarizer/half-wave plate, is used as actinic source. The pump pulse is reduced and aligned along the same axis through the sample as the probe beam. A computer controlled shutter controls admittance of the pump pulses into the spectrometer. After passage through the sample, the pump pulse is collected in a beam dump. The probe beam of the spectrometer is provided by a low noise 300 W xenon arc lamp (Hamamatsu, L2480). The broadband output of the lamp is reduced before entering a monochromator (Applied Photophysics, F-34) to create a probe beam with a bandwidth of approximately 25 nm. The probe beam is directed through a computer controlled shutter before entering the spectrometer housing. The probe beam shutter minimizes the time that the probe beam interacts with the sample. The probe beam is focused through a brass sample holder and into a small adjustable aperture before being passed through a second monochromator (Jobin Yvon, H-20). The second monochromator provides 1 nm resolution for the detection of the probe beam and contributes to eliminating any stray photons from the intense laser pulse. The probe beam is directed through two laser line reject filters (Omega Optical, 532 nm) and onto the active area of the photomultiplier tube (Hamamatsu model R928; range, 300-650 nm). The photomultiplier tube output is digitized and temporarily stored in a digital sampling oscilloscope (Tektronix, TDS 520B). The spectrometer sample holder is associated with a liquid flow system for temperature control, a computer controlled magnetic stirrer, and sample and sample space gas flow lines. The spectrometer’s pump and probe pulse timing sequences and data collection are controlled by a Macintosh computer with LabVIEW software (National Instruments) via a GPIB/IEEE-488.2 interface. Data fitting and analysis was performed by using Matlab (Natick, MA) routines that were run on PC computers.
Quantum Yield Calculation
Quantum yield values were obtained by using laser pulse energies of 1.0, 1.5, and 2.0 mJ. Quantum yield measurements as a function of pulse energies up to 10 mJ were performed to determine the pulse energy threshold for multiple photon absorption. The 470 nm probe beam was used to detect the formation of cob(II)alamin. Quantum yield measurements were made with a 0.2 μs dwell time and a time constant of 0.02 μs. The quantum yield (φ) is defined as the concentration of cob(II)alamin photoproduct formed by the laser pulse, divided by the absorbed photon (hν) concentration, [hν]abs. The concentration of photoproduct is found by using the previously measured difference extinction coefficients for adenosylcob(III)alamin to cob(II)alamin conversion, Δε (M-1cm-1), for AdoCbl (45). The concentration of photoproduct is calculated by using the following expression
| [Eq. 1] |
where, νinitial and νfinal are the initial (pre-laser pulse) and final (post-photolysis) detector voltages respectively, and the sample pathlength is 1 cm. The ratio, νinitial/νfinal, is directly proportional to the ratio of the initial and final transmittance (T) values. The concentration of incident photons, [hν]incident, is obtained from the following expression
| [Eq. 2] |
where Ephoton is the photon energy at 532 nm, Vbeam is the pump excitation volume (1.3×10-4 L), NA is Avogadro’s number, and Epulse is the laser pulse energy. The value of [hν]abs is obtained from [hν]incident after scaling by the probability of photon absorption at 532 nm, which is given by (1-T).
Temperature-dependence of the first-order rate constant
The temperature dependence of the first order rate constant, k, is given by the Arrhenius expression (46):
| [Eq. 3] |
where Ea is the activation energy, R is the gas constant, and A is a prefactor that represents the value of k as Ea→0. The value of A is typically approximated as , where kB is Boltzmann’s constant and h is Planck’s constant.
RESULTS
Pre- and post-photolysis absorption of AdoCbl in solution and in EAL
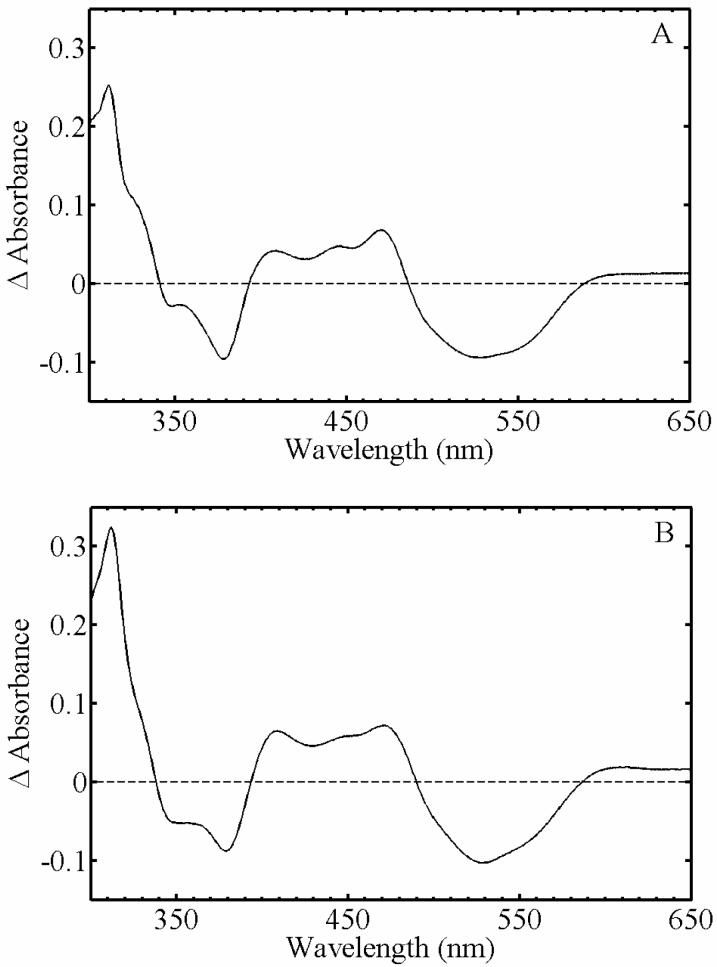
The photoproduct difference spectra (post-photolysis minus pre-photolysis) for AdoCbl in solution and in EAL are presented in Figure 3. The difference spectra in Figure 3A and 3B are characterized by the loss of the AdoCbl αβ-band maximum around 525 nm (negative feature) and the rise of long wavelength maximum of cob(II)alamin around 470 nm (positive feature). The comparable spectral features indicate that the difference spectrum for AdoCbl photolysis in EAL in aerobic solution in Figure 3A represents [AdoCbl minus cob(II)alamin], because it is the same as the reference [AdoCbl minus cob(II)alamin] difference spectrum obtained in anaerobic solution (Figure 3B). Small differences in the line shape and peak positions are caused by the different cofactor environments in aqueous solution and in EAL, as found for other AdoCbl-dependent enzymes (9, 10, 47).
Figure 3.
Absorption difference spectra of photolyzed minus pre-photolysis AdoCbl. (A) AdoCbl in EAL in aerobic solution. (B) AdoCbl in anaerobic solution.
Hogenkamp and coworkers have previously shown that aquocob(III)alamin arises from the reaction of cob(II)alamin with dioxygen (O2) (48). The results therefore show that the binding of the native AdoCbl cofactor in EAL leads to a state in which O2 is prevented from interacting with the photoproduct cob(II)alamin.
Quantum yield of cob(II)alamin formation following photolysis of AdoCbl in solution and in EAL
The quantum yield of photolysis of AdoCbl was determined on the 10-7 s time scale under different conditions by using low pulse energy (≤2 mJ) excitation from the 532 nm output of the pulsed-Nd-YAG laser, and a continuous-wave probe at 470 nm. The low energies were selected to prevent multiple photon absorption by AdoCbl, as described in Experimental Procedures. The ratio of the concentration of AdoCbl to EAL active sites was ≤0.5, to avoid interference from free cofactor. A dissociation constant (KD) for AdoCbl binding to EAL of 0.5 μM is estimated from the Michaelis constant (42), which indicates that, under the conditions of the experiments, the free cofactor represents 4% of the total cofactor in the sample. The quantum yield values are presented in Table 1. The measured quantum yield at 10-7 s for AdoCbl photolysis in 10 mM potassium phosphate buffer at pH 7.5 of 0.23 ± 0.01 is comparable to previously reported aqueous solution, room temperature values of 0.23± 0.04 (excitation wavelength,λex=532, 355 nm) (31) and 0.24 ±0.04 (λex=400, 520 nm) (20).
Table 1.
Quantum yield of cob(II)alamin at 10-7 s following photolysis of AdoCbl in anaerobic solution and in aerobic solutions of EAL in the absence and presence of substrate analogs.
| Cobalamin |
Substrate Analog |
Environment |
Quantum Yielda |
|---|---|---|---|
| Adenosylcobalamin | - | Solution | 0.23 ±0.01 |
| - | EAL | 0.08 ±0.01 | |
| (S)-1-Am-2-PrOH | EAL | 0.04 ±0.01 |
Average of N≥3 determinations, ±standard deviation.
Binding of AdoCbl by EAL leads to a 3-fold reduction in the quantum yield at 10-7 s relative to solution, to a value of 0.08 ±0.01. Reduction in the yield of cob(II)alamin at 9 ns has been previously reported for photolysis of AdoCbl bound in glutamate mutase, relative to solution (34). The yield at 9 ns in glutamate mutase was 0.05 ±0.03 (34). Table 1 shows that, in the presence of bound (S)-1-amino-2-propanol, the quantum yield of cob(II)alamin at 10-7 s is reduced by 50%, relative to the quantum yield in the holoenzyme.
Time-dependence of photoproduct cob(II)alamin following photolysis of AdoCbl in solution and in EAL
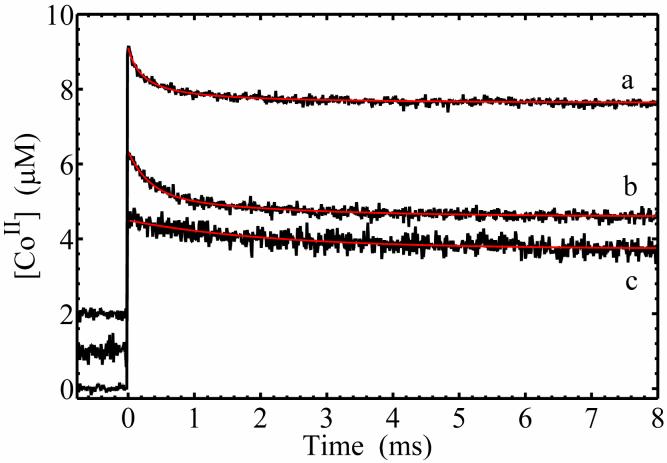
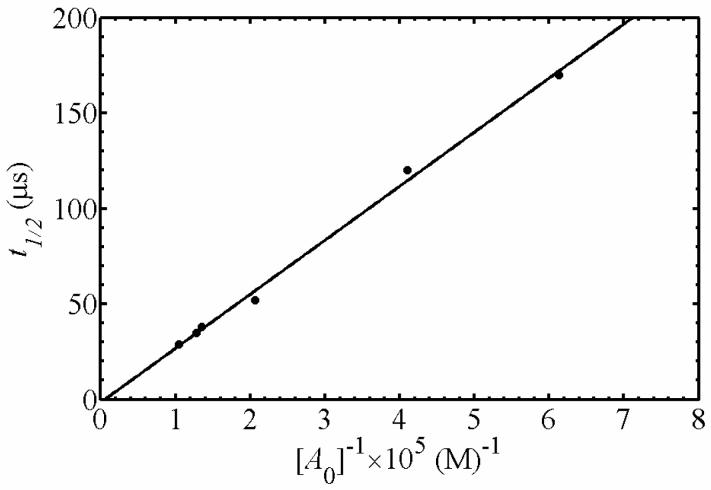
Figure 4 shows the transient kinetics of cob(II)alamin on the millisecond time scale, following photolysis of AdoCbl in the buffered aqueous solution, and in EAL in the absence and presence of bound (S)-1-amino-2-propanol. For these experiments, a relatively high laser pulse energy of 10 mJ/pulse was used to enhance the population of cob(II)alamin, by photolyzing AdoCbl that had undergone geminate recombination on the timescale of the 10 ns laser pulse width. Measurements of the decay on shorter time scales of at least 0.1 μs do not show additional kinetic transients. The kinetics in Figure 4 therefore represent the events that occur on the time scale of ≥0.1 μs for each condition. This is consistent with previous studies of AdoCbl photolysis in solution (19, 45). The time dependence of the cob(II)alamin photoproduct concentration in solution was fit by a second-order decay plus a constant function. The second-order character of the transient decay was confirmed by the linear dependence of the decay half-time on AdoCbl concentration (46), as shown in Figure 5. The linear fit in Figure 5 leads to a second-order rate constant of 3.5×109 M-1s-1, which is consistent with the reported diffusion-limited rate of cob(II)alamin recombination with the 5′-deoxyadenosyl radical (19). The constant component of the fit of the solution decay in Figure 4 represents cob(II)alamin “stranded” by the rapid radical-radical recombination reaction of 5′-deoxyadenosyl radicals. The radical-radical annihilation reaction proceeds with second-order rate constants of approximately 3×109 M-1s-1 (19), and therefore, competes favorably with recombination of cob(II)alamin with 5′-deoxyadenosyl radicals. The second-order and constant components of the time-dependent cob(II)alamin signal in solution confirm that the observed cob(II)alamin is the cage escape photoproduct.
Figure 4.
Time dependence of cob(II)alamin concentration following pulsed laser photolysis of AdoCbl, and overlaid best-fit functions (red). Cob(II)alamin concentration was obtained from the absorbance at 470 nm. (a) Anaerobic solution; second-order plus constant fit function. (b) In EAL in aerobic solution; biexponential plus constant fit function. (c) In EAL in aerobic solution, with (S)-1-amino-2-propanol bound; monoexponential plus constant fit function The decay has been multiplied by a factor of 2.0 to account for the two-fold higher concentration AdoCbl in (b) relative to (c). The fitting parameters are presented in Table 2.
Figure 5.
The half-time of cob(II)alamin decay as a function of inverse concentration of cob(II)alamin photoproduct following AdoCbl photolysis in anaerobic solution. The best-fit line corresponds to a second-order rate constant of 3.53×109 M-1s-1. Fitting parameters: Slope=2.83 ×10-10, ordinate intercept=-1.29×10-6, R2=0.9963.
Figure 4B shows that the decay of the long-lived cob(II)alamin photoproduct following photolysis of AdoCbl bound to EAL also displays transient and constant components. The transient decay was well fit by using a biexponential plus constant function. The single exponential plus constant function did not provide a satisfactory fit to the transient decay (Figure S1 and Figure S2, Supporting Information). A proportion of 19% of the long-lived cob(II)alamin decays with a lifetime of 0.46 ms and a second component (13%) decays with a lifetime of 2.4 ms. The remaining 68% of the cob(II)alamin does not recombine on the longest time scales observed by transient absorption, or by subsequent recording of the static spectra of the photolyzed samples (≤300 s). The kinetics and amplitudes of the decay are not influenced by changing the concentrations of AdoCbl or EAL. The presence of the spin trap, 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) in the solution with holo-EAL does not influence the decay kinetics, and no spin-trapped, paramagnetic DMPO-adduct species were detected in the solution around the protein following photolysis, by using electron paramagnetic resonance (EPR) spectroscopy. These results indicate that the observed cob(II)alamin recombines with the 5′-deoxyadenosyl radical within the protein interior. Therefore, the transients correspond to first-order kinetic processes.
Figure 4C shows the time dependence of the long-lived cob(II)alamin photoproduct state in EAL in the presence of bound (S)-1-amino-2-propanol. The transient decay can be fit by using a single exponential function plus a constant. A biexponential function did not lead to a significantly better fit (Figure S3, Supporting Information). The fit shows that the cob(II)alamin signal decays with a lifetime of 2.2 ms, which accounts for 27% of the long-lived cob(II)alamin. A proportion of 73% of the cob(II)alamin did not recombine at ≤300 s. The results show that (S)-1-amino-2-propanol selectively influences the cob(II)alamin photoproduct species, by eliminating the fast decay process, and by reducing the amplitude of the constant phase relative to the slow phase.
DISCUSSION
Model for the formation and reaction of cage escaped CoII-radical pair populations following AdoCbl photolysis in EAL
Three cage escape photoproduct states are identified from the different components of the decay of cob(II)alamin that are observed in the transient absorption measurements (Figure 4; Table 2), as follows: (a) fast transient (corresponding to photoproduct population, Pf), (b) slow transient (photoproduct population, Ps), and (c) constant amplitude (photoproduct population, Pc). Figure 6 shows the kinetic model that is proposed to account for the formation and time dependence of the cob(II)alamin photoproduct states in holo-EAL, in which three pathways from the geminate radical pair state lead to formation of cage escape photoproducts, which remain within the protein interior. We favor this branched reaction mechanism of separate paths from the geminate radical pair to the Pf, Ps, and Pc states, because it is the most simple mechanism that is consistent with the data. A mechanism in which cage escape leads to sequential formation of the Pf and then Ps states is not favored, because the decay rate constant assigned to Ps in holo-EAL is maintained in the absence of Pf in (S)-1-amino-2-propanol bound holo-EAL. This is not expected for a sequential mechanism, A1 ⇆ A2 ⇆ A3, where A1 denotes the geminate radical pair and A2, A3 are sequential cage escape states, because the two relaxation rate parameters,λ1 andλ2, for the linear two-step mechanism are functions of all four rate constants [λ= λ(kij, kji) where i=1, 2, 3, j=1,2,3, i≠j, and the kij and kji are the forward and backward rate constants for each step, respectively] (46). In the general case, this mechanism leads to the prediction that the single relaxation rate constant, which is measured upon elimination of the terminal state, is different fromλ1 andλ2.
Table 2.
Relative amplitudes and observed recombination rate constants for cage escape populations in holo-EAL and in holo-EAL with bound (S)-1-amino-2-propanol.
| Condition |
Population |
Rel. Amplitude |
kdecay (s-1) |
|---|---|---|---|
| EAL•AdoCbl | Pf | 0.19 ±0.05 | 2.2 (±0.4) ×103 |
| Ps | 0.13 ±0.05 | 4.2 (±1.5) ×102 | |
| Pc | 0.68 ±0.08 | - | |
| EAL•AdoCbl | Pf | 0 | - |
| •Substrate Analog | Ps | 0.27 ± 0.11 | 4.5 (±1.3) ×102 |
| Pc | 0.73 ±0.11 | - |
Figure 6.
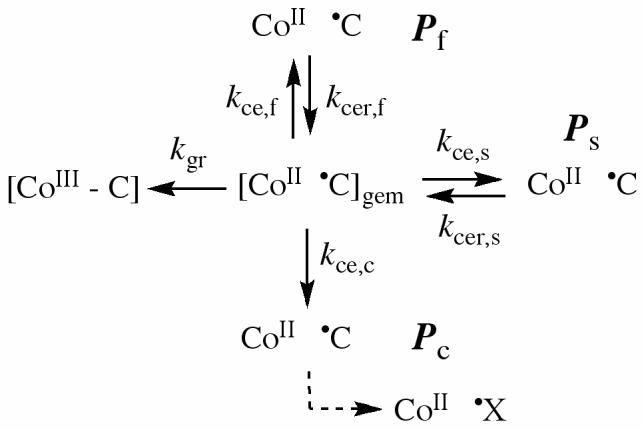
Proposed kinetic scheme for reactions of the cob(II)alamin-5′-deoxyadenosyl radical pair states following formation by photolysis in EAL.
Substrate analog binding influences the cage escape process
Table 3 shows the quantum yields for the states, Pf, Ps, and Pc in holo-EAL. These values were obtained by scaling the relative amplitudes in Table 2 by using the quantum yield value of 0.08 ±0.01 (Table 1). Table 3 also shows the quantum yields in holo-EAL in the presence of bound (S)-1-amino-2-propanol. The absence of the Pf state, and the decrease in amplitude of the Pc state relative to Ps (see Table 2) in the presence of (S)-1-amino-2-propanol, accounts for the substrate analog-induced decrease in the quantum yield of the cage escape population. Therefore, the decrease in quantum yield of the total cage escape population is caused by an effect of (S)-1-amino-2-propanol on the cage escape rate constant, rather than by an effect on the sub-nanosecond quantum yield of the geminate radical pair state, or by a change in the rate constant for geminate recombination. The latter two effects would cause a uniform decrease in the amplitudes of all three P states, with preservation of the relative amplitudes.
Table 3.
Absolute quantum yield value at 10-7 s and estimated rate constants and activation energies for cage escape cage for different escape populations in holo-EAL and in holo-EAL with bound (S)-1-amino-2-propanol.
| Condition |
Population Pi |
φi |
kce,i (s-1)a |
Ea,ce,i (kcal/mol)a |
|---|---|---|---|---|
| EAL•AdoCbl | Pf | 0.015 ±0.004 | 2×107 | 8 |
| Ps | 0.010 ±0.004 | 2×107 | 8 | |
| Pc | 0.054 ±0.009 | 6×107 | 7 | |
| EAL•AdoCbl | Pf | 0 | - | |
| •Substrate Analog | Ps | 0.011 ±0.005 | 1×107 | 8 |
| Pc | 0.029 ±0.009 | 3×107 | 7 |
kce calculated by assuming kgr = 1×10-9 s-1.
We speculate that Pf and Pc may represent states on non-productive pathways of the cob(II)alamin-5′-deoxyadenosyl radical pair reaction. If so, this would indicate that one role of protein changes induced by substrate binding is to suppress non-productive pathways of radical pair reaction (49). The Pc state may represent an intermediate in which the 5′-deoxyadenosyl radical has undergone internal radical rearrangement, or has reacted with the protein by hydrogen atom abstraction or radical substitution. These are reactions with chemical precedent in solution homolyses (50). Ps, whose yield is not suppressed by substrate analog binding, may thus resemble a productive radical pair state.
Estimation of cage escape rate constants and activation free energy barriers
The rate constants for cage escape in EAL can be estimated from the measured quantum yield values, if a value for the geminate recombination rate constant (kgr) is assumed. The work of Sension and coworkers has shown that the geminate recombination rate constants for the cob(II)alamin - 5′-deoxyadenosyl radical pair are comparable in a variety of solvents, as shown by the following values: Pure water (1.43×109 s-1) (23), ethylene glycol (1.34×109 s-1) (23) and the protein in glutamate mutase (1.08×109 s-1) (34). From these measurements, it was concluded that the rate constant for geminate recombination is “at most weakly dependent on solvent” (28), even for different alkyl axial ligands (28). If it is assumed that the geminate recombination rate constant in EAL is 1×109 s-1 (Arrhenius activation energy barrier, Ea,gr=5 kcal/mol), then the quantum yield values for Pf, Ps, and Pc (φf, φs, and φc, respectively) can be used in the following system of equations to solve for the corresponding cage escape rate constants (kce,f, kce,s, and kce,c, respectively), as follows:
| [Eq. 4] |
In Eq. 4, the index, i, corresponds to f, s, or c. The values of the three kce,i and the corresponding Arrhenius activation energies, Ea,i, are presented in Table 3.
The decays of Pf and Ps through the geminate radical pair each proceed by a two step mechanism, in which kce, kcer represent an equilibration step prior to the essentially irreversible geminate recombination (kgr). The estimated kce values are 16- to 90-fold slower than kgr, and therefore, kgr>>kce. Under these conditions, a steady-state approximation is appropriate for the geminate radical pair during the decay, and the observed monoexponential decay rate constant is equal to kcer to within approximately 1% (46). The kcer values in Table 2 correspond to Ea values of 13-14 kcal/mol. Therefore, if a cob(II)alamin-5′-deoxyadenosyl radical pair successfully escapes from the cage, then it is stabilized, by kinetic and/or thermodynamic factors, against recombination, for times that are commensurate with the lifetimes of hydrogen atom transfer reactions between carbon atoms (Ea≈13-14 kcal/mol) (51). Hydrogen atom transfer from the substrate C-H to the C5′ radical center follows radical pair separation in EAL.
Implications for the substrate trigger mechanism of cob(II)alamin-5′-deoxyadenosyl radical pair formation in EAL
The competition between geminate recombination and cage escape controls the microsecond quantum yield. The two reactions occur on a time scale of 10-9-10-7 s, and on the scale of relatively short cobalt-to-carbon distances [r(Co-C)]. These distances lie between the equilibrium Co-C bond length of 2.0 Å and approximately 4 Å, which corresponds to the diameter of one methylene carbon atom. Influences of substrate analog binding on cage escape are therefore characterized by changes in protein-coenzyme interactions that act at relatively short r(Co-C) values. (S)-1-amino-2-propanol binding effects cage escape at short r(Co-C) values, as demonstrated by the elimination of Pf and the decrease in Pc relative to Ps. However, substrate analog binding to EAL fails to increase the quantum yield, as predicted, based on the requirement of substrate binding for Co-C bond cleavage and radical pair separation (the “substrate trigger model”). We do not believe that this is because (S)-1-amino-2-propanol does not support the hydrogen atom transfer, because the time and r(Co-C) distance scales of the hydrogen atom transfer are ∼10-3 s (14, 52) and ∼7 Å (4, 5), respectively. Rather, the substrate analog may not fully induce the changes in protein-cofactor interactions that alter the cob(II)alamin-5′-deoxyadenosyl radical pair energetics (increased stabilization free energy, or decreased cage escape activation free energy barrier) that are characteristic of the true substrates, owing to steric disruption of substrate-protein interactions by the 1-methyl group. According to the mechanism in Figure 6, and the assumed value for kgr of 1×109 s-1 (28), a decrease in the value of Ea,s by 3 kcal/mol would increase the quantum yield of Ps to 0.5, from the observed value of 0.01. Thus, it is reasonable that a specific interaction of the protein with the true substrate would be reflected in an enhanced quantum yield. We will test this proposal by performing the photolysis measurements on the EAL·AdoCbl·substrate ternary complex, which is stabilized against turnover, in a recently developed cryosolvent system (52).
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Professor Thomas Netzel (Georgia State University, Atlanta) for helpful discussions regarding photomultiplier detection of optical absorption transients.
Abbreviations and Textual Footnotes
- AdoCbl
adenosylcobalamin
- EAL
ethanolamine ammonia-lyase
- EPR
electron paramagnetic resonance
- GluM
glutamate mutase
Footnotes
The project described was supported by Grant Number R01DK054514 from the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
SUPPORTING INFORMATION AVAILABLE
Monoexponential plus constant fit to the post-photolysis cob(II)alamin decay for AdoCbl in EAL, residuals for the biexponential and monoexponential fits to the post-photolysis cob(II)alamin decay for AdoCbl in EAL, and residuals for the biexponential and monoexponential fits to the post-photolysis cob(II)alamin decay for AdoCbl in EAL with bound (S)-1-amino-2-propanol.
REFERENCES
- 1.Toraya T. Radical catalysis in coenzyme B-12-dependent isomerization (eliminating) reactions. Chemical Reviews. 2003;103:2095–2127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R, Ragsdale SW. The many faces of vitamin B-12: Catalysis by cobalamin-dependent enzymes. Annual Review of Biochemistry. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 3.Brown KL. Chemistry and Enzymology of Vitamin B12. Chem. Rev. 2005;105:2075–2149. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- 4.Canfield JM, Warncke K. Geometry of reactant centers in the Co-II-substrate radical pair state of coenzyme B-12-dependent ethanolamine deaminase determined by using orientation-selection-ESEEM spectroscopy. Journal of Physical Chemistry B. 2002;106:8831–8841. doi: 10.1021/jp046167m. [DOI] [PubMed] [Google Scholar]
- 5.Canfield JM, Warncke K. Active site reactant center geometry in the Co-II-product radical pair state of coenzyme B-12-dependent ethanolamine deaminase determined by using orientation-selection electron spin-echo envelope modulation spectroscopy. Journal of Physical Chemistry B. 2005;109:3053–3064. doi: 10.1021/jp046167m. [DOI] [PubMed] [Google Scholar]
- 6.Hay BP, Finke RG. Thermolysis of adenosylcobalamin: A product, kinetic and Co-C5' bond dissociation energy study. Inorg. Chem. 1984;23:3041–3043. [Google Scholar]
- 7.Halpern J, Kim S-H, Leung TW. J. Am. Chem. Soc. 1984;106:8317–8319. [Google Scholar]
- 8.Hay BP, Finke RG. Thermolysis of the Co-C bond in adenosylcobalamin (Coenzyme B12) -IV. Products, kinetics and Co-C bond dissociation energy studies in ethylene glycol. Polyhedron. 1988;7:1469–1481. [Google Scholar]
- 9.Brooks AJ, Vlasie M, Banerjee R, Brunold TC. Spectroscopic and computational studies on the adenosylcobalamin-dependent methylmalonyl-CoA mutase: Evaluation of enzymatic contributions to Co-C bond activation in the Co+3 ground state. J. Am. Chem. Soc. 2004;126:8167–8180. doi: 10.1021/ja039114b. [DOI] [PubMed] [Google Scholar]
- 10.Brooks AJ, Vlasie M, Banerjee R, Brunold TC. Co-C bond activation in methylmalonyl-CoA mutase by stabilization of the post-homolysis product Co+2 cobalamin. J. Am. Chem. Soc. 2005;127:16522–16528. doi: 10.1021/ja0503736. [DOI] [PubMed] [Google Scholar]
- 11.Padmakumar R, Padmakumar R, Banerjee R. Evidence that cobalt-carbon bond homolysis is coupled to hydrogen atom abstraction from substrate in methylmalonyl-CoA mutase. Biochemistry-Us. 1997;36:3713–3718. doi: 10.1021/bi962503g. [DOI] [PubMed] [Google Scholar]
- 12.Marsh ENG, Ballou DP. Coupling of cobalt-carbon bond homolysis and hydrogen atom abstraction in adenosylcobalamin-dependent glutamate mutase. Biochemistry. 1998;37:11864–11872. doi: 10.1021/bi980512e. [DOI] [PubMed] [Google Scholar]
- 13.Licht SS, Booker S, Stubbe J. Studies on the catalysis of carbon-cobalt bond homolysis by ribonucleoside triphosphate reductase: Evidence for concerted carbon-cobalt bond homolysis and thiyl radical formation. Biochemistry-Us. 1999;38:1221–1233. doi: 10.1021/bi981885i. [DOI] [PubMed] [Google Scholar]
- 14.Bandarian V, Reed GH. Isotope effects in the transient phases of the reaction catalyzed by ethanolamine ammonia-lyase: Determination of the number of exchangeable hydrogens in the enzyme-cofactor complex. Biochemistry. 2000;39:12069–12075. doi: 10.1021/bi001014k. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard BK, Gulick AM, Babbitt PC, Rayment I, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: Mechanism, structure, and metabolic context of glucarate dehydratase from Escherichia coli. Faseb Journal. 1999;13:A1446–A1446. [Google Scholar]
- 16.Sun L, Warncke K. Comparative model of EutB from coenzyme B-12-dependent ethanolamine ammonia-lyase reveals a beta(8)alpha(8), TIM-barrel fold and radical catalytic site structural features. Proteins-Structure Function and Bioinformatics. 2006;64:308–319. doi: 10.1002/prot.20997. [DOI] [PubMed] [Google Scholar]
- 17.Bandarian V, Reed GH. Ethanolomine Ammonia-Lyase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley and Sons; New York: 1999. pp. 811–833. [Google Scholar]
- 18.Bradbeer C. Clostridial Fermentations of Choline and Ethanolamine .I. Preparation and Properties of Cell-Free Extracts. J Biol Chem. 1965;240:4669–4674. [PubMed] [Google Scholar]
- 19.Endicott JF, Netzel TL. Early Events and Transient Chemistry in the Photohomolysis of Alkylcobalamins. Journal of the American Chemical Society. 1979;101:4000–4002. [Google Scholar]
- 20.Shiang JJ, Walker LA, Anderson NA, Cole AG, Sension RJ. Time-resolved spectroscopic studies of B-12 coenzymes: The photolysis of methylcobalamin is wavelength dependent. Journal of Physical Chemistry B. 1999;103:10532–10539. [Google Scholar]
- 21.Shiang JJ, Cole AG, Sension RJ, Hang K, Weng Y, Trommel J, Marzilli L, Lian T. Ultrafast excited-state dynamics in vitamin B12 and related cob(III)alamins. J. Am. Chem. Soc. 2006;128:801–808. doi: 10.1021/ja054374+. [DOI] [PubMed] [Google Scholar]
- 22.Noyes RM. Effects of Diffusion Rates on Chemical Kinetics. Prog React Kinet Mec. 1961;1:129–160. [Google Scholar]
- 23.Yoder LM, Cole AG, Walker LA, Sension RJ. Time-resolved spectroscopic studies of B-12 coenzymes: Influence of solvent on the photolysis of adenosylcobalamin. Journal of Physical Chemistry B. 2001;105:12180–12188. [Google Scholar]
- 24.Walker LA, Shiang JJ, Anderson NA, Pullen SH, Sension RJ. Time-resolved spectroscopic studies of B-12 coenzymes: The photolysis and geminate recombination of adenosylcobalamin. Journal of the American Chemical Society. 1998;120:7286–7292. [Google Scholar]
- 25.Walker LA, Jarrett JT, Anderson NA, Pullen SH, Matthews RG, Sension RJ. Time-resolved spectroscopic studies of B-12 coenzymes, the identification of a metastable cob(III)alamin photoproduct in the photolysis of methylcobalamin. Journal of the American Chemical Society. 1998;120:3597–3603. [Google Scholar]
- 26.Shiang JJ, Cole AG, Sension RJ, Hang K, Weng YX, Trommel JS, Marzilli LG, Lian TQ. Ultrafast excited-state dynamics in vitamin B-12 and related Cob(III)alamins. Journal of the American Chemical Society. 2006;128:801–808. doi: 10.1021/ja054374+. [DOI] [PubMed] [Google Scholar]
- 27.Sension RJ, Walker LA, Shiang JJ. Ultrafast transient absorption studies of B-12 enzymes and coenzymes. Abstracts of Papers of the American Chemical Society. 1998;216:U684–U684. [Google Scholar]
- 28.Sension RJ, Harris DA, Cole AG. Time-resolved spectroscopic studies of B-12 coenzymes: Comparison of the influence of solvent on the primary photolysis mechanism and geminate recombination of methyl-, ethyl-, n-propyl-, and 5 ′-deoxyadenosylcobalamin. Journal of Physical Chemistry B. 2005;109:21954–21962. doi: 10.1021/jp053202w. [DOI] [PubMed] [Google Scholar]
- 29.Cole AG, Yoder LM, Shiang JJ, Anderson NA, Walker LA, Holl MMB, Sension RJ. Time-resolved spectroscopic studies of B-12 coenzymes: A comparison of the primary photolysis mechanism in methyl-, ethyl-, n-propyl-, and 5 ′-deoxyadenosylcobalamin. Journal of the American Chemical Society. 2002;124:434–441. doi: 10.1021/ja011628s. [DOI] [PubMed] [Google Scholar]
- 30.Cole AG, Anderson N, Shiang JJ, Sension RJ. Ultrafast spectroscopic studies of coenzyme B-12 derivatives and analogs. Abstracts of Papers of the American Chemical Society. 2000;220:U223–U223. [Google Scholar]
- 31.Chen EF, Chance MR. Nanosecond Transient Absorption-Spectroscopy of Coenzyme-B-12 - Quantum Yields and Spectral Dynamics. Abstracts of Papers of the American Chemical Society. 1990;200:196-INOR. [PubMed] [Google Scholar]
- 32.Brownawell AM, Chen E, Chance MR. Laser Photolysis of Alkyl-Cobinamides - Models for Homolytic and Heterolytic Cobalamin Enzymes. Biophysical Journal. 1993;64:A161–A161. [Google Scholar]
- 33.Lott WB, Chagovetz AM, Grissom CB. Alkyl radical geometry controls geminate cage recombination in alkylcobalamins. J. Am. Chem. Soc. 1995;117:12194–12201. [Google Scholar]
- 34.Sension RJ, Cole AG, Harris AD, Fox CC, Woodbury NW, Lin S, Marsh ENG. Photolysis and recombination of adenosylcobalamin bound to glutamate mutase. Journal of the American Chemical Society. 2004;126:1598–1599. doi: 10.1021/ja0396910. [DOI] [PubMed] [Google Scholar]
- 35.Sension RJ, Harris DA, Stickrath A, Cole AG, Fox CC, Marsh ENG. Time-resolved measurements of the photolysis and recombination of adenosylcobalamin bound to glutamate mutase. Journal of Physical Chemistry B. 2005;109:18146–18152. doi: 10.1021/jp052492d. [DOI] [PubMed] [Google Scholar]
- 36.Pett VB, Liebman MN, Murrayrust P, Prasad K, Glusker JP. Conformational Variability of Corrins - Some Methods of Analysis. J Am Chem Soc. 1987;109:3207–3215. [Google Scholar]
- 37.Masuda J, Shibata N, Morimoto Y, Toraya T, Yasuoka N. How a protein generates a catalytic radical from coenzyme B-12: X-ray structure of a dioldehydratase-adeninylpentylcobalamin complex. Structure. 2000;8:775–788. doi: 10.1016/s0969-2126(00)00164-7. [DOI] [PubMed] [Google Scholar]
- 38.Khoroshun DV, Warncke K, Ke SC, Musaev DG, Morokuma K. Internal degrees of freedom, structural motifs, and conformational energetics of the 5-deoxyadenosyl radical: Implications for function in adenosylcobalamin-dependent enzymes. A computational study. Journal of the American Chemical Society. 2003;125:570–579. doi: 10.1021/ja028393k. [DOI] [PubMed] [Google Scholar]
- 39.Gani D, Wallis CO, Young DW. Stereochemistry of the rearrangement of 2-aminoethanol by ethanolamine ammonia-lyase. Eur. J. Biochem. 1983;136:303–311. doi: 10.1111/j.1432-1033.1983.tb07742.x. [DOI] [PubMed] [Google Scholar]
- 40.Yan S-J, McKinnie BG, Abacherii C, Hill RK, Babior BM. Stereochemistry of the ethanolamine ammonia-lyase reaction with stereospecifically labeled [1-2H1]-2-aminoethanol. J. Am. Chem. Soc. 1984;106:2961–2964. [Google Scholar]
- 41.Faust LRP, Connor JA, Roof DM, Hoch JA, Babior BM. Cloning, Sequencing, and Expression of the Genes Encoding the Adenosylcobalamin-Dependent Ethanolamine Ammonia-Lyase of Salmonella-Typhimurium. Journal of Biological Chemistry. 1990;265:12462–12466. [PubMed] [Google Scholar]
- 42.Faust LP, Babior BM. Overexpression, Purification, and Some Properties of the Adocbl-Dependent Ethanolamine Ammonia-Lyase from Salmonella-Typhimurium. Archives of Biochemistry and Biophysics. 1992;294:50–54. doi: 10.1016/0003-9861(92)90135-j. [DOI] [PubMed] [Google Scholar]
- 43.Harkins TT, Grissom CB. The Magnetic-Field Dependent Step in Bit Ethanolamine Ammonia-Lyase Is Radical-Pair Recombination. Journal of the American Chemical Society. 1995;117:566–567. [Google Scholar]
- 44.Kaplan BH, Stadtman ER. Ethanolamine Deaminase a Cobamide Coenzyme-Dependent Enzyme .I. Purification Assay and Properties of Enzyme. Journal of Biological Chemistry. 1968;243:1787. [PubMed] [Google Scholar]
- 45.Chen E, Chance MR. Continuous-Wave Quantum Yields of Various Cobalamins Are Influenced by Competition between Geminate Recombination and Cage Escape. Biochemistry. 1993;32:1480–1487. doi: 10.1021/bi00057a011. [DOI] [PubMed] [Google Scholar]
- 46.Moore JW, Pearson RG. Kinetics and Mechanism. Wiley and Sons; New York: 1981. [Google Scholar]
- 47.Huhta MS, Chen H-P, Hemann C, Hille CR, Marsh ENG. Protein-coenzyme interactions in adenosylcobalamin-dependent glutamate mutase. Biochem. J. 2001;355:131–137. doi: 10.1042/0264-6021:3550131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogenkam Hp. Photolysis of Methylcobalamin. Biochemistry-Us. 1966;5:417. doi: 10.1021/bi00866a005. [DOI] [PubMed] [Google Scholar]
- 49.Retey J. Enzymic reaction selectivity by negative catalysis or how do enzymes deal with highly reactive intermediates. Angew. Chem. Int. Ed. Engl. 1990;29:355–361. [Google Scholar]
- 50.Hay BP, Finke RG. Thermolysis of the Co-C bond in adenosylcorrins. 3. Quantification of the axial base effect in adenosylcobalamin by the synthesis and thermolysis of axial base free adenosylcobinamide. Insights into the energetics of enzyme-assisted cobalt-carbon bond homolysis. J. Am. Chem. Soc. 1987;109:8012–8018. [Google Scholar]
- 51.Zavitsas AA, Chatgilialoglu C. Energies of activation. The paradigm of hydrogen abstraction by radicals. J. Am. Chem. Soc. 1995;117:10645–10654. [Google Scholar]
- 52.Wang M, Warncke K. Kinetic and Thermodynamic Characterization of CoII-Substrate Radical Pair Formation in Coenzyme B12-Dependent Ethanolamine Ammonia-Lyase in a Cryosolvent System by using Time-Resolved, Full-Spectrum Continuous-Wave Electron Paramagnetic Resonance Spectroscopy. J. Am. Chem. Soc. 2008;130:4846–4858. doi: 10.1021/ja710069y. [DOI] [PubMed] [Google Scholar]
- 53.Ke SC, Torrent M, Museav DG, Morokuma K, Warncke K. Identification of dimethylbenzimidazole axial coordination and characterization of N-14 superhyperfine and nuclear quadrupole coupling in Cob(II)alamin bound to ethanolamine deaminase in a catalytically-engaged substrate radical-cobalt(II) biradical state. Biochemistry-Us. 1999;38:12681–12689. doi: 10.1021/bi983067w. [DOI] [PubMed] [Google Scholar]
- 54.Abend A, Bandarian V, Nitsche R, Stupperich E, Retey J, Reed GH. Ethanolamine ammonia-lyase has a “base-on” binding mode for coenzyme B-12. Archives of Biochemistry and Biophysics. 1999;370:138–141. doi: 10.1006/abbi.1999.1382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.