Summary
Centrosome- and microtubule-associated proteins have been shown to be important for maintaining the neural progenitor pool during neocortical development by regulating the mitotic spindle. It remains unclear whether these proteins may control neurogenesis by regulating other microtubule-dependent processes such as nuclear migration. Here we identify Cep120, a novel centrosomal protein preferentially expressed in neural progenitors during neocortical development. We demonstrate that silencing Cep120 in the developing neocortex impairs both interkinetic nuclear migration (INM), a characteristic pattern of nuclear movement in neural progenitors, and neural progenitor self-renewal. Furthermore, we show that Cep120 interacts with transforming acidic coiled-coil proteins (TACCs) and that silencing TACCs also causes defects in INM and neural progenitor self-renewal. Our data suggest a critical role for Cep120 and TACCs in both INM and neurogenesis. We propose that sustaining INM may be a novel mechanism by which microtubule-regulating proteins maintain the neural progenitor pool during neocortical development.
Introduction
During neocortical devel opment neurons are generated from progenitors lining the lateral ventricles. The size of the progenitor pool is a key determinant of the total number of neurons produced during the entire neurogenesis period (Bond and Woods, 2006; Caviness et al., 1995; Rakic, 2006). Recent studies suggest that the centrosome and its associated microtubules play a major role in controlling the progenitor pool. First, four causative genes have been identified for human autosomal recessive primary microcephaly, a disorder characterized by an abnormally small brain that is likely caused by a reduced progenitor pool (Bond and Woods, 2006). Remarkably, three of the four genes, including ASPM, CDK5RAP2, and CENPJ, encode centrosomal proteins (Bond et al., 2002, 2005; Hung et al., 2000; Kouprina et al., 2005; Zhong et al., 2005). One of these centrosomal proteins, Aspm, has been proposed to promote the self-renewal of neural progenitors by adjusting the orientation of the mitotic spindle during mitosis (Fish et al., 2006). Next, mice lacking Nde1, a centrosomal protein important for mitotic spindle assembly and orientation, exhibit a reduction in both brain and neural progenitor pool size (Feng and Walsh, 2004). Finally, the microtubule-associated protein Doublecortin-like kinase (DCLK) has been shown to be important for the formation of mitotic spindle and for preventing premature differentiation of neural progenitors (Shu et al., 2006). These studies suggest that microtubule-regulating proteins may maintain the neocortical neural progenitor pool by controlling mitotic spindle assembly and positioning.
While previous studies have focused on mitotic spindle assembly and positioning, it remains unclear whether microtubule-regulating proteins may govern neocortical neurogenesis by controlling other microtubule-dependent processes such as nuclear migration. During neurogenesis, the nuclei of progenitors oscillate within the proliferative layer called the ventricular zone (VZ). They stay at the ventricular surface during mitosis, ascend to the basal side of the VZ during G1, and, after completing S-phase, descend back toward the ventricular surface during G2. This cell cycle-dependent nuclear movement is known as interkinetic nuclear migration (INM). Given that centrosome- and microtubule-associated proteins are critical for both nuclear migration and neocortical neurogenesis, it is striking that a role for INM in neocortical neurogenesis has not been investigated. One previous study treated chick embryonic retinas with the actin-depolymerizing agent cytochalasin B and found an inhibition of INM and a premature depletion of the neural progenitor pool (Murciano et al., 2002). However, since mitotic spindle positioning is also actin-dependent, it remains unclear whether the effect of cytochalasin B on neurogenesis in this system was due to impaired INM.
Although INM has been a hallmark of vertebrate neural progenitors for many decades (Gotz and Huttner, 2005; Sauer, 1935), the mechanism underlying INM is poorly understood. Studies using pharmacological inhibitors suggest that INM is dependent on the actin and microtubule cytoskeleton (Karfunkel, 1972; Messier and Auclair, 1973, 1974; Webster and Langman, 1978). The unique mode of nuclear movement during INM suggests that microtubule regulation in neural progenitors may exhibit distinct features. In other types of cells, the nucleus closely follows the centrosome during migration due to the tight association between the nucleus and the centrosome by microtubules (Reinsch and Gonczy, 1998; Tsai and Gleeson, 2005). In neural progenitors, however, the nucleus repeatedly migrates toward and away from the ventricular surface, where the centrosome remains throughout INM (Hinds and Ruffett, 1971; Astrom and Webster, 1991; Chenn et al., 1998). This drastic change in the relative position between the nucleus and the centrosome suggests that the microtubules coupling the nucleus and the centrosome must be dynamically regulated. Currently little is known about the molecular mechanisms underlying this regulation.
In this study, we show that INM is dependent on the regulation of centrosome-associated microtubules by Cep120, a novel centrosomal protein highly expressed in neural progenitors, and transforming acidic coiled-coil proteins (TACCs), centrosome- and microtubule-associated proteins that have previously been implicated in microtubule growth and nuclear migration. Furthermore, we provide evidence that Cep120 interacts with TACCs and regulates the localization of TACC3 to the centrosome. Finally, we demonstrate that both Cep120 and TACCs are essential for maintaining the neural progenitor pool during mouse neocortical development. These results suggest that microtubule-regulating proteins may promote the self-renewal of neocortical neural progenitors by sustaining INM.
Results
Identification and Expression of Cep120
In a search for novel regulators of centrosome-associated microtubules, we performed a yeast-two-hybrid screen to identify interactors of focal adhesion kinase (FAK), which is implicated in the organization of centrosome-associated microtubules (Xie et al., 2003). Five of the 42 isolated clones represent a previously uncharacterized gene designated as coiled-coil domain containing 100 (Ccdc100). This gene encodes a centrosomal protein with an apparent molecular mass of ~120KD (see below) and therefore, is named Cep120 (Centrosomal Protein of 120KD). Putative orthologs of Cep120 exist in species from ciliate protozoans to mammals (Figure S1).
The EST profile (UniGene ID: Mm.193678) indicates that Cep120 mRNA is ubiquitously expressed. Consistent with this, we were able to amplify the Cep120 gene by RT-PCR from all mouse tissues examined (Figure S2). To investigate the expression of Cep120 protein, we generated a rabbit antiserum against the C-terminal fragment of mouse Cep120. This antiserum recognized full-length exogenous Cep120 in COS7 cells and endogenous Cep120 in mouse brain lysates (Figure 1A). Immunoblot analysis of multiple tissues using this antiserum revealed that Cep120 protein is ubiquitously expressed in mouse embryos but with higher levels in the brain, lung, and kidney (Figure 1B). Interestingly, the expression of Cep120 in the brain is considerably higher in embryonic stages than in postnatal stages or in the adult (Figure 1C), suggesting that Cep120 may be important for embryonic brain development.
Figure 1.
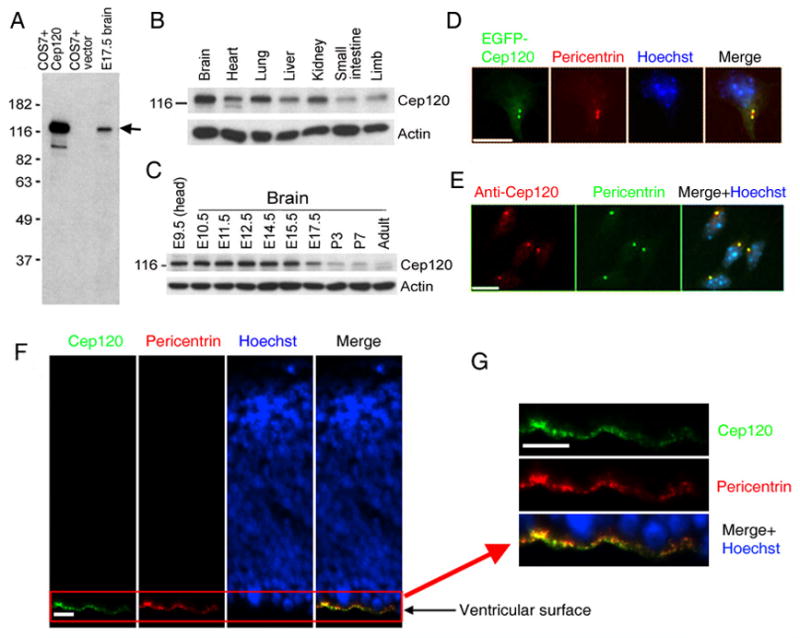
Expression and localization of Cep120. (A–C) Lysates prepared in RIPA buffer were resolved by SDS-PAGE and subjected to immunoblot analysis. (A) The Cep120 antiserum recognizes exogenous Cep120 expressed in COS7 cells and endogenous Cep120 in brain lysates. The arrow indicates the Cep120 protein. (B) Cep120 protein levels in E17.5 mouse tissues. (C) Cep120 protein levels in the mouse brain at different developmental stages. Each lane was loaded with 20 μg total proteins in (B) and (C). (D and E) Cep120 is a centrosomal protein. (D) EGFP-Cep120 colocalizes with pericentrin in cultured neocortical cells. (E) Affinity-purified Cep120 antibody labels the centrosome in cultured neocortical cells. (F and G) Expression of Cep120 in the mouse neocortex at E12.5. The Cep120 antibody prominently labels centrosomes (indicated by pericentrin co-staining) along the ventricular surface (bottom). The boxed area in (F) is shown in (G) at higher magnification to reveal the puncta labeled by both Cep120 and pericentrin antibodies along the ventricular surface. Scale bars: 10 μm in (D) and (E), 20 μm in (F) and (G).
Cep120 Is a Centrosomal Protein Preferentially Expressed in Neural Progenitors
To determine the subcellular localization of Cep120, we added an EGFP tag to the N-terminus of recombinant Cep120. When expressed at low to moderate levels, EGFP-Cep120 formed one or two puncta that colocalized with a centrosomal marker pericentrin in cultured neocortical cells (Figure 1D). Affinity-purified Cep120 antibody also prominently labeled the centrosome (Figure 1E). Thus, we conclude that Cep120 is a centrosomal protein.
The expression profile of Cep120 (Figure 1C) roughly correlates with the timing for proliferative and neurogenic divisions of neocortical neural progenitors. To determine whether Cep120 is expressed in neocortical neural progenitors, we performed immunohistochemistry using affinity-purified Cep120 antibody on cryosections of embryonic mouse brain. The Cep120 antibody prominently labeled pericentrin-positive puncta at the ventricular surface (Figure 1, F and G), whereas weak labeling of pericentrin-positive puncta at other areas of the neocortex was visible at higher exposure (data not shown). The Cep120 immunoreactive puncta at the ventricular surface also co-localized with a different centrosomal marker, RFP-CETN2, which was introduced into the neocortex via in utero electroporation (Figure S3). Since centrosomes at the ventricular surface belong to neural progenitors (Hinds and Ruffett, 1971; Astrom and Webster, 1991; Chenn et al., 1998), these data suggest that Cep120 is preferentially expressed in neural progenitors. Consistent with this, images from GENSAT database (GENSAT image 35740 and 35741) revealed that the somata of neural progenitors in the VZ exhibited higher levels of Cep120 mRNA than that of cells in other neocortical layers.
Cep120 Is Important for INM
Since the centrosome is a key organelle in nuclear migration, the preferential expression of Cep120 in neural progenitors suggests that Cep120 may play an important role in INM. To examine a potential role for Cep120 in INM, we designed pSilencer-based siRNA plasmids against the mouse Cep120 gene, designated as Cep120 i2968 and Cep120 i1265 (the numbers 2968 and 1265 indicate siRNA targeting regions in Cep120 mRNA represented by NM_178686). These plasmids can efficiently knock down Cep120 protein both in vitro and in vivo (Figure S4).
We then determined whether Cep120 siRNA affects INM. Previous studies have shown that nuclei of neural progenitors are elongated during migration and round up when they stop migrating (Chenn and McConnell, 1995; Haubensak et al., 2004). Thus a defect in INM may correlate with less elongated nuclei. To test this, we introduced Cep120 i2968 together with a Venus plasmid into the neocortex of mouse embryos at E11.5 by in utero electroporation and examined the nuclear morphology of transfected neural progenitors at E14.5. We analyzed the cells that exhibited an apical process contacting the ventricular surface because these cells express nestin (Figure 2A), a progenitor marker. We found that nuclei of the Cep120 i2968 group were generally less elongated along the apical-basal axis compared to that of the control or the rescue (Cep120 i2968 + siRNA resistant Cep120) group (Figure 2B). To quantify this difference, we measured the ratio of nuclear length to width and found that the ratio for the Cep120 i2968 group was significantly (P<0.01, one-way ANOVA) lower than that for the control or the rescue group (Figure 2C). This altered nuclear morphology is consistent with a potential nuclear migration defect.
Figure 2.
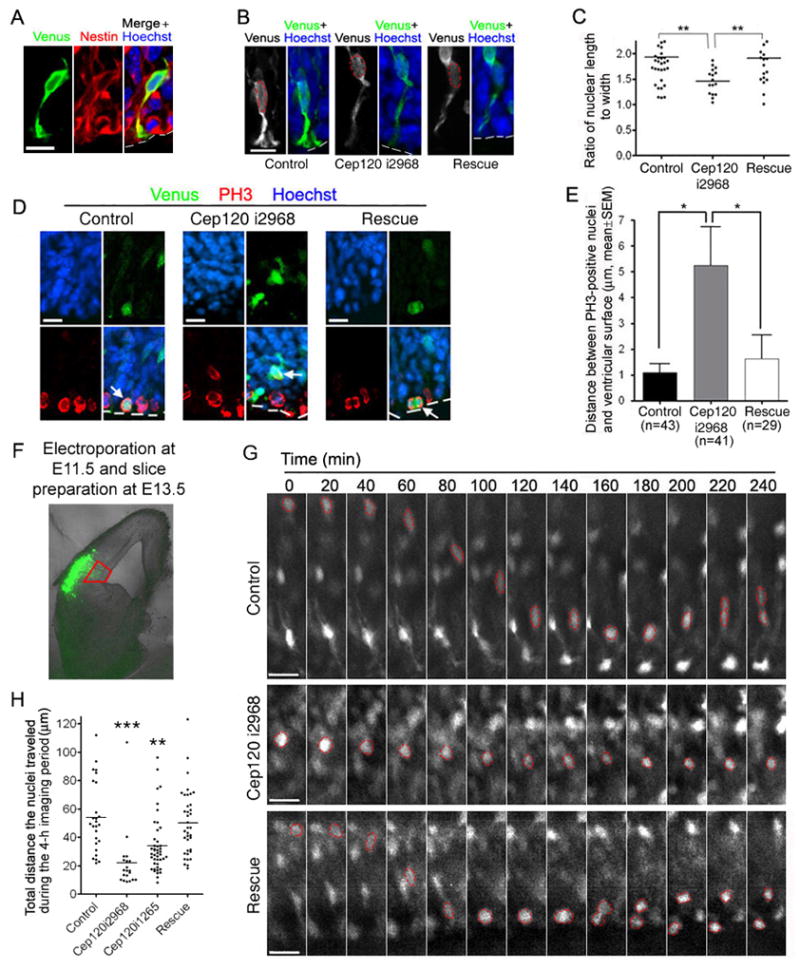
Cep120 is important for INM. (A–C) Mouse embryos were electroporated at E11.5 and sacrificed at E14.5. (A) A transfected cell with an apical process contacting the ventricular surface (outlined by dashed lines) expresses Nestin, a neural progenitor marker. (B and C) The nuclei of Cep120 i2968-expressing neural progenitors are less elongated along the apical-basal axis compared to the control or rescue (Cep120 i2968 + Cep120 i2968 resistant Cep120 plasmid). (B) Representative neural progenitors. The nuclei and the ventricular surface are outlined by red and white dashed lines, respectively. (C) Quantification of the ratio of nuclear length to width. ** P<0.01, one-way ANOVA. (D and E) Ectopic PH3-positive nuclei in Cep120 i2968-expressing neural progenitors. Mouse embryos were electroporated at E11.5 and sacrificed at E13.5. (D) Representative transfected PH3-positive cells (indicated by arrows). The ventricular surface is indicated by white dashed lines. (E) Quantification of the distance between PH3-positive nuclei and the ventricular surface. * P<0.05, one-way ANOVA. (F–H) Silencing Cep120 impairs INM in brain slice preparations. Mouse embryos were electroporated at E11.5 and sacrificed at E13.5. Acute brain slices of 150-μm were then prepared and imaged for 4 h. (F) A representative imaging area at the VZ (boxed region, transfected cells are green). (G) Examples of transfected cells in which the nuclei (outlined by red dashed lines) moved toward the ventricular surface (bottom). The control and the rescue cell divided before the end of the imaging period. (H) Quantification of the total distance the nuclei traveled during the 4-h imaging period. ** P<0.01, *** P<0.001, one-way ANOVA. Scale bars: 10 μm in (A), (B) and (D), 20 μm in (G).
When nuclear movement toward the ventricle is impaired, cells may enter mitosis before their nuclei migrate back to the ventricular surface. To determine if Cep120 regulates nuclear movement toward the ventricle, we transfected Cep120 i2968 into E11.5 mouse embryos and analyzed the position of transfected mitotic nuclei relative to the ventricular surface at E13.5. The mitotic nuclei were identified by a phospho-histone H3 (PH3) antibody. We found that the overall distance between PH3-positive nuclei and the ventricular surface was significantly greater (P<0.05, one-way ANOVA) in the Cep120 i2968 group compared to the control or the rescue group (Figure 2, D and E). This data suggests that Cep120 i2968 may impair the migration of the nuclei toward the ventricular surface.
To directly assess the role of Cep120 in INM, we electroporated Cep120 i2968 into mouse embryos at E11.5 and prepared acute brain slices at E13.5 for time-lapse imaging. At the time of slice preparation and imaging, the ventricular zone only contained cells with weak Venus fluorescence (Figure 2F), possibly due to the dilution of the plasmids by repeated division of the neural progenitors. At this low fluorescence intensity, we were able to identify the nucleus, which usually exhibits stronger Venus fluorescence than the cytoplasm does in live brain slices. However, we were generally unable to identify the processes and therefore, could not distinguish neural progenitors from newborn neurons based on cell morphology. Since neurons may be generated both near the ventricular surface and at the basal side of the VZ (Buchman and Tsai, 2007), we also could not distinguish neural progenitors from newborn neurons based on cell position. However, because newborn neurons migrate away from, but not toward, the ventricle at early developmental stages (Takahashi et al., 1999; Bielas et al., 2004; Kriegstein and Noctor, 2004), we were able to exclude neurons by restricting our analysis to ventricle-directed nuclear migration.
The migration of the nuclei toward the ventricle was severely impaired in Cep120 i2968-expressing cells. Overall there were significantly fewer cells in which the nuclei migrated toward the ventricle in the Cep120 i2968 group (11.67 ± 5.31%, mean ± SD, of total transfected VZ cells) compared to the control (27.54 ± 1.75%; P<0.01, one-way ANOVA) or the rescue group (33.07 ± 6.23%; P<0.001, one-way ANOVA). Of the cells in which the nuclei were migrating toward the ventricle, the total distance the nuclei traveled during the 4-h imaging period was considerably shorter (P<0.001, one-way ANOVA) in the Cep120 i2968 group (21.96 ± 4.82 μm, mean ± SEM, n = 20) than in the control (53.74 ± 4.88, n = 26) or the rescue group (50.16 ± 3.81, n = 37) (Figure 2, G and H; Movies S1–S3). In addition to the distance of migration, Cep120 i2968 also apparently altered the nuclear morphology. The nuclei in the control and the rescue group were quite elongated during migration (Figure 2G; Movies S1 and S3). In the Cep120 i2968 group, however, the nuclei were generally more round during migration (Figure 2G; Movie S2). A different Cep120 siRNA construct, Cep120 i1265, also significantly impaired INM (Figure 2H), suggesting that INM defects associated with Cep120 siRNA constructs were due to specific knockdown of Cep120.
Cep120 Is Required for Maintaining the Neural Progenitor Pool
To investigate whether INM defects associated with Cep120 knockdown might affect neurogenesis in the developing mouse neocortex, we electroporated mouse embryos at E11.5, administered BrdU to label S-phase cells at E13.5, and analyzed progenitor proliferation at E14.5. First, we measured the mitotic index, i.e. the percentage of transfected cells that were PH3-positive in the VZ. This index was significantly (P<0.01, one-way ANOVA) decreased in Cep120 i2968 and Cep120 i1265 groups compared to the control (Figure 3, A and B), suggesting that silencing Cep120 reduced the percentage of M-phase progenitors at E14.5. Next, we measured the BrdU labeling index, i.e. the percentage of transfected cells that were BrdU-positive. This index was about two-fold lower in Cep120 i2968 and Cep120 i1265 groups compared to the control (Figure 3, C and D), suggesting that silencing Cep120 reduced the percentage of proliferating S-phase cells at E13.5. Finally, we measured the percentage of transfected cells that were positive for BrdU but negative for Ki67, a proliferating cell marker. This percentage represents the fraction of cells that were proliferating at E13.5 but exited the cell cycle at E14.5 and thus, is designated as the cell cycle exit index. This index was significantly increased (P<0.001, one-way ANOVA) in both Cep120 i2968 and Cep120 i1265 groups compared to the control (Figure 3, C and E), suggesting that progenitors expressing Cep120 siRNA exited the cell cycle more rapidly. Taken together, the indices of BrdU labeling, mitosis, and cell cycle exit indicate that silencing Cep120 causes a premature depletion of proliferating progenitors.
Figure 3.
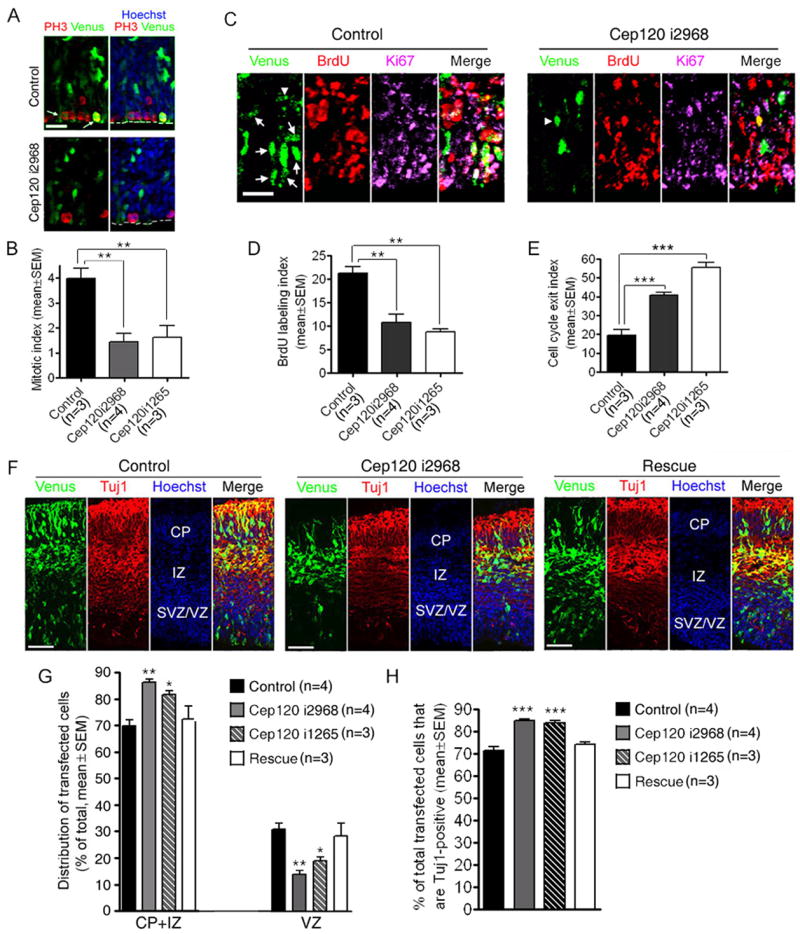
Cep120 is required for maintaining the neural progenitor pool. (A–E) Progenitors expressing Cep120 siRNA prematurely exit the cell cycle. Mouse embryos were electroporated at E11.5, exposed to BrdU at E13.5, and sacrificed at E14.5. The proliferation of progenitors in the neocortex was analyzed by immunostaining using antibodies against PH3, BrdU, and Ki67. (A) Representative images showing PH3-positive in the VZ of electroporated embryos. White arrows indicate transfected PH3-positive cells in the control group. (B) Quantification of the mitotic index, i.e. the percentage of transfected cells in the VZ that are PH3-positive. (C) Representative images showing transfected cells in neocortices co-stained with BrdU and Ki67 antibodies. White arrows indicate transfected cells that are positive for both BrdU and Ki67. White arrowheads indicate transfected cells that are positive for BrdU but negative for Ki67. (D) Quantification of the BrdU labeling index, i.e. the percentage of transfected cells that are BrdU-positive. (E) Quantification of the cell cycle exit index, i.e. the percentage of transfected BrdU-positive cells that do not express Ki67. Compared to the control, Cep120 i2968 and Cep120 i1265 groups exhibit a significant decrease in the mitotic index and BrdU labeling index and a significant increase in the cell cycle exit index. ** P<0.01, *** P<0.001, t-test. (F–H) Cep120 siRNA causes increased neuronal production. Mouse embryos were electroporated at E11.5 and sacrificed at E14.5. (F) Representative images showing the distribution of transfected cells across neocortical layers. (G) Quantification of the distribution of transfected cells. A higher percentage of transfected cells in Cep120 siRNA groups are located in the IZ or CP compared to the control or rescue group. * P<0.05, ** P<0.01, one-way ANOVA. (H) Quantification of the percentage of transfected cells that express Tuj1. The percentage is significantly higher in Cep120 siRNA groups compared to the control or rescue group. *** P<0.001, one-way ANOVA. CP, cortical plate; IZ, intermediate zone; SVZ/VZ, subventricular zone/ventricular zone. Scale bars: 20 μm in (A) and (C), 50 μm in (F).
To determine whether the depletion of proliferating progenitors corresponds to increased number of postmitotic neurons, we analyzed the neuronal pool in E14.5 mouse embryos that were electroporated with Cep120 siRNA at E11.5 by the distribution of transfected cells across neocortical layers and by the expression of a neuronal marker Tuj1. At this stage the somata of neural progenitors are located in the VZ or subventricular zone (SVZ), whereas the somata of postmitotic neurons migrate toward the cortical plate (CP) through the intermediate zone (IZ). Compared to the control or the rescue group, both Cep120 i2968 and Cep120 i1265 groups exhibited a significant increase in the percentage of transfected cells in the IZ/CP (Figure 3, F and G), a significant decrease in the percentage of transfected cells in the SVZ/VZ (Figure 3, F and G), and a significant increase in the percentage of transfected cells that areTuj1-positive (Figure 3, F and H). These data suggest that more neurons were present in Cep120 siRNA groups. We have attempted to determine whether the increase in Tuj1-positive cells corresponds to a decrease in nestin-positive progenitors. However, because many neurons are attached to nestin-positive radial glial fibers, which are highly crowded and span the entire thickness of the neocortex, we could not accurately identify transfected nestin-positive cells in some areas of the neocortex. Nevertheless, the indices of mitosis, BrdU labeling, and cell cycle exit (Figure 3, A–E) suggest that the total number of progenitors are likely decreased in Cep120 siRNA groups.
Interestingly, silencing Cep120 also resulted in mispositioning of neurons at deep layers of the neocortex (Figure 3F and data not shown). When mouse embryos were electroporated at E14.5 and the distribution of transfected neurons was examined at E18.5, the mispositioning defect was more pronounced (Figure S5). These data suggest that, in addition to neurogenesis, Cep120 may also play an important role in neuronal migration.
Cep120 Interacts with TACCs
To elucidate the molecular mechanisms by which Cep120 regulates INM, we performed a yeast-two-hybrid screen to identify potential Cep120-interacting centrosome-associated proteins. We found that a number of centrosome-associated proteins interact with Cep120 in the yeast-two-hybrid system, including TACCs (Figure 4A), ninein, Cep290, Cep164, myomegalin, and KIAA1009 (data not shown). However, of these interactors, only TACCs, which are associated with both the centrosome and microtubules (Gergely, 2002), have previously been linked to nuclear migration (Bellanger and Gonczy, 2003; Le Bot et al., 2003; Srayko et al., 2003; Gergely et al., 2000a). Thus, TACCs are potential candidates that mediate the function of Cep120 in INM.
Figure 4.
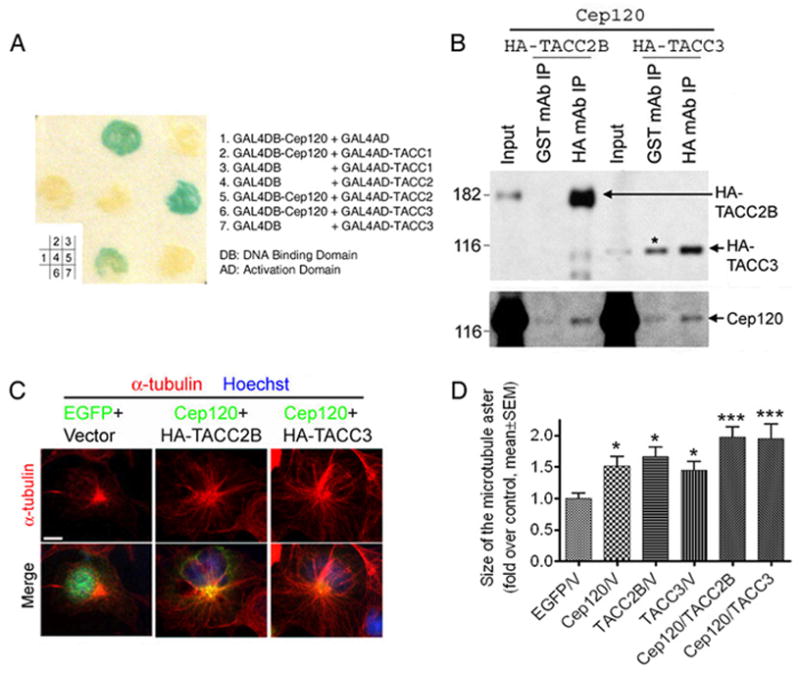
Cep120 interacts with TACCs. (A) β-Gal assay was performed using yeast cells expressing the coiled-coil domain of Cep120 and/or the C-terminus of TACCs. Positive results (blue color) were obtained only when Cep120 and TACCs were co-expressed. (B) Lysates from COS7 cells expressing exogenous Cep120 and either HA-TACC2B or HA-TACC3 were subjected to anti-HA immunoprecipitation. A fraction of Cep120 was co-immunoprecipitated. The asterisk indicates that a significant amount of HA-TACC3 was pulled down nonspecifically by sepharose beads. (C and D) Cep120 and TACCs operate together to increase the size of the microtubule aster emanating from the centrosome in transfected COS7 cells. (C) Examples of transfected cells immunostained with an α-tubulin antibody. (D) Measurement of the size of the microtubule aster. * P<0.05, *** P<0.001, one-way ANOVA. Scale bars: 10 μm.
From the embryonic mouse brain, we were able to amplify a short isoform of TACC1 (TACC1s), TACC2B, and TACC3 by RT-PCR (Figure S6), suggesting that they are expressed during cortical development. From these amplified genes we constructed plasmids for the expression of HA-tagged TACCs (Figure S7). HA-TACC1s formed large aggregates throughout the cytoplasm when overexpressed (data not shown); thus, in the following experiments we only analyzed HA-TACC2B and HA-TACC3. Consistent with the Cep120-TACC interaction in the yeast-two-hybrid system, a fraction of Cep120 co-immunoprecipitated with HA-TACCs (Figure 4B).
Interestingly, overexpression of Cep120 or TACCs in COS7 cells resulted in an increase in the size of the microtubule aster emanating from the centrosome (Figure 4, C and D). This increase was more apparent when Cep120 and TACCs were co-expressed (Figure 4, C and D). This data suggests that Cep120 and TACCs may operate together to promote the assembly of microtubules from the centrosome.
TACCs Are Important for INM
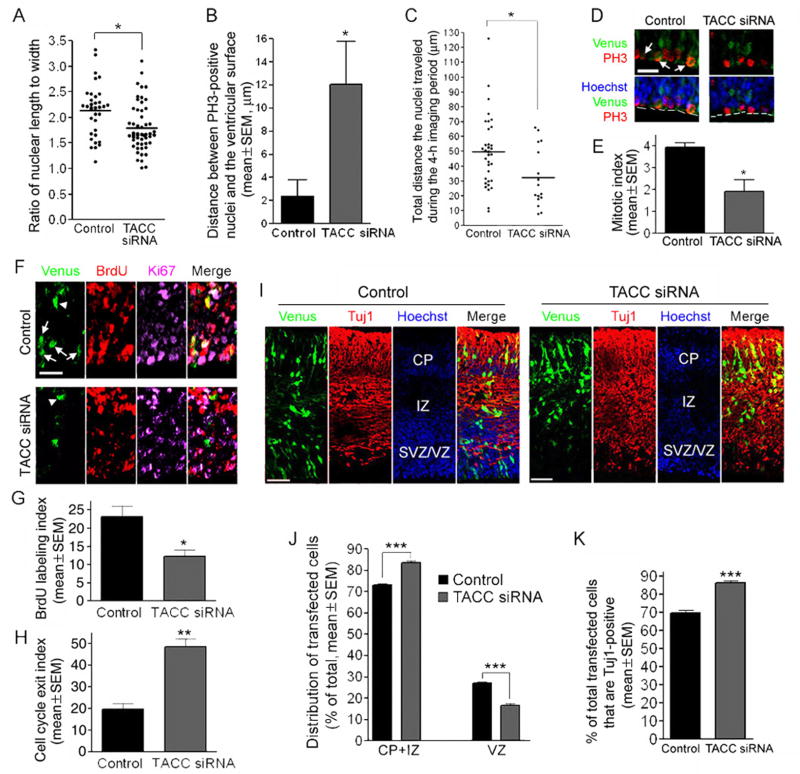
To investigate whether TACCs might regulate INM, we designed siRNA plasmids against individual TACCs that are expressed in the embryonic mouse brain (Figure S7). We found that a mixture of these siRNA plasmids (hereafter referred to as TACC siRNA) affected INM similarly as Cep120 siRNA did. First, quantification of the ratio of nuclear length to width revealed that TACC siRNA rendered the nuclei less elongated along the apical-basal axis (Figures 5A and S8A). Next, TACC siRNA significantly (P<0.05; t-test) increased the distance between PH3-positive mitotic nuclei and the ventricular surface (Figures 5B and S8B). Finally, in our time-lapse imaging experiment the total distance the nuclei traveled toward the ventricular surface during the 4-h imaging period was significantly less (P<0.05; t-test) in the TACC siRNA group than in the control group (Figures 5C and S8C; movies S4 and S5). Taken together, these data suggest that TACCs are important for INM.
Figure 5.
TACC siRNA impairs INM and neural progenitor self-renewal. (A–C) TACC siRNA impairs INM. (A) The nuclei of TACC siRNA-expressing neural progenitors are less elongated along the apical-basal axis compared to the control. Mouse embryos were electroporated at E11.5 and sacrificed at E14.5, and the ratio of nuclear length to width in transfected neural progenitors was quantified. ** P<0.01, t-test. (B) Ectopic PH3-positive nuclei in TACC siRNA-expressing neural progenitors. Mouse embryos were electroporated at E11.5 and sacrificed at E13.5, and the distance between PH3-positive nuclei and the ventricular surface was quantified. * P<0.05, t-test. (C) TACC siRNA impairs INM in brain slice preparations. Mouse embryos were electroporated at E11.5 and acute brain slices of 150-μm were prepared at E13.5 for imaging. The total distance the nuclei traveled during the 4-h imaging period was quantified. * P<0.05, t-test. (D–H) Progenitors expressing TACC siRNA prematurely exit the cell cycle. Mouse embryos were electroporated at E11.5 and exposed to BrdU at E13.5, and PH3-, BrdU-, and Ki67-immunostaining was performed at E14.5. (D) Representative images showing PH3-positive cells in the VZ of electroporated embryos. White arrows indicate transfected PH3-positive cells in the control group. White dashed lines outline the ventricular surface. (E) Quantification of the mitotic index. (F) Representative images showing transfected cells in neocortices co-stained with BrdU and Ki67 antibodies. White arrows indicate transfected cells labeled by both BrdU and Ki67. White arrowheads indicate transfected cells labeled by BrdU but not Ki67. (G) Quantification of the BrdU labeling index. (H) Quantification of the cell cycle exit index. TACC siRNA caused a significant decrease in the mitotic index and BrdU labeling index, and a significant increase in the cell cycle exit index. * P<0.05, ** P<0.01, t-test. (I–K) TACC siRNA causes increased neuronal production. Mouse embryos were electroporated at E11.5 and sacrificed at E14.5. (I) Representative images showing transfected cells in the neocortex immunostained with a Tuj1 antibody. (J) Quantification of the distribution of transfected cells. Compared to the control, TACC siRNA increased the percentage of transfected cells in the CP or IZ and decreased the percentage of transfected cells in the SVZ or VZ. *** P<0.001, t-test. (K) Quantification of the percentage of transfected cells that are Tuj1-positive. The percentage is significantly higher in the TACC siRNA group. *** P<0.001, t-test. Scale bars: 20 μm in (D) and (F), 50 μm in (I).
TACCs Are Required for Maintaining the Neural Progenitor Pool
In addition to INM defects, TACC siRNA also recapitulated neurogenesis defects caused by Cep120 siRNA. Compared to the control, the TACC siRNA group exhibited a significant decrease in the mitotic index (P<0.05, t-test) (Figure 5, D and E) and the BrdU labeling index (P<0.05, t-test) (Figure 5, F and G), and a significant increase in the cell cycle exit index (P<0.01, t-test) (Figure 5, F and H). Furthermore, TACC siRNA caused more cells to exit VZ and increased the percentage of cells that were Tuj1-positive (Figure 5, I–K). These data suggest that, similar to Cep120 siRNA, TACC siRNA also leads to premature differentiation of neural progenitors into neurons.
Cep120 and TACCs Regulate the Integrity of Microtubules Coupling the Centrosome and the Nucleus
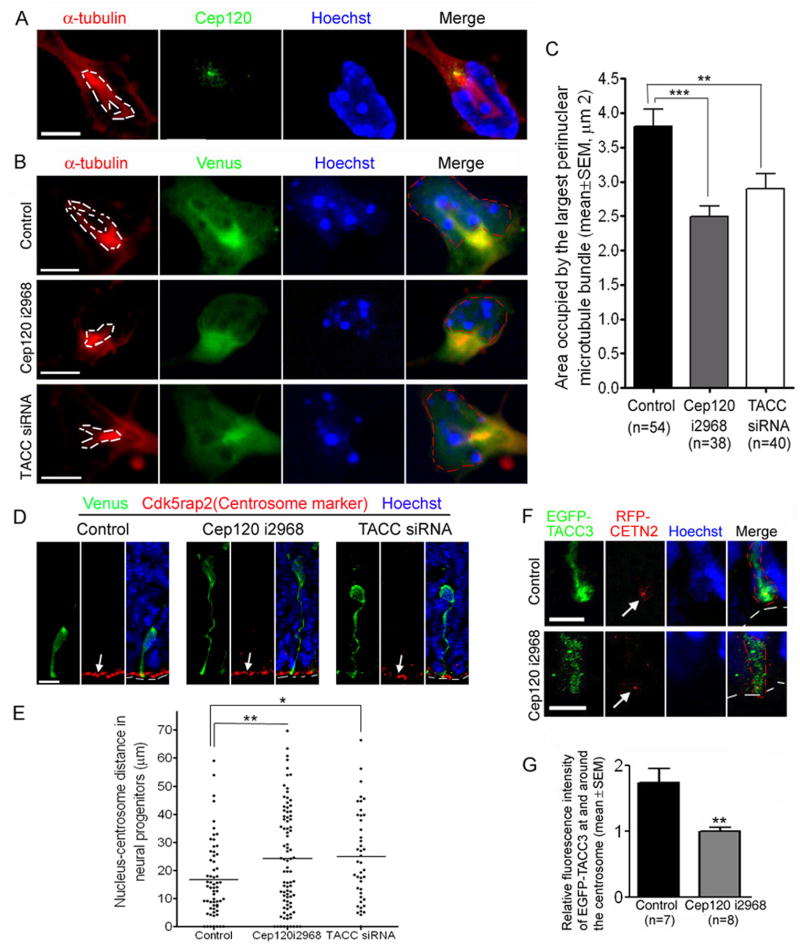
To investigate the mechanisms of how Cep120 and TACCs regulate INM, we determined whether siRNA plasmids against Cep120 or TACCs affect centrosome- and nucleus-associated microtubules, which generally sustain nuclear migration by coupling the centrosome and the nucleus (Reinsch and Gonczy, 1998; Tsai and Gleeson, 2005). Because of the difficulty in distinguishing the microtubules in transfected cells in brain sections, we developed an assay to analyze microtubules in dissociated neocortical cells (see Experimental Procedures). Under this condition, most of the cells express the progenitor marker nestin (Figure S9). Interestingly, these cells display a distinct microtubule structure resembling a previously described microtubule “fork” that couples the centrosome and the nucleus in cultured neurons (Xie et al., 2003). This structure consists of usually two, and occasionally more, prominent microtubule bundles that are associated with both the centrosome and the nucleus (Figure 6A). Notably, these microtubule bundles were less prominent in cells expressing Cep120 i2968 or TACC siRNA than in control cells (Figure 6B). To quantify this difference, we measured the area occupied by the largest microtubule bundle of the “fork”-like structure. The area was significantly less in cells expressing Cep120 i2968 or TACC siRNA compared to the control (Figure 6C). This data suggests that both Cep120 and TACCs are important for the integrity of microtubules coupling the centrosome and the nucleus.
Figure 6.
An important role for Cep120 and TACCs in the integrity of microtubules coupling the centrosome and the nucleus and for Cep120 in the centrosomal localization of TACC3. (A–C) Cep120 and TACCs regulate microtubules coupling the centrosome and the nucleus. Neocortical cells lining the lateral ventricle of mouse embryos were co-transfected with Venus and siRNA plasmids via electroporation and dissociated for α-tubulin-immunostaining analysis (see Experimental Procedures). (A) A microtubule “fork”-like structure (indicated by α-tubulin immunostaining) associated with both the centrosome (indicated by Cep120 immunostaining) and the nucleus (indicated by Hoechst labeling) in a non-transfected cell. Several Z-series images were merged to show the entire “fork”-like structure (outlined by white dashed lines). (B) Representative images showing the microtubule “fork”-like structure (outlined by white dashed lines) in transfected cells. Several Z-series images were merged to show the entire “fork”-like structure. The nuclei are outlined by red dashed lines. The microtubule “fork”-like structure is smaller in cells expressing Cep120 i2968 or TACC siRNA compared to the control. (C) Quantification of the area occupied by the largest microtubule bundle of the “fork”-like structure. ** P<0.01, *** P<0.001, one-way ANOVA. (D and E) Cep120 i2968 and TACC siRNA increase nucleus-centrosome distance in neural progenitors in vivo. Mouse embryos were electroporated with Venus and siRNA plasmids at E11.5 and cryosections were prepared at E14.5. Transfected cells were identified by immunostaining using a chicken anti-GFP antibody. Centrosomes were visualized by Cdk5rap2 immunostaining. (D) Examples of transfected neural progenitors in which centrosomes (indicated by white arrows) were visualized by Cdk5rap2 labeling. The ventricular surface is outlined by white dashed lines. (E) Quantification showing that the nucleus-centrosome distance is significantly increased in neural progenitors expressing Cep120 i2968 or TACC siRNA. * P<0.05, ** P<0.01, one-way ANOVA. (F and G) Cep120 regulates centrosomal localization of EGFP-TACC3 in neural progenitors in vivo. Mouse embryos were co-electroporated with Cep120 i2968, EGFP-TACC3, and RFP-CETN2 at E12.5. The localization of EGFP-TACC3 in transfected neural progenitors was examined about 30h after electroporation by anti-GFP immunostaining. (F) Representative images showing the enrichment of EGFP-TACC3 at and around the centrosome in the control cell (upper panels) but not in the Cep120 i2968-expressing cell (lower panels). Only the apical processes of transfected progenitors were shown. RFP-CETN2 labels the centrosome (indicated by white arrows). White and red dashed lines outline the ventricular surface and apical processes of transfected progenitors, respectively. (G) Quantification of the effect of Cep120 i2968 on centrosomal localization of EGFP-TACC3. The relative intensity of EGFP-TACC3 fluorescence at and around the centrosome (i.e. the ratio of the mean intensity of EGFP-TACC3 fluorescence at and around the centrosome to that in a nearby segment of the apical process) was significantly lower in progenitors expressing Cep120 i2968 compared to the control. ** P<0.01, t-test. Scale bars: 5 μm in (A), (B), and (F), 10 μm in (D).
Previous studies have suggested that a defect in nucleus-centrosome coupling correlates with a general increase in nucleus-centrosome distance in migrating neurons (Shu et al., 2004; Tanaka et al., 2004). To determine whether this was the case in neural progenitors, we used an antibody against a centrosomal protein Cdk5rap2 to immunostain E14.5 brain sections prepared from mouse embryos electroporated at E11.5 and then measured the nucleus-centrosome distance in transfected neural progenitors (Figure 6D). We found that nucleus-centrosome distance was significantly increased in both Cep120 i2968 (P<0.01, one-way ANOVA) and TACC siRNA (P<0.05) groups compared to the control (Figure 6, D and E). This data suggests that silencing Cep120 or TACCs impairs nucleus-centrosome coupling in neural progenitors undergoing INM.
Cep120 Regulates Centrosomal Localization of TACC3 in Neural Progenitors
We tested the possibility that Cep120 and TACCs may affect each other’s localization to the centrosome. To investigate whether Cep120 regulates the centrosomal localization of TACCs, we co-electroporated E12.5 mouse embryos with Cep120 or control siRNA, EGFP-TACC3 or EGFP-TACC2B, and a centrosomal marker RFP-CETN2 (Shu et al., 2004; Tanaka et al., 2004), and examined the localization of EGFP-TACC3 or EGFP-TACC2B in neural progenitors about 30h after electroporation. In control-transfected progenitors, EGFP-TACC3 was often found enriched at and around RFP-CETN2 puncta (Figure 6, F and G), which were localized at the tip of the apical process adjacent to the ventricular surface. In progenitors transfected with Cep120 i2968, RFP-CETN2 localization was similar to that of the control; however, the enrichment of EGFP-TACC3 around the RFP-CETN2 puncta was abolished (Figure 6, F and G). This data suggests that Cep120 is important for the recruitment of TACC3 to the centrosome in neural progenitors in vivo. In progenitors expressing EGFP-TACC2B, we were unable to identify the centrosome by either pericentrin immunostaining or RFP-CETN2 co-transfection (data not shown), possibly due to partial disruption of the centrosome by exogenous TACC2B. Thus, we could not determine the effect of silencing Cep120 on centrosomal localization of TACC2B. In contrast to the important role of Cep120 in centrosomal localization of TACC3, knockdown of TACCs did not affect the centrosomal localization of Cep120 in neural progenitors (data not shown).
Discussion
Centrosome- and microtubule-associated proteins play a major role in maintaining the neural progenitor pool during neocortical development. Previous studies on the mechanisms by which these proteins control neurogenesis have focused on mitotic spindle assembly and orientation. However, it remains unclear whether these proteins may control neocortical neurogenesis by regulating other microtubule-dependent processes such as nuclear migration. Here we identify Cep120, a novel centrosomal protein preferentially expressed in neural progenitors, and show that it interacts with TACCs and is important for the recruitment of TACC3 to the centrosome. We provide evidence that both Cep120 and TACCs play an essential role in INM during neocortical development by regulating microtubules coupling the centrosome and the nucleus. Importantly, INM defects resulting from the knockdown of Cep120 or TACCs were accompanied by premature differentiation of neocortical progenitors into neurons, resulting in a depletion of the proliferating progenitor pool. Thus, our data have uncovered a critical role for the regulation of microtubules by Cep120 and TACCs in INM. Furthermore, our data suggest that, in addition to controlling mitotic spindle, sustaining INM may also be an important mechanism by which microtubule-regulating proteins maintain the neural progenitor pool during neocortical development.
Cep120 Is a Novel Centrosomal Protein
A critical role for the centrosome in neocortical neurogenesis is underscored by the fact that three of the four identified causative genes for autosomal recessive primary microcephaly, a disorder likely caused by abnormal neurogenesis, encode centrosomal proteins (Bond et al., 2002, 2005; Hung et al., 2000; Kouprina et al., 2005; Zhong et al., 2005). Thus, the elucidation of centrosomal signaling may provide key information regarding the mechanisms underlying neocortical neurogenesis. In this study, we identify Cep120, a novel centrosomal protein preferentially expressed in neural progenitors during neocortical development (Figure 1). Cep120 showed a distinct localization to the centrosome in all types of cells we examined and at all phases of the cell cycle (Figure 1, D and E; data not shown). Moreover, Cep120 interacts with a number of established centrosomal proteins in the yeast-two-hybrid system, including TACCs, myomegalin, Cep290, ninein, Cep164, and KIAA1009 (Figure 4A and data not shown). These data suggest that Cep120 is a resident centrosomal protein.
Putative orthologs of Cep120 were identified from ciliate protozoans to mammals (Figure S1), suggesting that Cep120 may play a conserved role in key cellular processes that are dependent on the centrosome. Interestingly, all of the putative Cep120 orthologs contain a coiled-coil domain (Figure S1), which shares significant homology to many other centrosomal proteins. For instance, the coiled-coil domain of mouse Cep120 is homologous to coiled-coil domains of maskin (the Xenopus TACC3, E value: 2e-04), mouse ninein (E value: 6e-04), human Cep290 (E value: 2e-05), and microcephaly gene products CDK5RAP2 (E value: 1e-04) and CENPJ (E value: 5e-04). Thus, this domain may be critical for the function of Cep120 and other centrosomal proteins.
Cep120 Regulates Centrosomal Localization of TACC3
The orthologs of TACCs have been identified from nematodes to mammals and are important for the growth or stabilization of centrosome-associated microtubules (Barros et al., 2005; Bellanger and Gonczy, 2003; Chen et al., 2000; Gergely et al., 2000a, 2000b, 2003; Kinoshita et al., 2005; Le Bot et al., 2003; O'Brien et al., 2005; Peset et al., 2005; Srayko et al., 2003; Still et al., 1999a, 1999b). Interestingly, while vertebrate TACCs display prominent localization to both the centrosome and microtubules (Chen et al., 2000; Gergely et al., 2000b; Kinoshita et al., 2005; Peset et al., 2005; Piekorz et al., 2002), their orthologs in C. elegans and Drosophila embryos are localized more specifically to the centrosome (Bellanger and Gonczy, 2003; Gergely et al., 2000a; Le Bot et al., 2003; Srayko et al., 2003). Studies using the Xenopus egg extract system suggest that the regulation of microtubules by TACCs may be dependent on their localization to the centrosome (Kinoshita et al., 2005; Peset et al., 2005). In the present study, we show that in neural progenitors in the developing neocortex, Cep120 is important for recruiting TACC3 to the centrosome (Figure 6, D and E). This recruitment likely affects centrosome-associated microtubules and cellular processes that are dependent on these microtubules such as nuclear migration.
All of the three members of mouse TACCs, including TACC1, TACC2, and TACC3, are expressed in the embryonic mouse brain (Figure S6). Previous studies suggest that TACC1 and TACC3 may be expressed in the brain at relatively higher levels in early embryonic stages than in later stages (Aitola et al., 2003; Lauffart et al., 2006). Mice lacking TACC3 display increased apoptosis of hematopoietic stem cells and a reduced size of most organs including the brain (Piekorz et al., 2002); mice lacking TACC2 are apparently normal (Schuendeln et al., 2004). However, it remains possible that the loss of a single TACC may be partially compensated for by other TACC family members. Thus in our experiments we simultaneously knocked down all of the three TACCs to assess potential defects in INM and neurogenesis.
Cep120 and TACCs Control INM by Regulating Centrosome-Associated Microtubules
Although INM was reported seven decades ago (Sauer, 1935), the mechanism underlying INM remains an enigma. Strikingly, while the nucleus and the centrosome (or its equivalent structure) are tightly associated during most other types of nuclear migration, the distance between the nucleus and the centrosome dynamically changes and can span the entire thickness of the VZ during INM. Currently little is known about the molecular mechanisms sustaining this unique mode of nuclear movement.
In this study we show that both Cep120 and TACCs play a critical role in INM (Figures 2; Figure 5, A–C; Figure S8). Consistent with the premise that centrosomal proteins control nuclear migration through microtubules (Reinsch and Gonczy, 1998; Tsai and Gleeson, 2005), Cep120 or TACC knockdown impaired microtubule-dependent coupling of the nucleus and the centrosome (Figure 6, A–E). Based on these data and the finding that the centrosome remains adjacent to the ventricular surface throughout INM (Hinds and Ruffett, 1971; Astrom and Webster, 1991; Chenn et al., 1998; Figure 6, D and F), we propose a model in which the dynamic regulation of microtubules by Cep120 and TACCs plays a key role in sustaining INM (Figure 7). In this model Cep120 recruits TACCs to the centrosome to promote the growth of long microtubules from the centrosome, which is localized adjacent to the ventricular surface. These microtubules sustain INM by maintaining the coupling between the centrosome and the nucleus.
Figure 7.
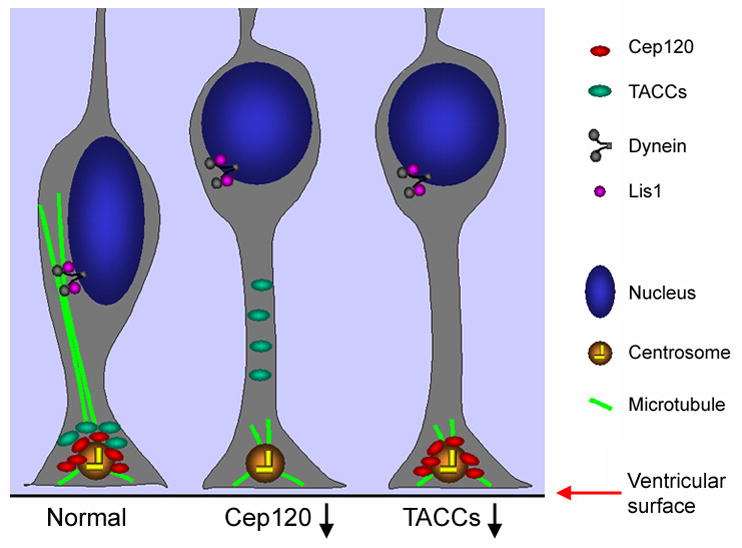
A tentative model for INM. The centrosome is localized at the tip of the apical process adjacent to the ventricular surface. Cep120 recruits TACCs to the centrosome and together with TACCs promotes the assembly of long microtubules from the centrosome. The nucleus may be transported toward the centrosome along these microtubules by dynein or be pulled toward the centrosome due to the shortening of these microtubules. Lis1 may sustain the migration of the nucleus by regulating the activity of dynein. Upon depletion of Cep120 or TACCs, microtubules emanating from the centrosome are too short to reach the nucleus when the nucleus is located deep within the ventricular zone. Consequently, the transport or pulling of the nucleus toward the ventricular surface is impaired.
How do microtubules mediate nuclear migration during INM? One possible mechanism is that the nucleus may be transported along microtubules toward and away from the centrosome by the minus end-directed motor dynein and the plus end-directed motor kinesin, respectively. Interestingly, a dynein-regulating protein Lis1 has been shown to regulate INM (Gambello et al., 2003; Tsai et al., 2005), although the underlying mechanism is unclear. Alternatively, the nucleus may be pulled toward and pushed away from the centrosome by depolymerizing and polymerizing microtubules, respectively. In either scenario, the capacity of the centrosome to assemble long microtubules is critical.
Due to technical limitations, we have not analyzed INM of neural progenitors at stages earlier than E11.5. Interestingly, Cep120 is already expressed at high levels in the brain before E11.5 (Figure 1C). Since the cell morphology and INM pattern of neural progenitors at stages before E11.5 (known as neuroepithelial cells) are similar to that of progenitors arising at later stages (known as radial glial cells) (Gotz and Huttner, 2005), Cep120 and TACCs may also be important for INM at stages before E11.5.
Cep120 and TACCs Are Essential for Maintaining the Neural Progenitor Pool
We found that knockdown of either Cep120 or TACCs resulted in a decrease in the mitotic index and BrdU labeling index, an increase in the cell cycle exit index, and a greater percentage of transfected cells that migrated into the CP or IZ and that expressed the neuronal marker Tuj1 (Figures 3; Figure 5, D–K). These data suggest that Cep120- or TACC-knockdown causes a premature depletion of proliferating progenitors and an increase in neuronal production. Since we rarely detected pyknotic or necrotic transfected cells in the electroporated embryos (data not shown), the depletion of proliferating progenitors and the increase in neuronal production are likely caused by premature differentiation of progenitors into neurons.
The critical role for both Cep120 and TACCs in INM and neocortical progenitor self-renewal suggests that a major role of INM may be to maintain the neural progenitor pool in the developing neocortex. We investigated whether cellular defects other than impaired INM may underlie the depletion of neural progenitors. First, we found that silencing Cep120 or TACCs did not alter the cleavage plane of neural progenitors that were at anaphase or telophase (data not shown). Second, we did not observe an M-phase block in neural progenitors expressing Cep120 siRNA or TACC siRNA plasmids (Figure 3, A and B; Figure 5, D and E; and data not shown), suggesting that silencing of Cep120 or TACCs did not severely impair the organization of the mitotic spindle. Third, our BrdU pulse-labeling experiments suggest that silencing Cep120 or TACCs does not affect cell cycle length (Figure S10). Lastly, apical polarity seems to be normal in neural progenitors expressing Cep120 siRNA or TACC siRNA. This is indicated by the normal contact of their apical processes with the ventricular surface (Figure 6D and data not shown), the normal positioning of the centrosome (Figure 6, D and F), and the intact ventricular surface around these cells (data not shown). Therefore, our hypothesis is that INM defects directly underlie the neurogenesis phenotype caused by the knockdown of Cep120 or TACCs.
In summary, we have identified a novel centrosomal protein, Cep120, and shown that it is important for recruiting TACC3 to the centrosome in neural progenitors. Furthermore, we have demonstrated an essential role for Cep120 and TACCs in sustaining INM and maintaining the neural progenitor pool in the developing neocortex. Our data suggest that controlling INM may be a novel mechanism by which microtubule-regulating proteins govern neocortical neurogenesis.
Experimental Procedures
Vector-Based siRNA Constructs
The pSilencer-based siRNA plasmids were generated by ligating annealed oligonucleotides containing hairpin sequences against specific mRNA targets into BamH I/Hind III sites of pSilencer 2.0-U6. The oligonucleotides for siRNA constructs include the following pairs: i2968f (5′ gatcccgttatgcggtcctc atggttatttgat atccgataaccatgaggaccg cataattttttccaaa 3′) and i2968r (5′ agcttttggaaaaaa ttatgcggtcctc atggttatc ggatatcaaata accatgaggaccgcataacgg 3′) for Cep120 i2968; i1265f (gatccgccaaggatgatgcaacagattcaag agatctgttgcatcatccttggttttttggaaa) and i1265r (agcttttccaaaaaaccaaggatgatgcaacagatctcttg aatctgttgcatcatccttggcg) for Cep120 i1265; t1s_2f (5′ gatcccgttatctctt ctctgatcaacgttgata tccgcgttgatcagagaagagataa ttttttccaaa 3′) and t1s_2r (5′ agcttttggaaaaaat tatctcttctctgatcaa cgcg gatatcaacgttgatcaga gaagagataacgg 3′) for siRNA against TACC1s; t2_5f (5′ gatcccgtcataggagtttctgtagtccttgatatccgggactacagaaa ctcctatgattttttccaaa 3′) and t2_5r (5′ agct tttggaaaaaatcataggagtttctgtagtcccggatatcaaggac tacagaaactcctatgacgg 3′) for siRNA against TACC2B; t3_1f (5′ gatcccatttcacgtacaaagact gctttgatatccgagcagtctttgta cgtgaaatttttttccaaa 3′) and t3_1r (5′ agcttttggaaaaaaatttcacgtaca aagactgctcggatatcaaagcag tctttgtacgtgaaatgg 3′) for siRNA against TACC3.
Production of Antibodies
Rabbit Cep120 antiserum was produced by Pocono Rabbit Farm & Laboratory, Inc (Canadensis, PA) using residues 660-988 of mCep120 as an immunogen. Rabbit Cdk5rap2 antibody was raised against the N-terminus of mouse Cdk5rap2 by Pocono Rabbit Farm & Laboratory, Inc.
In Utero Electroporation
Avertin was used to anesthetize pregnant Swiss Webster mice. Five pulses of current (50msec on/950msec off) were delivered across the head of the embryos (Voltages: 22–30V for E11.5; 32–33V for E12.5; 35–40V for E14.5). In the DNA mixture, the concentration of the siRNA plasmid was 2-3 fold higher than that of the Venus plasmid. In BrdU-labeling experiments, mouse embryos were electroporated at E11.5 and BrdU (1 ml, 2 mg/ml in saline) was injected intraperitoneally into dams at E13.5.
Brain Slice Culture and Imaging
To assess INM, mouse embryos were electroporated at E11.5 and acute brain slices (coronal, 150 μm) were prepared at E13.5. The slices were cultured in a Millicell culture insert system (Millipore) in Neurobasal medium supplemented with B27, N2, 0.5mM glutamine, 1% penicillin /streptomycin, and 5% horse serum for one hour before time-lapse imaging on a DeltaVision system. During imaging, the slices were kept in the Millicell system and the temperature and PH of the medium were maintained by a 37°C water bath and 5% CO2, respectively.
Analysis of Microtubules in Transfected Dissociated Neocortical Cells
A mixture of Venus plasmid and siRNA plasmid was injected into lateral ventricles of E13.5 mouse embryos. The DNA plasmids were then introduced into neocortical cells lining the lateral ventricle by electroporation. Immediately following electroporation, the neocortex was dissected out and cultured as explants (pial surface facing up) on a Millicell culture insert system in Neurobasal medium supplemented with B27, N2, 0.5mM glutamine, 1% penicillin /streptomycin, and 5% horse serum. After two days of explant culture, the neocortex was trypsinized, dissociated, plated onto coverslips pre-coated with poly-D-lysine and laminin, and cultured overnight (the medium was the same as that for explant culture) before fixation. The microtubules were visualized by anti-α-tubulin immunostaining. Several Z-series confocal images of transfected cells were merged to show the entire microtubule “fork”-like structure.
Confocal Microscopy
Confocal images were obtained using a Zeiss LSM 510 system.
Supplementary Material
Acknowledgments
We thank Dr. M. E. Hatten for providing Venus vectors and Dr. J.M. Ihle for providing TACC3 antibodies. We also thank Dr. B.A. Samuels and X. Ge for critical reading of the manuscript. Z.X. was supported by a Taplin postdoctoral fellowship and is a postdoctoral fellow of the Howard Hughes Medical Institute. K.S. held a fellowship from the Naito Foundation and was supported by a JSPS Postdoctoral Fellowship for Research Abroad. L-H.T. is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant #NS37007 to L.-H.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitola M, Sadek CM, Gustafsson JA, Pelto-Huikko M. Aint/Tacc3 is highly expressed in proliferating mouse tissues during development, spermatogenesis, and oogenesis. J Histochem Cytochem. 2003;51:455–469. doi: 10.1177/002215540305100407. [DOI] [PubMed] [Google Scholar]
- Astrom KE, Webster HD. The early development of the neopallial wall and area choroidea in fetal rats. A light and electron microscopic study. Adv Anat Embryol Cell Biol. 1991;123:1–76. [PubMed] [Google Scholar]
- Barros TP, Kinoshita K, Hyman AA, Raff JW. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol. 2005;170:1039–1046. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Gonczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol. 2004;20:593–618. doi: 10.1146/annurev.cellbio.20.082503.103047. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. doi: 10.1038/nrn2058. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- Chen HM, Schmeichel KL, Mian IS, Lelievre S, Petersen OW, Bissell MJ. AZU-1: a candidate breast tumor suppressor and biomarker for tumor progression. Mol Biol Cell. 2000;11:1357–1367. doi: 10.1091/mbc.11.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000a;19:241–252. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci U S A. 2000b;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F. Centrosomal TACCtics. BioEssays. 2002;24:915–925. doi: 10.1002/bies.10162. [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Ruffett TL. Cell proliferation in the neural tube: an electron microscopic and Golgi analysis in the mouse cerebral vesicle. Z Zellforsch Mikrosk Anat. 1971;115:226–264. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20:7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karfunkel P. The activity of microtubules and microfilaments in neurulation in the chick. J Exp Zool. 1972;181:289–301. doi: 10.1002/jez.1401810302. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J, Mochida GH, Risinger JI, Goldsmith P, Gunsior M, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet. 2005;14:2155–6215. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Lauffart B, Dimatteo A, Vaughan MM, Cincotta MA, Black JD, Still IH. Temporal and spatial expression of TACC1 in the mouse and human. Dev Dyn. 2006;235:1638–1647. doi: 10.1002/dvdy.20724. [DOI] [PubMed] [Google Scholar]
- Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol. 2003;13:1499–1505. doi: 10.1016/s0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- Messier PE, Auclair C. Inhibition of nuclear migration in the absence of microtubules in the chick embryo. J Embryol Exp Morphol. 1973;30:661–671. [PubMed] [Google Scholar]
- Messier PE, Auclair C. Effect of cytochalasin B on interkinetic nuclear migration in the chick embryo. Dev Biol. 1974;36:218–223. doi: 10.1016/0012-1606(74)90206-1. [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- O’Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol Biol Cell. 2005;16:2836–2847. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekorz RP, Hoffmeyer A, Duntsch CD, McKay C, Nakajima H, Sexl V, Snyder L, Rehg J, Ihle JN. The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. EMBO J. 2002;21:653–664. doi: 10.1093/emboj/21.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. A century of progress in corticoneurogenesis: from silver impregnation to genetic engineering. Cereb Cortex . 2006;16(Suppl 1):i3–17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- Schuendeln MM, Piekorz RP, Wichmann C, Lee Y, McKinnon PJ, Boyd K, Takahashi Y, Ihle JN. The centrosomal, putative tumor suppressor protein TACC2 is dispensable for normal development, and deficiency does not lead to cancer. Mol Cell Biol. 2004;24:6403–6409. doi: 10.1128/MCB.24.14.6403-6409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Shu T, Tseng HC, Sapir T, Stern P, Zhou Y, Sanada K, Fischer A, Coquelle FM, Reiner O, Tsai LH. Doublecortin-like kinase controls neurogenesis by regulating mitotic spindles and M phase progression. Neuron. 2006;49:25–39. doi: 10.1016/j.neuron.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–1511. doi: 10.1016/s0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- Still IH, Hamilton M, Vince P, Wolfman A, Cowell JK. Cloning of TACC1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene. 1999a;18:4032–4038. doi: 10.1038/sj.onc.1202801. [DOI] [PubMed] [Google Scholar]
- Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999b;58:165–170. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS., Jr Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Webster W, Langman J. The effect of cytochalasin B on the neuroepithelial cells of the mouse embryo. Am J Anat. 1978;152:209–221. doi: 10.1002/aja.1001520204. [DOI] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Zhong X, Liu L, Zhao A, Pfeifer GP, Xu X. The abnormal spindle-like, microcephaly-associated (ASPM) gene encodes a centrosomal protein. Cell Cycle. 2005;4:1227–1229. doi: 10.4161/cc.4.9.2029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.