Abstract
In this study, we report the characterization of a novel genotoxic and nongenotoxic stress-regulated gene that we had previously named as SKNY. Our results indicate that SKNY encodes the recently identified neutral sphingomyelinase-3 (nSMase3; hereafter SKNY is referred to as nSMase3). Examination of nSMase3 subcellular distribution reveals nSMase3 to localize to the endoplasmic reticulum (ER), and deletion of a COOH-terminal region containing its putative transmembrane domain and ER targeting signal partly alters its compartmentalization to the ER. Treatment with genotoxic Adriamycin and nongenotoxic tumor necrosis factor-α up-regulates endogenous nSMase3 expression, albeit with different kinetics. Tumor necrosis factor-α up-regulates nSMase3 expression within 2 h that lasts beyond 24 h and declines to control levels by 36 h. Adriamycin up-regulation of nSMase3 is transient, occurs within 30 min, and declines to control levels by 120 min. Prolonged treatment with Adriamycin by 24 h and beyond, however, causes a down-regulation in nSMase3 expression. Activation of wild-type p53 also down-regulates nSMase3 expression, suggesting that DNA damage-mediated nSMase3 down-regulation seems to occur partly through the tumor suppressor p53. Overexpression of exogenous nSMase3 sensitizes cells to Adriamycin-induced cell killing, a finding consistent with the proposed proapoptotic role of nSMase enzymes and nSMase-generated ceramide. We further investigated nSMase3 expression in various human malignancies and found its expression to be deregulated in several types of primary tumors when compared with their matching normal tissues. Collectively, our results have identified nSMase3 to be an important molecule that is linked to tumorigenesis and cellular stress response.
Introduction
Long been known to serve as basic structural components of cellular membranes, sphingolipids are becoming increasingly recognized as critical modulators of cell signaling events (1, 2). Ceramide is a central hub for sphingolipid metabolism with its breakdown or modification resulting in the formation of a host of sphingolipids such as sphingosine, sphingosine-1-phosphate, and ceramide-1-phosphate that, like ceramide itself, can potentially serve as both structural and/or cell signaling molecules (1–4). It has been well established that ceramide mostly functions to potentiate cell death; however, some lines of evidence suggest that it may also serve to promote cell growth and differentiation, thus making it a pleiotropic molecule (5). Complimentary to ceramide catalysis is its synthesis, which can occur through multiple mechanisms, most notably via (a) de novo ceramide synthesis or (b) the hydrolysis of sphingomyelin into ceramide and phosphorylcholine (1–4). The former occurs at the cytosolic surface of the endoplasmic reticulum (ER) and is followed by vesicular or CERT-mediated nonvesicular transport of ceramide to the Golgi apparatus (6, 7). The generation of ceramide that occurs through the breakdown of sphingomyelin is facilitated by sphingomyelinases, of which there are multiple classes categorized by the pH at which they achieve optimal activity (8).
In the context of human malignancy, increased ceramide production by either ceramide-synthesizing pathway is usually synonymous with the suppression of tumor cell growth and proliferation. This is evidenced by numerous cell-based and animal studies showing the ability of ceramide to induce cell cycle arrest or apoptosis (9–11). Furthermore, ceramide has been shown to function as a key mediator of apoptosis brought on by stress-inducing therapeutic agents, such as anthracyclines (11–14), ionizing radiation (15, 16), tumor necrosis factor (TNF)-related apoptosis-inducing ligand (17), and paclitaxel (18). Reciprocally, the inhibition of ceramide production by compounds such as the ceramide synthase inhibitor fumonisin B1 or by small interfering RNA approaches has been shown to abrogate stress-induced apoptosis (11, 19–21). Collectively, these findings have outlined (a) a critical inhibitory role for ceramide during tumorigenesis and (b) its ability to sensitize malignant cells to the effects of radiation and chemotherapeutic agents.
The modulation of ceramide levels (a) by stress-inducing agents (11–18) or (b) as noted in multiple primary tumor types (reviewed in refs. 3, 22) is likely to result due to multiple changes in the regulation of ceramide-metabolizing enzymes or pathways; however, the exact molecular mechanisms responsible for these alterations remain unclear. Many initial studies implicated acid sphingomyelinase (aSMase) as a critical mediator of the stress response. For example, activation of death receptors TNF receptor 1 and Fas (CD95) is known to stimulate ceramide production through activation of aSMase (23, 24). Similarly, the activation of aSMase by ionizing radiation is well established (16, 25) and increased aSMase activity following the DNA-damaging drug Adriamycin (doxorubicin) has also been reported (26). The importance of aSMase-mediated ceramide production is exemplified in aSMase−/− mice and in lymphoblasts from Niemann-Pick patients that lack aSMase activity, where generation of ceramide and induction of apoptosis following exposure to ionizing radiation is abrogated (16).
More recently, the neutral sphingomyelinases (nSMase) have also been implicated in cell stress-induced growth arrest and apoptosis. For example, nSMase1 and nSMase2 activation has been reported by TNF-α (27, 28), and thus, ceramide produced following TNF-α is likely to result due to coordinate increases in the activities of both aSMases and nSMases. In addition to TNF-α, nSMase2 is also activated by oxidative stress (29) and seems to be involved in inducing growth arrest in MCF-7 human breast cancer cells (30). Furthermore, nSMase activity is enhanced by ionizing radiation (15, 31), the DNA-damaging agents Adriamycin, daunorubicin, and etoposide (12, 32, 33), hypoxia (34), and UV radiation (35), although the relative contribution of specific nSMase enzymes in mediating the response to these stressors remains unclear. The creation of nSMase1−/− and nSMase2−/− mice has provided some insights into the functions of these enzymes (36, 37); however, more studies are needed to fully examine the independent effects of each in cellular stress-induced ceramide generation and their role in tumorigenesis. Adding another layer to the complexity of ceramide metabolism, Krut et al. (38) have very recently reported the identification of nSMase3 as a third neutral sphingomyelinase. Remarkably, several years ago, using an unbiased technique, we had discovered a novel genotoxic and nongenotoxic stress-regulated gene named SKNY, which was recently identified to encode nSMase3. Given that very little is known about the biological functions of nSMase3, here we report further characterization of nSMase3 and its potential role in chemotherapy-induced growth inhibition in human cancer cells and its deregulation in various human tumor tissues.
Results
Identification of nSMase3 as a Novel Stress-Regulated Gene
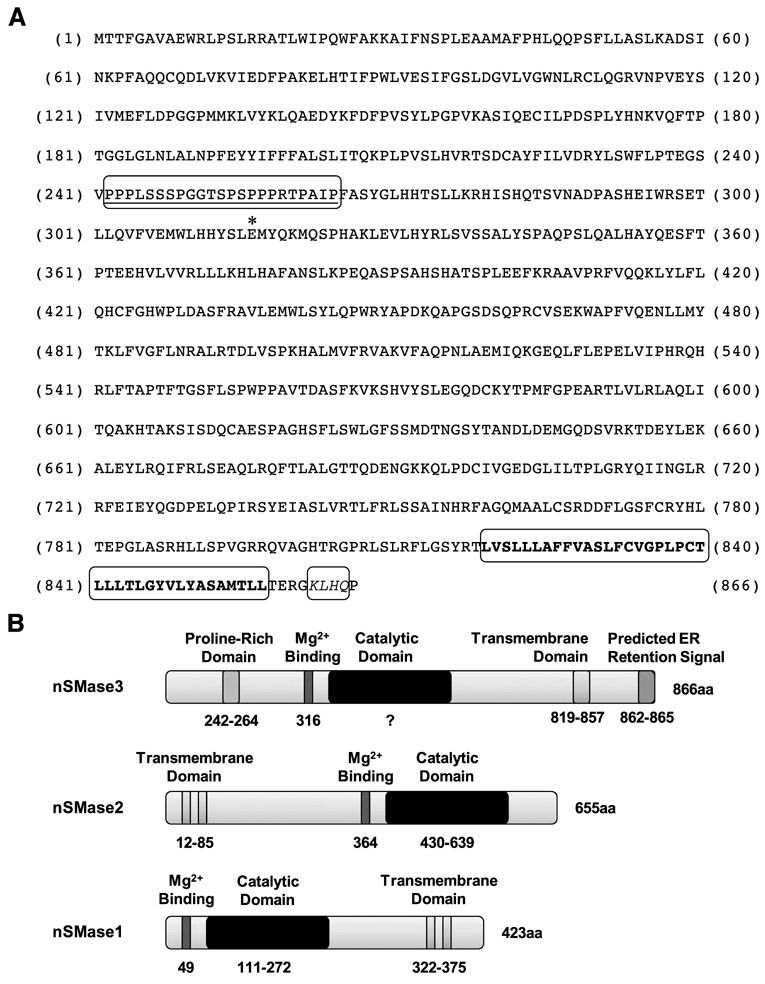
Previously, we reported the cloning and characterization of DNA damage-regulated genes (39, 40) whose partial-length cDNAs were first identified by subtraction hybridization approach (41). Recently, we used computer-based approaches to aligning redundant expressed sequence tags in the databases to search for sequences that contained novel open reading frames (ORF), and identified multiple novel ORFs exhibiting regulation in response to genotoxic (DNA damage) and nongenotoxic stresses. This approach enabled us to identify and clone several novel stress-regulated genes, including the recently reported PDRG (42) and PIQ (43). One of the novel stress-regulated cDNAs (temporarily named SKNY) was found to encode a hypothetical protein of 866 amino acids with predicted molecular mass of 98 kDa. More recently, Krut et al. (38) reported the identification of a nSMase3; our sequence comparison of nSMase3 and SKNY revealed both to be identical (hereafter SKNY will be referred to as nSMase3). Interestingly, nSMase3 does not display significant sequence homology with nSMase1 and nSMase2 or any other known sphingomyelinases, which is in keeping with our inability to identify SKNY as a novel neutral sphingomyelinase. Figure 1 shows the amino acid sequence of nSMase3 and a schematic illustration of its putative motifs. The putative structural motifs were searched using multiple protein prediction programs, including ELM, EMBL-EBI, Prosite, PSORTII, SOSUI, and TMPed. These predictions revealed a putative proline-rich region, which is believed to mediate protein-protein interactions, and, depending on the program, either a one or two transmembrane-spanning region residing at amino acids 819 to 857 at the COOH terminus of nSMase3 (Fig. 1). Several of these programs predicted a putative KKXX-like ER retention signal also at the nSMase3 COOH terminus (Fig. 1).
FIGURE 1. nSMase3 protein sequence and schematic illustration of nSMase functional motifs.
A. Amino acid sequences of human nSMase3. The predicted proline-rich (242–264) region is underlined. The putative glutamic acid (Glu316) involved in Mg2+ binding is denoted by an asterisk. The predicted transmembrane-spanning region (819–857) is in bold letters and the predicted KKXX-like ER retention signal KLHQ (862–865) is italicized. B. Schematic illustration of putative nSMase3 domains; schematic illustration of known nSMase1 and nSMase2 domains is also shown for comparison.
nSMase3 Is Localized to the ER
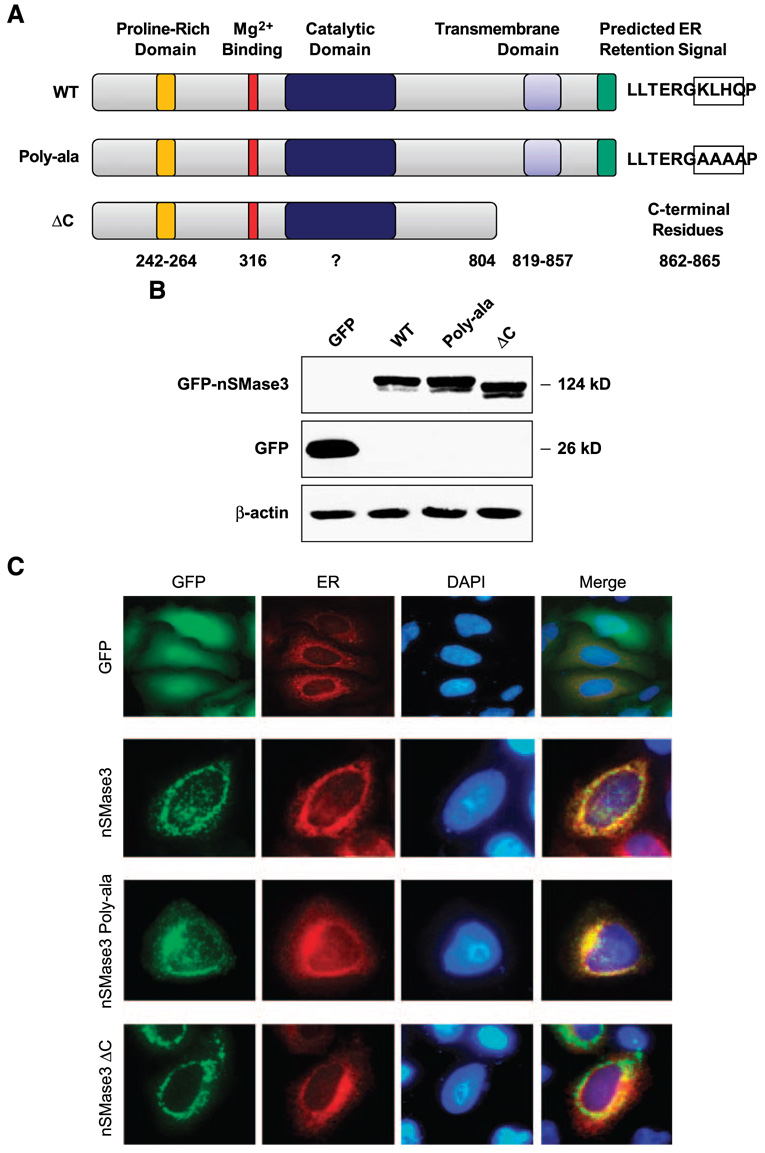
Based on subcellular localization predictions, we did a series of experiments to investigate whether nSMase3 indeed resides at the ER. First, we generated the expression construct pEGFPnSMase3 in which green fluorescent protein (GFP) was tagged to the NH2 terminus of nSMase3. HEK293T cells were transiently transfected with the pEGFP-nSMase3, and Western blot analyses done on cell lysates confirmed the expression as well as the correct size of the GFP-tagged nSMase3 (Fig. 2A and B). HeLa human cervical cancer cells were then transiently transfected with expression constructs pEGFP or pEGFP-nSMase3 in combination with the expression vector pDsRed2-ER. The expression vector pDsRed2-ER expresses a red fluorescent protein directed specifically to the ER by an NH2-terminal ER localization signal and a COOH-terminal ER retention signal. As shown in Fig. 2C, GFP-tagged nSMase3 exhibited colocalization with ER-localized red fluorescent protein, indicating that nSMase3 was in fact directed to the ER. Similar results were obtained when GFP was tagged to the COOH terminus of nSMase3 (data not shown), suggesting that the localization of nSMase3 was not influenced by the presence of GFP tag.
FIGURE 2. Subcellular distribution of GFP-tagged nSMase3.
A. Schematic illustration of full-length and mutation/deletion constructs of nSMase3. Numbers represent the amino acid residues corresponding to putative functional domains. Predicted KKXX-like signal (KLHQ) in WT nSMase3 and those residues changed to alanine in nSMase3 (Poly-ala) are boxed. B. Western blotting was done using an anti-GFP antibody to detect GFP and GFP-nSMase3 expression. The same blot was then probed for β-actin to show equal protein loading. C. HeLa cells were cotransfected with expression vectors carrying GFP or GFP-nSMase3 with pDsRed2-ER using Lipofectamine 2000. Cells were washed with PBS, fixed with 4% paraformaldehyde, and incubated with 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclei. Cells were analyzed with an Olympus AX70 fluorescent microscope and photographs were captured using a digital camera. Photomicrographs of GFP-transfected cells are not to scale with those transfected with GFPnSMase3 expression vectors.
We also sought to investigate the significance of the predicted KKXX-like ER retention signal present at position 862 to 865 (KLHQ) in nSMase3. To that end, we did site-directed mutagenesis and replaced the KLHQ sequence with alanine residues and assessed the subcellular localization of the mutant nSMase3. As shown in Fig. 2C, mutation of the putative ER retention signal “KLHQ” had no apparent effect on nSMase3 localization, suggesting that this sequence does not seem to play a role in the ER localization of nSMase3. It may be possible that nSMase3 belongs to the C-tail–anchored protein family that lacks the consensus ER targeting and retention signals (38); however, whether nSMase3 is in fact a C-tail–anchored protein remains to be investigated. C-tail–anchored proteins are believed to harbor their ER or mitochondrial targeting signals within their COOH-terminal transmembrane domains (TMD; refs. 44, 45) We therefore reasoned that if nSMase3 also harbored ER targeting and retention signal in the TMD, then deletion of the TMD should significantly alter the localization of nSMase3 to the ER. To investigate this possibility, we did site-directed mutagenesis to delete the COOH terminus of nSMase3 (nSMase3 ΔC) and analyzed its subcellular distribution. As shown in Fig. 2C, deletion of the putative nSMase3 TMD decreased its colocalization with the ER marker protein (red fluorescent); however, the majority of nSMase3 seemed to maintain a punctuate perinuclear distribution pattern, suggesting most of the nSMase3 ΔC to still localize to the ER (Fig. 2C).
TNF-α and Adriamycin Regulate nSMase3 mRNA Expression
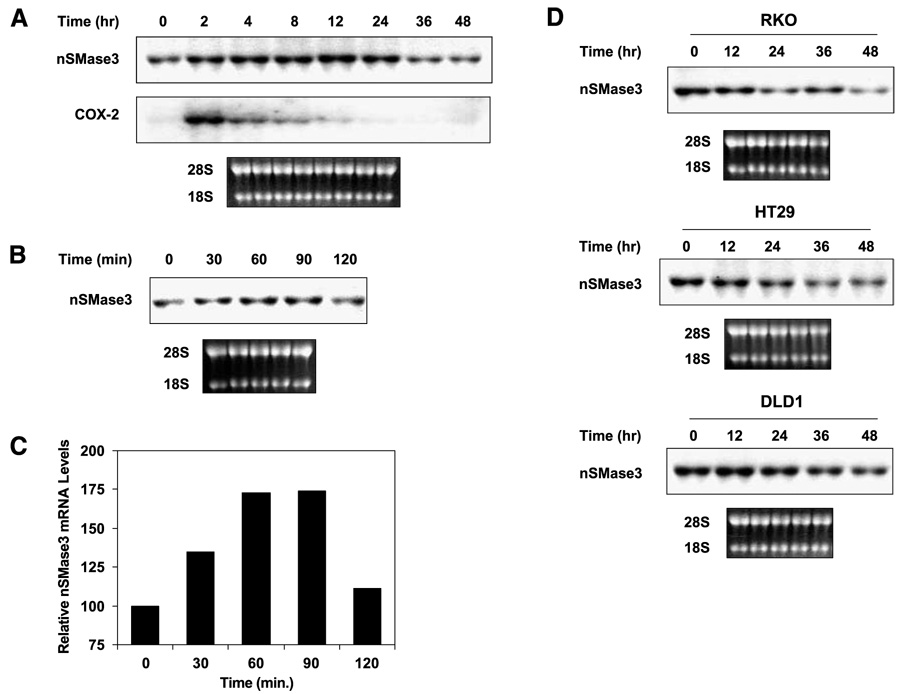
Figure 3A shows nSMase3 regulation by the nongenotoxic agent TNF-α, and as is shown, TNF-α treatment of HT29 human colon cancer cells up-regulated nSMase3 mRNA expression as a function of time and its effect was blunted by 36 h and thereafter. It is well established that TNF-α potently enhances the expression of cyclooxygenase-2 during the inflammatory response (46). Therefore, as a control, we probed the same blot for cyclooxygenase-2 expression, and as expected, TNF-α up-regulated cyclooxygenase-2 mRNA levels (Fig. 3A). The genotoxic agent Adriamycin also up-regulated nSMase3 expression that was noted at very early time points. Figure 3B shows a representative Northern blot indicating nSMase3 up-regulation by Adriamycin in RKO human colon cancer cells, and as is shown, Adriamycin-mediated up-regulation of nSMase was noted within 30 min of Adriamycin treatment and declined by 120 min. Next, we sought to investigate the effect of Adriamycin on nSMase3 expression following longer-term treatment, and as shown in Fig. 3D, prolonged treatment with Adriamycin caused a decrease in nSMase3 levels in various human colon cancer cell lines, including RKO, HT29, and DLD1.
FIGURE 3. TNF-α and DNA-damaging agents regulate nSMase3 mRNA expression.
A. A representative Northern blot showing the effect TNF-α treatment on endogenous nSMase3 mRNA levels in HT29 cells. Cells were either left untreated or treated with 50 ng/mL TNF-α for 2, 4, 8, 12, 24, 36, or 48 h. COX-2, cyclooxygenase-2. B. A representative Northern blot showing the effect of short-term Adriamycin treatment on endogenous nSMase3 mRNA levels in RKO cells. Cells were either left untreated or treated with 1 µmol/L Adriamycin for 30, 60, 90, or 120 min. C. A graphic representation of nSMase3 levels depicted in B. Relative nSMase3 mRNA levels were determined by normalizing nSMase3 mRNA to 18S rRNA. D. A representative Northern blot showing the effect of long-term Adriamycin treatment on endogenous nSMase3 mRNA levels. Cells were either left untreated or treated with 0.50 µmol/L (RKO), 1 µmol/L (DLD1), or 5 µmol/L (HT29) of Adriamycin for 12, 24, 36, or 48 h. A, B, and D. Total RNA was harvested and then subjected to Northern blot analysis as described in Materials and Methods. The blots were sequentially probed with 32P-labeled nSMase3 and cyclooxygenase-2 or p21 probes; ethidium bromide staining of the 28S and 18S rRNA is also shown to illustrate loading and RNA integrity.
p53 Down-Regulates nSMase3 mRNA Expression
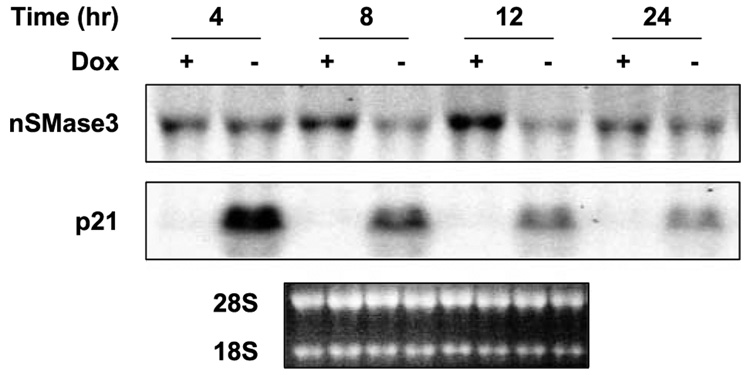
The tumor suppressor p53 is a major regulator of the cellular response to genotoxic stress (47–49). One way in which p53 modulates the cellular stress response is by functioning as a transcription factor to regulate the expression of genes important to cell growth arrest and apoptosis, such as p21WAF1, PUMA, DR5, and BAX. Because genotoxic stress regulated nSMase3 expression, we therefore sought to investigate p53 regulation of nSMase3 expression. To that end, we used DLD1 human colon cancer cells harboring a “bidirectional tet-off expression vector” system controlling the inducible expression of exogenous wild-type (WT) p53 (50). In these cells, the expression of p53 is repressed when they are grown in the presence of doxycycline but induced when the antibiotic is removed from the growth medium. Figure 4 shows that induction of p53 expression in these cells was associated with an obvious decrease in the endogenous nSMase3 mRNA levels within 8 h. To confirm the functionality of WT p53, we also analyzed the expression of p21WAF1, a well-established downstream target of p53. As is shown in Fig. 4, p21WAF1 levels were increased following p53 induction, thereby confirming the functionality of exogenous WT p53. Our results therefore suggest that p53 seems to be partly involved in the down-regulation of nSMase3 expression during prolonged genotoxic stress.
FIGURE 4. nSMase3 is down-regulated by p53.
A representative Northern blot showing the effect of inducible WT p53 on endogenous nSMase3 mRNA levels in DLD1 cells. Cells grown in the presence (p53 repressed) or absence (p53 induced) of 40 ng/mL doxycycline (Dox) for indicated periods of time were harvested, RNA extracted, and subjected to Northern blot analysis as described in Materials and Methods. The blot was sequentially probed with 32P-labeled nSMase3 and p21WAF1 probes; ethidium bromide staining of the 28S and 18S rRNA is also shown for RNA loading and integrity.
nSMase3 Overexpression Sensitizes Cells to DNA Damage-Induced Apoptosis
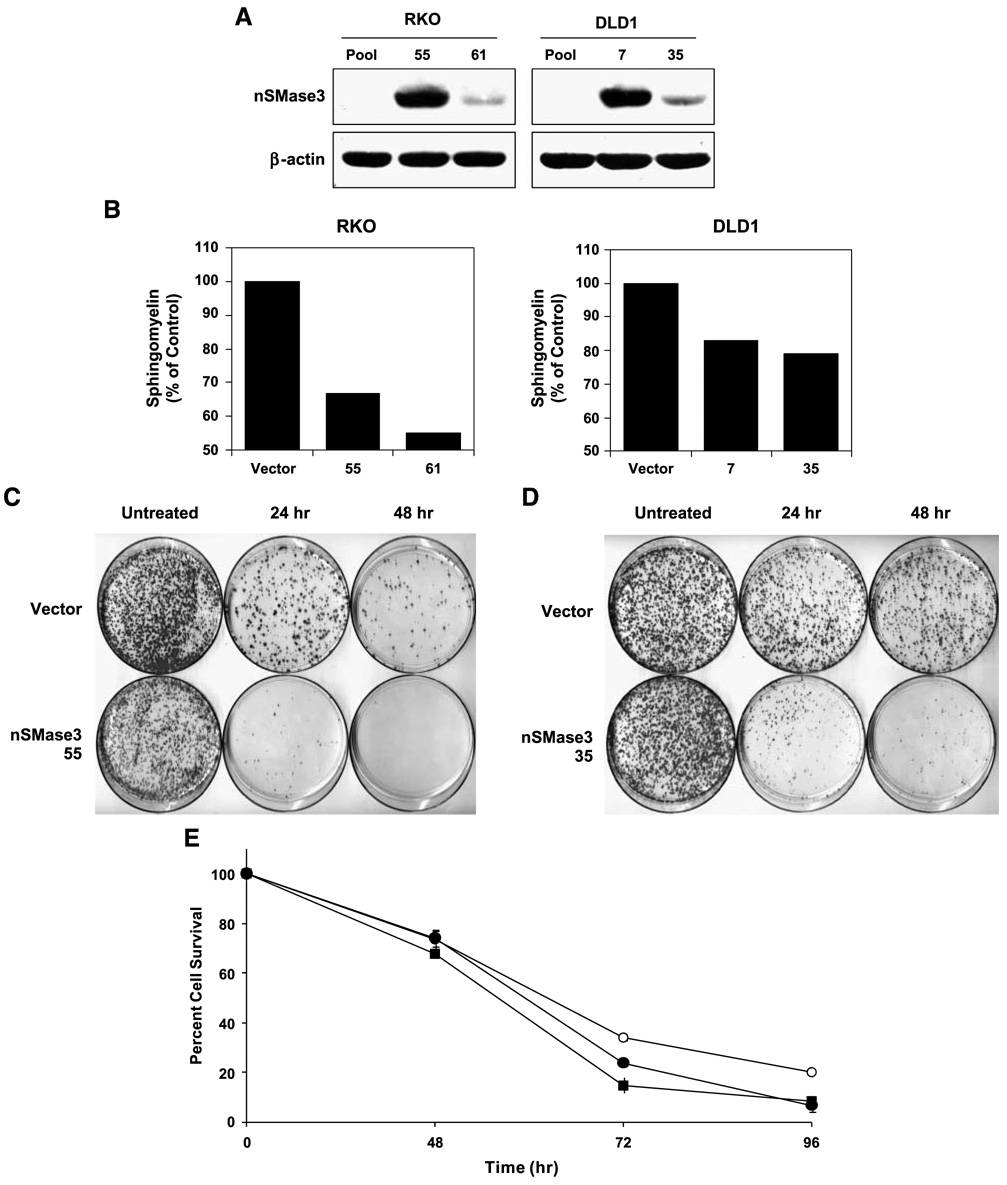
Given that nSMase3 is a newly discovered member of the neutral sphingomyelinase family, its role in modulating cell growth or apoptosis has not been investigated. Thus, to gain further insight as to what role nSMase3 may play in these processes, we first generated cell lines stably expressing exogenous nSMase3. RKO and DLD1 human colon cancer cells were used for transfections with an expression vector carrying Myc-tagged nSMase3. Stable clones expressing nSMase3 were identified by Western blotting using anti-Myc antibodies. We selected two clones for each cell line for further studies, and the relative expression of exogenous nSMase3 in these stable transfectants is shown in Fig. 5A. Cellular sphingomyelin levels were also measured in these stable transfectants to confirm that the exogenous nSMase3 was enzymatically active, and as shown in Fig. 5B, both RKO and DLD1 expressing exogenous nSMase3 displayed reduced levels of total sphingomyelin compared with vector-only–transfected cells. The major species were C24-sphingomyelin and C16-sphingomyelin, whose levels were 13.7 and 20.4 or 25.5, 9.4, and 16.9 pmol/nmol Pi in vector-transfected RKO or DLD1 cells, respectively. The overexpression of nSMase3 resulted in average about 45% and 20% or 26% and 32% reduction in the levels of C24-sphingomyelin and C16-sphingomyelin levels in RKO or DLD1 cells, respectively. These results indicated that the exogenous nSMase3 in these cells was functional. Interestingly, measurement of C14-ceramide to C26-ceramide levels did not show any significant up-regulation in response to nSMase3 overexpression compared with controls (data not shown), suggesting a rapid metabolic turnover of ceramide after hydrolysis of sphingomyelin by nSMase3 in these cells.
FIGURE 5. nSMase3 sensitizes cells to DNA damage-induced apoptosis.
A. A representative Western blot showing the stable expression of exogenous Myc-tagged nSMase3 in DLD1 or RKO cells. Pooled vector (pcDNA3.1-Myc) transfectants and isolated single clones stably expressing exogenous nSMase3 (pcDNA3.1-Myc-nSMase3) were harvested and subjected to SDS-PAGE. Western blotting was done using an anti-Myc antibody. The same blot was then probed for β-actin to show equal protein loading. B. Membrane fractions from RKO and DLD1 vector and nSMase3 stable cell lines in two independent stable clones named 55 and 61, and 7 and 35, respectively. C and D. Vector and nSMase3 stable cell lines were treated with 20 nmol/L (RKO) or 40 nmol/L (DLD1) of Adriamycin for 24 or 48 h and clonogenic survivals were assessed as described in Materials and Methods. Representative plates from an RKO (C) and DLD1 (D) experiment are shown. E. DLD1 vector (○) and nSMase3-clone 7 (●) and nSMase3-clone 35 (■) stable transfectants were treated continuously for 48, 72, or 96 h with 200 nmol/L Adriamycin and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell viability assays were done as described in Materials and Methods. The experiment was done in triplicate. Points, average of triplicates; bars, SE.
Next, we treated vector-transfected or nSMase3-transfected cells with Adriamycin and did clonogenic survival assays. As shown in Fig. 5C and D, overexpression of nSMase3 in RKO as well as DLD1 cells sensitized cells to Adriamycin-mediated killing as was evident by reduced clonogenic survival by nSMase3-expressing cells (Table 1 and Table 2).We also investigated the role of exogenous nSMase3 in modulating the acute effect of genotoxic stress, and to that end, we did 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays using vector-only–transfected or nSMase3-transfected DLD1 cells. As shown in Fig. 5E, nSMase3 also sensitized DLD1 cells to acute Adriamycin-induced cell killing, a finding that is consistent with results obtained from clonogenic survival assays. Similar results were also obtained using RKO vector and nSMase3 stable transfectants (data not shown). Collectively, the results indicate that nSMase3 is involved in sensitizing cells to genotoxic stress-induced cell death.
Table 1.
nSMase3 Overexpression Sensitizes RKO Cells to Adriamycin-Induced Apoptosis
| Stable Cell Line | Colonies per Plate |
|||||
|---|---|---|---|---|---|---|
| Experiment 1 |
Experiment 2 |
Experiment 3 |
||||
| Control | Adriamycin | Control | Adriamycin | Control | Adriamycin | |
| Vector | 3,460 | 303 | 2,550 | 162 | 3,090 | 379 |
| nSMase3 55 | 2,930 | 30 | 2,590 | 11 | 3,080 | 78 |
| nSMase3 61 | 3,550 | 180 | 2,790 | 104 | 3,640 | 280 |
NOTE: RKO human colon cancer cells stably expressing pcDNA3.1-Myc or pcDNA3.1-Myc-nSMase3 were treated with 20 nmol/L Adriamycin for 24 h. Cells were then cultured in medium without Adriamycin for 10 to 14 d and the numbers of colonies were counted.
Table 2.
nSMase3 Overexpression Sensitizes DLD1 Cells to Adriamycin-Induced Apoptosis
| Stable Cell Line | Colonies per Plate |
|||||
|---|---|---|---|---|---|---|
| Experiment 1 |
Experiment 2 |
Experiment 3 |
||||
| Control | Adriamycin | Control | Adriamycin | Control | Adriamycin | |
| Vector | 2,430 | 168 | 2,430 | 464 | 3,170 | 636 |
| nSMase3 7 | 2,410 | 88 | 2,790 | 400 | 3,010 | 148 |
| nSMase3 35 | 2,920 | 6 | 2,220 | 57 | 2,750 | 60 |
NOTE: DLD1 human colon cancer cells stably expressing pcDNA3.1-Myc or pcDNA3.1-Myc-nSMase3 were treated with 40 nmol/L Adriamycin for 48 h. Cells were then cultured in medium without Adriamycin for 10 to 14 d and the numbers of colonies were counted.
nSMase3 Expression Is Increased in Primary Human Cancers
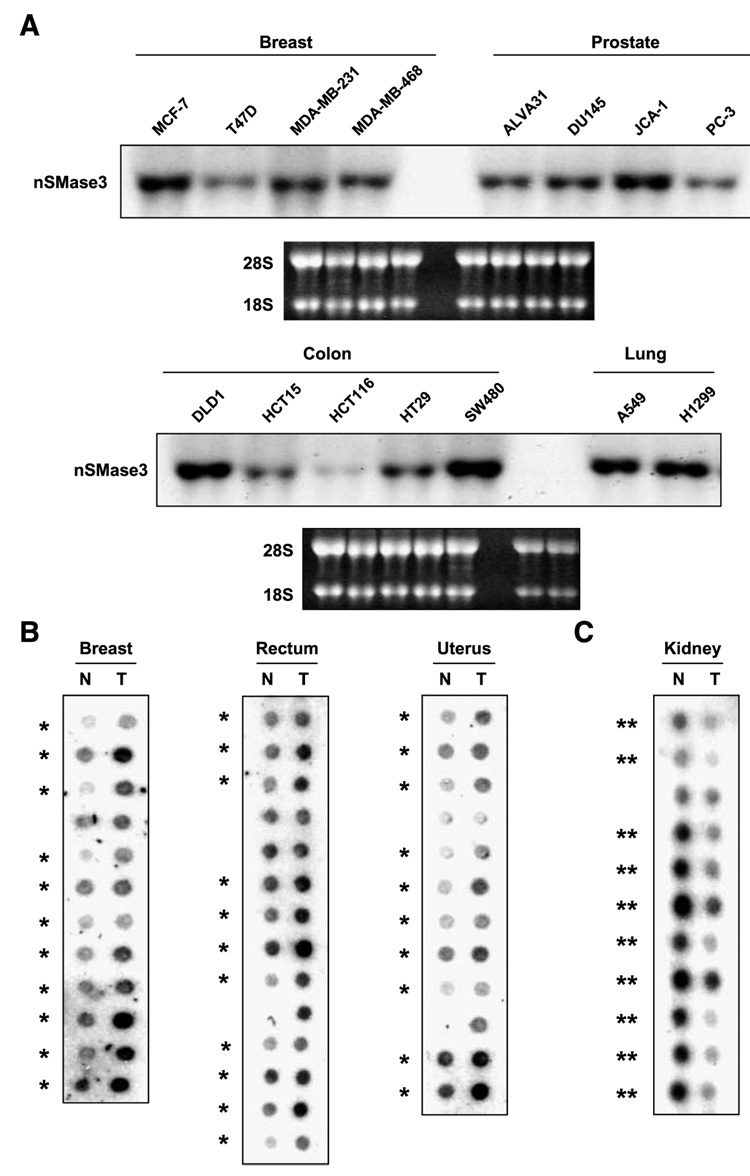
Our results indicate that nSMase3 is regulated by DNA damage and it is also regulated by the tumor suppressor p53. p53 is activated by DNA damage and is frequently inactivated in human malignancies. Consequently, we found it relevant to investigate the expression status of nSMase3 in human malignancies. First, we analyzed nSMase3 expression in established cell lines representing various human malignancies, including cancers of the colon, lung, breast, and prostate. Figure 6A shows that nSMase3 expression was easily detectable in all cancer cell lines tested. Next, we examined its expression in primary matched normal and tumor specimens representing 241 patients and 13 tissue types. Figure 6B shows representative results of nSMase3 expression in samples representing breast, rectum, and uterus. The overall results, summarized in Table 3, show that nSMase3 expression was increased in ~58% (29 of 50) of breast, 56% (19 of 34) of colon, and 72% (13 of 18) of rectal tumors when compared with their matched normal tissues. Interestingly, however, nSMase expression was found to be down-regulated in cancers of the lung (42%, 9 of 21) and kidney (70%, 14 of 20) when compared with their matching normal tissues (Fig. 6C; Table 3). Deregulation of nSMase3 expression in human malignancies seems to be tissue specific, although for some malignancies such as pancreas, prostate, small intestine, and thyroid the sample size was too small to draw a meaningful conclusion. The clinicopathologic features of the tumors exhibiting changes in nSMase3 regulation are presented in Supplementary Tables S1 to S4.
FIGURE 6. nSMase3 is differentially expressed in matched normal and tumor samples.
A. Representative Northern blots showing nSMase3 expression in breast, prostate, colon, and lung cancer cell lines. The blots were probed with 32P-labeled nSMase3 probes; ethidium bromide staining of the 28S and 18S rRNA is also shown to illustrate loading and RNA integrity. JCA-1 cells were previously considered as prostate cancer cells; now, they are believed to represent bladder cancer. B and C. Cancer profiling arrays containing normalized cDNA from tumor (T) and corresponding normal (N) tissue samples from individual patients were probed with nSMase3 probe as described in Materials and Methods. *, matched pairs showing higher nSMase3 mRNA levels in tumor versus matched normal tissue samples; **, matched pairs showing lower nSMase3 mRNA levels in tumor versus matched normal tissue samples.
Table 3.
nSMase3 Expression Is Deregulated in Human Malignancy
| Tissue | % Increased nSMase3 Expression |
|---|---|
| Breast | 58.0 (29/50) |
| Uterus | 52.4 (22/42) |
| Colon | 55.9 (19/34) |
| Stomach | 39.3 (11/28) |
| Ovary | 50.0 (7/14) |
| Rectum | 72.2 (13/18) |
| Tissue |
% Decreased nSMase3 Expression |
| Lung | 42.9 (9/21) |
| Kidney | 70.0 (14/20) |
NOTE: Summary of the cancer array results indicating that nSmase3 mRNA expression is up-regulated in cancers of the breast, uterus, colon, stomach, ovary, and rectum and down-regulated in lung and kidney versus matching normal tissues.
Discussion
A large body of evidence has implicated nSMases and nSMase-derived ceramide as important modulators of the cellular response not only to TNF-α but also to genotoxic agents such as γ-radiation and Adriamycin (12, 31–33). nSMase3 is a new member of the neutral sphingomyelinase family, and before this report, its role in cell growth modulation and apoptosis was not known. Here, we report that nSMase3 gene expression is regulated by nongenotoxic TNF-α and genotoxic Adriamycin. Our results further show that expression of exogenous nSMase3 sensitizes cells to DNA damage-induced cell killing, a novel finding that is consistent with the expected growth-inhibitory and proapoptotic role of other nSMase enzymes. It is therefore conceivable that endogenous nSMase3 would also serve to sensitize cells to DNA damage-mediated negative growth regulation. Interestingly, DNA damage-induced up-regulation of endogenous nSMase3 occurs only at early (<2 h) time points followed by a decrease in its expression after ~24 h. Given that DNA damage-mediated down-regulation of nSMase3 at later time points would likely result in decreased nSMase activity, conceivably it is the early up-regulation of endogenous nSMase3 that may be responsible, at least in part, for its ability to sensitize cells to DNA damage-induced cell killing.
Although speculative, our results also tend to suggest that nSMase3 may not be responsible for lengthened periods of ceramide production that occur in hours to days following treatment with γ-radiation or Adriamycin (11, 13, 25). The possibility therefore exists that one of several other ceramide-generating pathways may be responsible for the extended production of ceramide. Ceramide synthase, for example, is activated by daunorubicin and γ-radiation resulting in prolonged ceramide production (11, 51), suggesting that de novo ceramide synthesis rather than a sphingomyelinase-mediated mechanism may be involved. Alternatively, it has been proposed that elevated ceramide levels following DNA damage is partly the result of the p53-mediated down-regulation of sphingosine-kinase-1, which is involved in the catabolism of ceramide to sphingosine-1-phosphate (52). The result of sphingosine-kinase-1 down-regulation by these means would therefore be through ceramide accumulation versus alterations in ceramide synthesis. Further studies are therefore needed to fully elucidate the relative contributions of the different ceramide-metabolizing pathways during both acute and long-term phases of the genotoxic stress.
Our results indicate that RKO and DLD1 cells expressing exogenous nSMase3 are more sensitive to Adriamycin-induced cell death. In these cells, we also showed that exogenous nSMase3 was functional, as nSMase3-expressing cells displayed reduced levels of total sphingomyelin compared with vector control cells. Interestingly, measurement of C14-ceramide to C26-ceramide levels did not show any significant up-regulation in response to nSMase3 overexpression compared with controls. These results suggest a rapid metabolic turnover of ceramide after hydrolysis of sphingomyelin by nSMase3 in these cells. As ceramide is believed to be a central hub for sphingolipid metabolism, several pathways could essentially be acting to catabolize or convert nSMase3-generated ceramide in these cells. For instance, it has been proposed that ceramidase activity may be great enough in some cells to essentially convert ceramide in a stoichiometric fashion to sphingosine such that increases in ceramide levels are not observed (53). In this regard, increases in sphingosine levels are indeed elevated in nSMase3-expressing DLD1 cells (but not in RKO cells) compared with vector-only cells (data not shown). Alternatively, ceramide could be converted to acylceramide or glucosylceramide by the actions of acyl or glucosyl ceramide synthases (53). Of note, the contribution of nSMase3-dependent decreases in sphingomyelin levels to the sensitization to Adriamycin must also be carefully considered. Future in-depth studies evaluating the contribution of nSMase3 enzymatic activity will be valuable in better defining its apparent proapoptotic role.
In light of our finding that DNA damage regulates nSMase3 expression, we also evaluated whether the tumor suppressor p53, a major regulator of the response of the cell to genotoxic stress, could do the same. We have found that WT p53 down-regulates nSMase3 mRNA expression in p53-inducible DLD1 cells; however, whether p53 mediates this effect directly or indirectly remains to be elucidated. Because nSMase3 expression is down-regulated in both RKO cells (WT p53) and HT29 and DLD1 cells (mutant p53) by prolonged Adriamycin treatment, it is likely that nSMase3 expression can also occur in a p53-independent manner. Whether WT p53 is responsible, at least in part, for increased nSMase3 levels in the early time frame of Adriamycin treatment also warrants further investigation. Because RKO cells harbor WT p53, it is possible that p53 can in fact mediate both nSMase3 up-regulation at very early time points as well as the repression of nSMase3 expression that occurs in these cells with prolonged Adriamycin treatment.
Nonetheless, our results support a role for WT p53 in genotoxic stress-mediated down-regulation of nSMase3. The idea that genotoxic stress and p53 would down-regulate nSMase3 seems incongruous to the well-established role of p53 as a promoter of growth arrest and apoptosis. To the best of our knowledge, this is the first report of p53 modulating the expression of an individual nSMase; however, the notion that p53 facilitates decreased ceramide levels is not without precedence. For instance, Hara et al. (25) have reported that γ-radiation causes a p53-dependent increase in acid ceramidase, an enzyme that hydrolyzes proapoptotic ceramide to form growth-promoting sphingosine. Reciprocally, previous studies have shown the p53-dependent increases in ceramide levels via down-regulation of sphingosine-kinase-1, thus forming the concept that p53 promotes ceramide generation rather than promoting its breakdown or abrogating its synthesis (52). Thus, the role p53 seems to play in ceramide metabolism is currently unclear and therefore warrants further examination.
To gain additional insight into the function of nSMase3, we analyzed the nSMase3 protein sequence using multiple publicly available protein prediction programs and these predictions revealed either a one or two transmembrane-spanning region as well as a putative KKXX-like ER retention signal at the nSMase3 COOH terminus (Fig. 1). Our results showed that mutation of the putative KKXX-like motif (KLHQ) did not abolish compartmentalization of nSMase3 to the ER. Given that C-tail–anchored proteins are believed to retain their ER targeting signals within their COOH-terminal TMD (44, 45), we also deleted the COOH-terminal TMD. However, our deletion of the TMD from nSMase3 resulted in only partial disruption of nSMase3 localization to the ER. nSMase3 localization to the trans-Golgi apparatus has also been shown (38), and thus, deletion of the nSMase3 TMD may allow for partial migration from the ER to the nearby Golgi network. Alternatively, nSMase3 may be targeted or retained in the ER by additional signals still present within the protein or perhaps through protein-protein interactions. Further in-depth studies examining nSMase3 protein structure and its potential binding partners would help to elucidate the exact mechanisms that lead to nSMase3 ER compartmentalization.
Given that nSMase3 is regulated by DNA damage and it is also regulated by the tumor suppressor p53, we found it relevant to investigate the expression status of nSMase3 in human malignancies. Our results indicate that in tumors of the colon, rectum, stomach, breast, ovary, and uterus, nSMase3 mRNA levels were increased when compared with their respective matching normal tissues. Assuming that increased mRNA levels to be coupled with increased nSMase3 activity, the increased levels of nSMase3 in these tumor types would seem incongruous because ceramide is a well-established growth-inhibitory molecule and we have shown nSMase3 to sensitize cells to Adriamycin. Interestingly, however, it has been reported that in some malignancies, for example, in head and neck squamous cell carcinoma, total ceramide levels are actually elevated in tumor tissues when compared with corresponding normal tissues (22). Thus, it is possible that nSMase3 expression and activity may be enhanced in some tumor types as we have shown but it is also possible that tumors could lose their sensitivity to the effects of nSMase3-generated ceramide. Furthermore, up-regulation of growth-inhibitory molecules in human malignancies is not without precedence. For example, p21WAF1 (54), death receptors DR4 and DR5 (55, 56), and death receptor ligands FasL (55) and TNF-related apoptosis-inducing ligand (55, 56) are up-regulated in renal (54), cervical (55), and colon (56) cancers, and in some cases, their increased expression has been associated with higher-grade tumors and poor prognosis.
It is also possible that up-regulation of nSMase3 expression noted in primary tumors could be a positive compensatory response by the cell in an effort to make up for decreased ceramide production or enhanced ceramide breakdown by other ceramide-metabolizing proteins. For instance, sphingosine-kinase-1 mRNA and protein expression are elevated in a variety of malignancies, which would increase ceramide to sphingosine-1-phosphate turnover (57). Thus, by enhancing nSMase3 expression, the cell would attempt to correct ceramide production, which is abrogated in primary tumor specimens (3, 22). We have also shown that p53 down-regulates nSMase3 expression, and thus, elevated nSMase3 expression in primary tumors could in some cases also be the result of p53 inactivation, which is known to occur in 50% of all cancers and 70% of colorectal cancers, specifically (48, 49). Future studies are certainly needed to evaluate these and other possibilities that might lead to nSMase3 up-regulation in primary tumors. We also noted that tumors of the lungs and kidneys displayed decreased expression of nSMase3 when compared with their respective normal tissues. Currently, the reason for the differential regulation of nSMase in tumors of the lungs and kidneys is not clear and analyses of additional tissue specimens are needed to draw a firm conclusion. Additionally, future long-term studies aimed at characterizing the phenotype of cancer cells in which nSMase3 expression is abrogated will be helpful to further define its role in tumorigenesis, particularly in cancers of the lungs and kidneys where nSMase3 expression seems to be down-regulated.
In summary, our present study has provided valuable information about the regulation and potential function of nSMase3 in cellular response to stress and during tumorigenesis. These findings have laid the groundwork for future in-depth studies to further dissect the molecular mechanisms regulating the functions of nSMase3 and to better our understanding of its role in physiologic and pathophysiologic processes.
Materials and Methods
Cell Culture and Reagents
DMEM, McCoy’s 5A, Ham’s F-12, and RPMI 1640 medium were from Cellgro/Mediatech. HEK293T (embryonic kidney); HS578T, MCF-7, MDA-MB-231, MDA-MB-468, and T47D (breast); HeLa (cervical); HT29, RKO, and SW480 (colon); H1299 and A549 (lung); and ALVA31, DU145, and JCA-1 (prostate/bladder) human cancer cells were regularly maintained in DMEM. PC-3 human prostate cancer cells were cultured in a 1:1 mixture of DMEM and Ham’s F-12 medium. HCT15 and HCT116 human colon cancer cells were cultured in McCoy’s 5A medium. DLD1 human colon cancer cells were regularly maintained in RPMI 1640. p53-inducible DLD1 cells (kindly provided by Dr. Bert Vogelstein, Johns Hopkins, Baltimore, MD) were kept in DMEM and 40 ng/mL doxycycline was added to suppress p53 expression. All media were supplemented with 10% fetal bovine serum (Gemini Bioproducts, Inc.), 100 units/mL penicillin (Cellgro), 100 µg/mL streptomycin (Cellgro), and 2 mmol/L l-glutamine (Cellgro). Adriamycin, cycloheximide, and doxycycline were purchased from Sigma-Aldrich.
Plasmids
The plasmid containing an expressed sequence tag corresponding to SKNY/nSMase3 was purchased from the American Type Culture Collection. pEGFP-C1, pEGFP-N1, and pDsRed2-ER were purchased from Clontech. To create an NH2-terminal GFP-tagged nSMase3 expression vector, the nSMase3 ORF was amplified using the following primers: forward, 5′-CCGCTCGAGGCATGACGACTTTCGGCGCCGT-3′; reverse, 5′-GGAATTCTCAGGGCTGGTGCAGCTTCC-3′. The amplified products were digested with XhoI and EcoRI and subcloned into pEGFP-C1 such that the nSMase3 ORF is in frame with that of GFP. A COOH-terminal GFP-tagged nSMase3 expression vector was generated similarly (forward, 5′-CCGCTCGAGGATGACGACTTTCGGCGCCGT-3′; reverse, 5′-CGGAATTCGACAGGGCTGGTGCAGCTTCC-3′) and subcloned into the pEGFP-N1 expression vector with GFP in frame with nSMase3. NH2-terminal GFP-tagged nSMase3 mutation and deletion constructs were generated by using QuikChange Site-Directed Mutagenesis kit (Stratagene). Primers used for the mutation and deletion of nSMase3 domains are as follows: GFP-nSMase3 poly-ala, 5′-GCTGACCGAGCGGGGGGCGGCGGCCGCGCCCTGAGAATTCTGC-3′ (forward) and 5′-GCAGAATTCTCAGGGCGCGGCCGCCGCCCCCCGCTCGGTCAGC-3′ (reverse). Deletion of the COOH-terminal 63-amino acid residues of GFP-nSMase3 was accomplished by excising a SacII fragment corresponding to base 2,412 of the nSMase3 ORF to a SacII site in the COOH-terminal pEGFP-C1 multiple cloning site. pcDNA3.1(+) was obtained from Invitrogen. pcDNA3.1-Myc was generated by introducing a Myc-tag plus linker into pcDNA3.1(+) using NheI and NotI restriction sites. The primers that were annealed and ligated into pcDNA3.1(+) are as follows: forward, 5′-CTAGCCACCATGGAAGAGCAAAAACTCATCTCAGAAGAGGATCTGTTTAAACGAATTCAAGCTTGC-3′; reverse, 5′-GGCCGCAAGCTTGAATTCGTTTAAACAGATCCTCTTCTGAGATGAGTTTTTGCTCTTCCATGGTGG-3′. To create an NH2-terminal Myc-tagged nSMase3 expression vector, the nSMase3 ORF was amplified using the following primers: forward, 5′-CGGAATTCGTTTAAAATGACGACTTTCGGCGCCGT-3′; reverse, 5′-CCGCTCGAGTCAGGGCTGGTGCAGCTTCC-3′. The amplified products were digested with EcoRI and XhoI and subcloned into pcDNA3.1-Myc such that the nSMase3 ORF was in frame with that of the Myc tag. All vectors were sequenced to confirm the correct in-frame subcloning or mutation/deletion.
Transfections
All transfections were done using Lipofectamine 2000 reagent (Invitrogen) per the manufacturer’s instructions. For GFP-nSMase3 protein expression analysis, HEK293T cells were transiently transfected for 6 h on 60-mm plates. For GFP-nSMase3 localization experiments, HeLa cells were transiently cotransfected with pEGFP-nSMase3 constructs and pDsRed2-ER in a 3:1 ratio for 6 h on LabTek II chamber slides. Both HEK293T and HeLa cells were then washed with medium following transfection and allowed to express for ~24 h.
To establish DLD1 and RKO cells stably expressing nSMase3, cells were plated at approximately 60% to 70% on 60-mm plates. Each cell line was transfected with either empty pcDNA3.1-Myc or pcDNA3.1-Myc-nSMase3 for 6 h. Twenty-four hours after transfection, DLD1 and RKO cells were harvested and reseeded onto 150-mm plates in the presence of 400 µg/mL or 1 mg/mL of G418 sulfate (Cellgro), respectively. At 12 to 14 d, several colonies were isolated and screened for the expression of nSMase3 by Western blotting analysis.
Northern Blot Analysis
Cells were harvested and total RNA was extracted with Trizol reagent (Invitrogen) per the manufacturer’s instructions. Approximately 7.5 or 15 Ag of total RNA per sample were resolved onto 1.2% agarose-formaldehyde gel, transferred to Nytran SuperCharge membranes (Schleicher & Schuell), and cross-linked via UV irradiation at 1,200 J/m2. Membranes were probed and signals were visualized using standard procedures as described previously (42). A 1,449-bp KpnI restriction fragment from the human nSMase3 ORF was generated for use as a nSMase3 probe. Full-length ORFs were used as probes for cyclooxygenase-2 and p21 analyses. To detect nSMase3 expression in paired human normal and cancer samples, Cancer Profiling Array I, purchased from Clontech, was probed with the nSMase3 probe as described above.
Western Blot Analysis
Cells were harvested and lysed on ice for 20 min in buffer containing 20 mmol/L HEPES (pH 7.4), 2 mmol/L EGTA, 50 mmol/L β-glycerophosphate, 1 mmol/L DTT, 1 mmol/L Na3VO4, 5 mmol/L NaF, 1% Triton X-100, 10% glycerol, 10 µg/mL aprotinin, 2 µg/mL pepstatin A, 2 µg/mL chymostatin, 2 µg/mL leupeptin, 0.1 µg/mL okadaic acid, and 400 µmol/L phenylmethylsulfonyl fluoride. Extracted protein samples were resolved on an 8% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Bio-Rad). Anti-GFP antibodies were purchased from Roche Applied Science, anti-Myc (9E10) from Santa Cruz Biotechnology, Inc., and anti-β-actin from Sigma-Aldrich. Horseradish peroxidase–conjugated secondary antibodies against mouse IgG were obtained from Vector Laboratories.
Measurement of Sphingomyelin Levels
RKO and DLD1 vector and nSMase3 stable cell lines were plated at a density of 2.0 × 106 cells in 150-mm plates and allowed to adhere for 48 to 72 h before harvesting. After washes with PBS, lipids were extracted from cell pellets, and measurement of sphingolipids, including sphingomyelin and ceramides, with different fatty acid chain lengths was done using liquid chromatography-tandem mass spectrometry, as described previously (22). The sphingolipid levels were normalized to Pi concentrations as described (22).
Cell Viability Assay
Approximately 5.0 × 104 DLD1 cells stably expressing empty pcDNA3.1-Myc or pcDNA3.1-Myc-nSMase3 were split into each well of a six-well plate. The cells were allowed 24 h to attach before being treated continuously with 100 nmol/L Adriamycin. At the time points indicated, cells were incubated with 1 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide for 30 to 60 min, depending on cell density. Absorbance was read at 570 and 650 nm, with 650 nm serving as a background.
Clonogenic Survival Assay
DLD1 or RKO cells (5.0 × 103) stably expressing empty pcDNA3.1-Myc or pcDNA3.1-Myc-nSMase3 were split onto 100-mm plates and allowed 36 h to attach. To allow for the counting of untreated control plates, 5.0 × 102 cells were simultaneously plated and subsequently counted colonies were multiplied by a factor of 10. DLD1 and RKO cells were treated with 20 and 40 nmol/L of Adriamycin, respectively, for 24 or 48 h. Plates were then rinsed with medium and cultured in drug-free medium for 10 to 15 d before they were fixed with ethanol, stained with crystal violet, and counted.
Supplementary Material
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Acknowledgments
We thank Dr. Bert Vogelstein (Johns Hopkins University) for generously providing the p53-inducible DLD1 cells.
Grant support: NIH grants ES014489 and ES005633 (M.S. Sheikh), CA113868 (Y. Huang), and DE016572 (B. Ogretmen).
Footnotes
The measurement of sphingolipids was done in the Lipidomics Core Facility at the Department of Biochemistry and Molecular Biology, Medical University of South Carolina.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zheng W, Kollmeyer J, Symolon H, et al. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Gulbins E, Li PL. Physiological and pathophysiological aspects of ceramide. Am J Physiol Regul Integr Comp Physiol. 2006;290:R11–R26. doi: 10.1152/ajpregu.00416.2005. [DOI] [PubMed] [Google Scholar]
- 3.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 4.Taha TA, Mullen TD, Obeid LM. A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim Biophys Acta. 2006;1758:2027–2036. doi: 10.1016/j.bbamem.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathias S, Pena LA, Kolesnick RN. Signal transduction of stress via ceramide. Biochem J. 1998;335:465–480. doi: 10.1042/bj3350465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funato K, Riezman H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol. 2001;155:949–959. doi: 10.1083/jcb.200105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada K, Kumagai K, Yasuda S, et al. Molecular machinery for nonvesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 8.Clarke CJ, Snook CF, Tani M, Matmati N, Marchesini N, Hannun YA. The extended family of neutral sphingomyelinases. Biochemistry. 2006;45:11247–11256. doi: 10.1021/bi061307z. [DOI] [PubMed] [Google Scholar]
- 9.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis WD, Kolesnick RN, Fornari FA, Traylor RS, Gewirtz DA, Grant S. Induction of apoptotic DNA damage and cell death by activation of the sphingomyelin pathway. Proc Natl Acad Sci U S A. 1994;91:73–77. doi: 10.1073/pnas.91.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82:405–414. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- 12.Jaffrézou JP, Levade T, Bettaïeb A, et al. Daunorubicin-induced apoptosis: triggering of ceramide generation through sphingomyelin hydrolysis. EMBO J. 1996;15:2417–2424. [PMC free article] [PubMed] [Google Scholar]
- 13.Lucci A, Han TY, Liu YY, Giuliano AE, Cabot MC. Modification of ceramide metabolism increases cancer cell sensitivity to cytotoxics. Int J Oncol. 1999;15:541–546. doi: 10.3892/ijo.15.3.541. [DOI] [PubMed] [Google Scholar]
- 14.Uchida Y, Itoh M, Taguchi Y, et al. Ceramide reduction and transcriptional up-regulation of glucosylceramide synthase through doxorubicin-activated Sp1 in drug-resistant HL-60/ADR cells. Cancer Res. 2004;64:6271–6279. doi: 10.1158/0008-5472.CAN-03-1476. [DOI] [PubMed] [Google Scholar]
- 15.Haimovitz-Friedman A, Kan CC, Ehleiter D, et al. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J Exp Med. 1994;180:525–535. doi: 10.1084/jem.180.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santana P, Pena LA, Haimovitz-Friedman A, et al. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86:189–199. doi: 10.1016/s0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- 17.Voelkel-Johnson C, Hannun YA, El-Zawahry A. Resistance to TRAIL is associated with defects in ceramide signaling that can be overcome by exogenous C6-ceramide without requiring down-regulation of cellular FLICE inhibitory protein. Mol Cancer Ther. 2005;4:1320–1327. doi: 10.1158/1535-7163.MCT-05-0086. [DOI] [PubMed] [Google Scholar]
- 18.Swanton C, Marani M, Pardo O, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Dbaibo GS, El-Assaad W, Krikorian A, et al. Ceramide generation by two distinct pathways in tumor necrosis factor α-induced cell death. FEBS Lett. 2001;503:7–12. doi: 10.1016/s0014-5793(01)02625-4. [DOI] [PubMed] [Google Scholar]
- 20.Luberto C, Hassler DF, Signorelli P, et al. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem. 2002;277:41128–41139. doi: 10.1074/jbc.M206747200. [DOI] [PubMed] [Google Scholar]
- 21.Neumeyer J, Hallas C, Merkel O, et al. TNF-receptor I defective in internalization allows for cell death through activation of neutral sphingomyelinase. Exp Cell Res. 2006;312:2142–2153. doi: 10.1016/j.yexcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Koybasi S, Senkal CE, Sundararaj K, et al. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J Biol Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 23.Schutze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Kronke M. TNF activates NF-κB by phosphatidylcholine-specific phospholipase C-induced “acidic” sphingomyelin breakdown. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 24.Cifone MG, De Maria R, Roncaioli P, et al. Apoptotic signaling through CD95 (Fas/Apo-1) activates an acidic sphingomyelinase. J Exp Med. 1994;180:1547–1552. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara S, Nakashima S, Kiyono T, et al. p53-independent ceramide formation in human glioma cells during γ-radiation-induced apoptosis. Cell Death Differ. 2004;11:853–861. doi: 10.1038/sj.cdd.4401428. [DOI] [PubMed] [Google Scholar]
- 26.Andrieu-Abadie N, Jaffrezou JP, Hatem S, Laurent G, Levade T, Mercadier JJ. L-carnitine prevents doxorubicin-induced apoptosis of cardiac myocytes: role of inhibition of ceramide generation. FASEB J. 1999;13:1501–1510. doi: 10.1096/fasebj.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 27.Tomiuk S, Hofmann K, Nix M, Zumbansen M, Stoffel W. Cloned mammalian neutral sphingomyelinase: functions in sphingolipid signaling? Proc Natl Acad Sci U S A. 1998;95:3638–3643. doi: 10.1073/pnas.95.7.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchesini N, Luberto C, Hannun YA. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J Biol Chem. 2003;278:13775–13783. doi: 10.1074/jbc.M212262200. [DOI] [PubMed] [Google Scholar]
- 29.Levy M, Castillo SS, Goldkorn T. nSMase2 activation and trafficking are modulated by oxidative stress to induce apoptosis. Biochem Biophys Res Commun. 2006;344:900–905. doi: 10.1016/j.bbrc.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchesini N, Osta W, Bielawski J, Luberto C, Obeid LM, Hannun YA. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J Biol Chem. 2004;279:25101–25111. doi: 10.1074/jbc.M313662200. [DOI] [PubMed] [Google Scholar]
- 31.Bruno AP, Laurent G, Averbeck D, et al. Lack of ceramide generation in TF-1 human myeloid leukemic cells resistant to ionizing radiation. Cell Death Differ. 1998;5:172–182. doi: 10.1038/sj.cdd.4400330. [DOI] [PubMed] [Google Scholar]
- 32.Maestre N, Tritton TR, Laurent G, Jaffrezou JP. Cell surface-directed interaction of anthracyclines leads to cytotoxicity and nuclear factor κB activation but not apoptosis signaling. Cancer Res. 2001;61:2558–2561. [PubMed] [Google Scholar]
- 33.Sawada M, Nakashima S, Kiyono T, et al. p53 regulates ceramide formation by neutral sphingomyelinase through reactive oxygen species in human glioma cells. Oncogene. 2001;20:1368–1378. doi: 10.1038/sj.onc.1204207. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura S, Banno Y, Nakashima S, et al. Ceramide formation leads to caspase-3 activation during hypoxic PC12 cell death. Inhibitory effects of Bcl-2 on ceramide formation and caspase-3 activation. J Biol Chem. 1998;273:6921–6927. doi: 10.1074/jbc.273.12.6921. [DOI] [PubMed] [Google Scholar]
- 35.Magnoni C, Euclidi E, Benassi L, et al. Ultraviolet B radiation induces activation of neutral and acidic sphingomyelinases and ceramide generation in cultured normal human keratinocytes. Toxicol In Vitro. 2002;16:349–355. doi: 10.1016/s0887-2333(02)00024-3. [DOI] [PubMed] [Google Scholar]
- 36.Zumbansen M, Stoffel W. Neutral sphingomyelinase 1 deficiency in the mouse causes no lipid storage disease. Mol Cell Biol. 2002;22:3633–3638. doi: 10.1128/MCB.22.11.3633-3638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoffel W, Jenke B, Blöck B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci U S A. 2005;102:4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krut O, Wiegmann K, Kashkar H, Yazdanpanah B, Kronke M. Novel tumor necrosis factor-responsive mammalian neutral sphingomyelinase-3 is a C-tail-anchored protein. J Biol Chem. 2006;281:13784–13793. doi: 10.1074/jbc.M511306200. [DOI] [PubMed] [Google Scholar]
- 39.Sheikh MS, Carrier F, Papathanasiou MA, et al. Identification of several human homologs of hamster DNA damage-inducible transcripts. Cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J Biol Chem. 1997;272:26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 40.Sheikh MS, Fernandez-Salas E, Yu M, et al. Cloning and characterization of a human genotoxic and endoplasmic reticulum stress-inducible cDNA that encodes translation initiation factor 1[eIF1(A121/SUI1)] J Biol Chem. 1999;274:16487–16493. doi: 10.1074/jbc.274.23.16487. [DOI] [PubMed] [Google Scholar]
- 41.Fornace AJ, Jr, Alamo I, Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo X, Huang Y, Sheikh CV. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene. 2003;22:7247–7257. doi: 10.1038/sj.onc.1207010. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, He Q, Huang Y, Sheikh MS. Cloning and characterization of a p53 and DNA damage down-regulated gene PIQ that codes for a novel calmodulin-binding IQ motif protein and is up-regulated in gastrointestinal cancers. Cancer Res. 2005;65:10725–10733. doi: 10.1158/0008-5472.CAN-05-1132. [DOI] [PubMed] [Google Scholar]
- 44.Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 46.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 47.Sheikh MS, Fornace AJ., Jr Role of p53 family members in apoptosis. J Cell Physiol. 2000;182:171–181. doi: 10.1002/(SICI)1097-4652(200002)182:2<171::AID-JCP5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–181. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Kastan MB. Wild-type p53: tumors can’t stand it. Cell. 2007;128:837–840. doi: 10.1016/j.cell.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 50.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc Natl Acad Sci U S A. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao WC, Haimovitz-Friedman A, Persaud RS, et al. Ataxia telangiectasia-mutated gene product inhibits DNA damage-induced apoptosis via ceramide synthase. J Biol Chem. 1999;274:17908–17917. doi: 10.1074/jbc.274.25.17908. [DOI] [PubMed] [Google Scholar]
- 52.Taha TA, Osta W, Kozhaya L, et al. Down-regulation of sphingosine kinase-1 by DNA damage: dependence on proteases and p53. J Biol Chem. 2004;279:20546–20554. doi: 10.1074/jbc.M401259200. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds CP, Maurer BJ, Kolesnick RN. Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett. 2004;206:169–180. doi: 10.1016/j.canlet.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 54.Weiss RH, Borowsky AD, Seligson D, et al. p21 is a prognostic marker for renal cell carcinoma: implications for novel therapeutic approaches. J Urol. 2007;177:63–68. doi: 10.1016/j.juro.2006.08.073. [DOI] [PubMed] [Google Scholar]
- 55.Reesink-Peters N, Hougardy BM, van den Heuvel FA, et al. Death receptors and ligands in cervical carcinogenesis: an immunohistochemical study. Gynecol Oncol. 2005;96:705–713. doi: 10.1016/j.ygyno.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 56.van Geelen CM, Westra JL, de Vries EG, et al. Prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand and its receptors in adjuvantly treated stage III colon cancer patients. J Clin Oncol. 2006;24:4998–5004. doi: 10.1200/JCO.2006.06.8809. [DOI] [PubMed] [Google Scholar]
- 57.French KJ, Schrecengost RS, Lee BD, et al. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 2003;63:5962–5969. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).