Abstract
A previously unknown basidiomycetous yeast is described for which the name Sympodiomycopsis lanaiensis is proposed. The type strain, LM418T, was isolated from driftwood collected on a beach on Lana‘i (Hawai‘i). On the basis of ribosomal DNA sequence analysis [large subunit (LSU), internal transcribed spacer (ITS) 1 & 2, and 18S], LM418T belongs to the order Microstromatales, which includes the genera Sympodiomycopsis, Rhodotorula, Microstroma, Volvocisporium and Quambalaria. The strain is described as a new species in the genus Sympodiomycopsis on the basis of morphological and physiological characteristics and the phylogenetic relationship to Sympodiomycopsis paphiopedili. Sexual reproduction was not observed in LM418T. GenBank accession numbers for nucleotide sequences of regions of the LSU, ITS and 18S regions of the ribosomal operon in LM418T are DQ990016, DQ990017 and DQ990018, respectively. LM418T has been deposited in the DSMZ as DSM 18755, in the ATCC as MYA-4092, in the Agricultural Research Service Culture Collection as NRRL Y-48466, and the Centraalbureau voor Schimmelcultures as CBS 10858.
Keywords: Sympodiomycopsis, yeast, Hawai‘i, wood
Introduction
The Hawaiian Archipelago hosts thousands of unique plant and animal species (Amadon, 1947; Carr et al., 1989). Although the region is widely acknowledged as a ‘biodiversity hotspot’, few novel cultivated microorganisms from the archipelago have been described (Kohlmeyer, 1969, 1985; Dring et al., 1971; Barr & Hodges, 1987; Kohlmeyer & Volkmann-Kohlmeyer, 1989; Myers et al., 2000). In this respect, one-third of the bacteria species (each ‘species’ shared ≤ 97% 16S rRNA gene nucleotide sequence identity) cultivated from Hawaiian lakes and at the submarine Lō‘ihi volcano were potentially new species or genera, four of which have been described to date (Donachie et al., 2003, 2004a, b, 2005, 2006). Only a fraction of the effort devoted globally to cultivating marine bacteria has been applied to cultivating marine filamentous fungi and yeasts; the latter two groups are thus poorly represented in culture collections and less so in the literature.
Although fungi and fungi-like organisms are ubiquitous in the ocean, the mycobiota off Hawai‘i have rarely been considered in terms of abundance, ecology, or phylogeny (Fell, 1976, 2001; Donachie & Zdanowski, 1998; Nagahama et al., 2001; Liu et al., 2003; Tsukamoto et al., 2004). Over 30 years ago, however, Kohlmeyer (1969) stated that, ‘… the marine fungal flora of Hawai‘i is rich in species and deserves further thorough investigation.’ Kohlmeyer (1985) also made an excellent case for novelty among the archipelago’s mycobiota by isolating Nimbospora octonae during two collections in the state 15 years apart. This species was not found anywhere else during Kohlmeyer’s 4000 collections over a 25-year period (Kohlmeyer, 1985). Considering Kohlmeyer’s work in Hawai‘i and that phylogenetically novel taxa appear to be abundant among cultivated Hawaiian Bacteria (Donachie et al., 2005, 2006), we believe that a dedicated isolation effort directed towards Hawaii’s marine fungi will yield new species. As part of a project to describe phylogenetic diversity and secondary metabolite production in Hawaiian marine fungi, we sampled marine habitats throughout Hawai‘i. Cultivation techniques were followed by DNA sequencing methods to rapidly enable tentative phylogenetic assignment of isolated strains. Molecular evidence of phylogenetic novelty was tested through morphological and physiological observations. We describe here Sympodiomycopsis lanaiensis LM418T, a previously unknown basidiomycetous yeast isolated from driftwood collected on Lana‘i, Hawai‘i.
Materials and methods
Sample collection
In November 2005, a piece of driftwood was collected in the intertidal zone of ‘Shipwreck Beach’ on the island of Lana‘i, Hawai‘i (20°55′15″N, 156°54′30″W). The sample was placed directly into a 50-mL sterile polypropylene tube with 4.5mL of sterile seawater, and processed in the laboratory at the University of Hawai‘i at Mānoa for cultivation of yeasts and filamentous fungi.
Yeast isolation
The sample (c. 2.5 cm × 2.5 cm × 0.2 cm) in 4.5 mL sterile seawater was vortexed vigorously for 30 s; 200 µL subsamples of the ‘diluent’ were spread on potato dextrose agar (PDA) (Difco) containing sodium chloride (20 g L−1), 300 mg L−1 of penicillin G and 250 mg L−1 of streptomycin sulfate. Plates were incubated at 25 °C. Representatives of colonies that arose after 7 days of incubation were streaked for isolation on PDA and incubated at 25 °C. The purity of isolated strains was checked by consistency of colony morphology and uniformity of cells in wet mount preparations. Strain LM418Twas characterized by standard methods (Yarrow, 1998). Cells were stored at 4 °C on potato dextrose slants and in potato dextrose broth (PDB) with 2% NaCl and 60% glycerol at −20 °C.
DNA-based analysis
Cells of LM418T grown in 10mL PDB (with 2% NaCl) for 72 h in an orbital shaker at 120 r.p.m. and room temperature were collected by centrifugation (313 g for 30 s). One microliter of the pellet was transferred to each of three PCR tubes and suspended in each in 41.75 µL of sterile distilled water. PCR reagents added to each tube were 1 µL dNTP, 5 µL 10 × PCR Reaction Buffer, 0.25 µL Taq DNA Polymerase (Roche PCR Core Kit), along with one of the following primer pairs, 1 µL per primer: (1) F63 forward (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and R635 reverse (5′CCCGTCTTGAAACACG-3′) for the D1/D2 region; (2) NS7 forward (5′-GAGGCAATAACAGGTCTGTGATGC-3′) and LR6 reverse (5′-CGCCAGTTCTGCTTACC-3′) for the internal transcribed spacer (ITS) region; (3) 18S forward (5′-TACCTGGTTGATCCTGCCAGT-3′) and 18S reverse (3′-TTGATCCTTCTGCAGGTTCACCTAC-5′) (Medlin et al., 1988; Fell et al., 2000; Fell, 2001). The PCR cycle was as follows: 94 °C (hot start), followed by 30 cycles of denaturation (94 °C for 45 s), annealing (55 °C for 45 s), extension (72 °C for 90 s) and a final extension step (72 °C for 7 min). PCR products were cleaned in the MoBio Ultraclean PCR Clean-Up kit before being sequenced in core laboratories at the University of Hawai‘i or the University of Miami.
Sequence chromatograms were manually checked for quality and edited. Consensus sequences were compared with other sequences in GenBank at the NCBI through a blastn search (Altschul et al., 1997). Sequences were initially aligned using the program muscle (Edgar, 2004) and manually edited in bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html). Maximum-likelihood, maximum parsimony and neighbour-joining phylogenetic analysis including bootstrapping were performed in the phylip software package (Felsenstein, 2005). The optimal model of nucleotide substitution was identified using the program modelgenerator (Keane et al., 2006). Bayesian analysis, as implemented in mrbayes 3.1.2 (Ronquist & Huelsenbeck, 2003), was performed for 500 000 generations using four independent chains and the GTR model. Substitution-rate variation among sites was modeled by a discrete approximation of the gamma-distribution with a proportion of invariable sites (I+Γ). The resultant trees were sampled every 100 generations with trees sampled during the first 50 000 generations discarded as burn-in (the burn-in period was estimated by plotting the likelihood of the sampled trees). Relationships among the remaining trees were summarized using a majority-rule consensus method with clade probabilities determined using the sumt command of mrbayes 3.1.2. Phylogenetic trees were annotated and refined for publication in Adobe Illustrator 9.0 (Adobe Systems Incorporated).
Morphological and physiological analyses
LM418T colony morphology was determined on malt extract agar (MEA) plates containing NaCl (2% w/v) after incubation at 22 °C for 3 days and 1 month. Cell size and shape were recorded after 7 days and 1 month in ME broth with NaCl (2% w/v). The assimilation of single carbon and nitrogen sources by LM418T was determined in liquid media in 2-mL microfuge tubes on a rotary shaker, which is a modification of the method described by Yarrow (1998). Fermentation of glucose was investigated in Durham tubes with a 2% sugar mixture and Brom Cresol purple, read after 3 days and then weekly for up to 1 month. The presence or absence of ballistoconidia was determined on inverted ME plates after 7 days and 30 days.
Results and discussion
A yeast strain designated LM418 was isolated from driftwood on Shipwreck Beach, Lana‘i. Our initial analysis of the strain, which was based on partial 18S rRNA gene sequence, indicated (data not shown) a relationship within the Microstromatales, an order that includes the filamentous, tree-associated genera Quambalaria, Microstroma and Volvocisporium. de Beer et al. (2006) placed these genera in three families (Quambalariaceae, Microstomataceae and Volvocisporiaceae) based on sequence analysis [large subunit (LSU) and ITS rRNA] and septal morphology. Yeast-like species in the order include three species of Rhodotorula (Rhodotorula bacarum, Rhodotorula phylloplana and Rhodotorula hinnulea) and Sympodiomycopsis paphiopedili. The Rhodotorula species are phylogenetically related to the genus Microstroma, although teleomorphic/anamorphic relationships have not been determined (de Beer et al., 2006). The monotypic, anamorphic genus Sympodiomycopsis was erected by Sugiyama et al. (1991) to accommodate a basidiomycete with a dominant yeast form and a less prevalent formation of true hyphae. Cell reproduction was enteroblastic-annellidic or rarely by holoblastic-sympodial budding. Sympodial budding was also observed on the hyphae (Sugiyama & Suh, 1998). Bootstrap and Baysian Markov analyses of the ITS region placed S. paphiopedili on a separate branch and incertae sedis within the Microstromatales (de Beer et al., 2006).
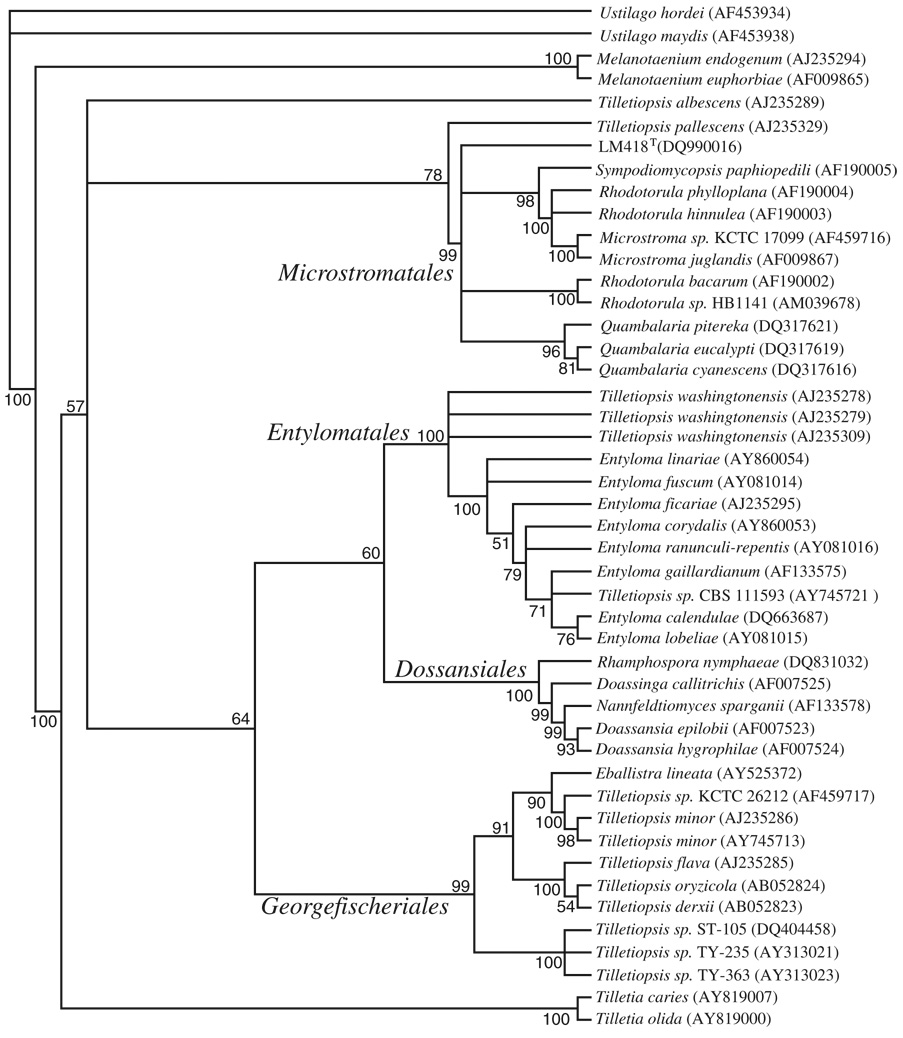
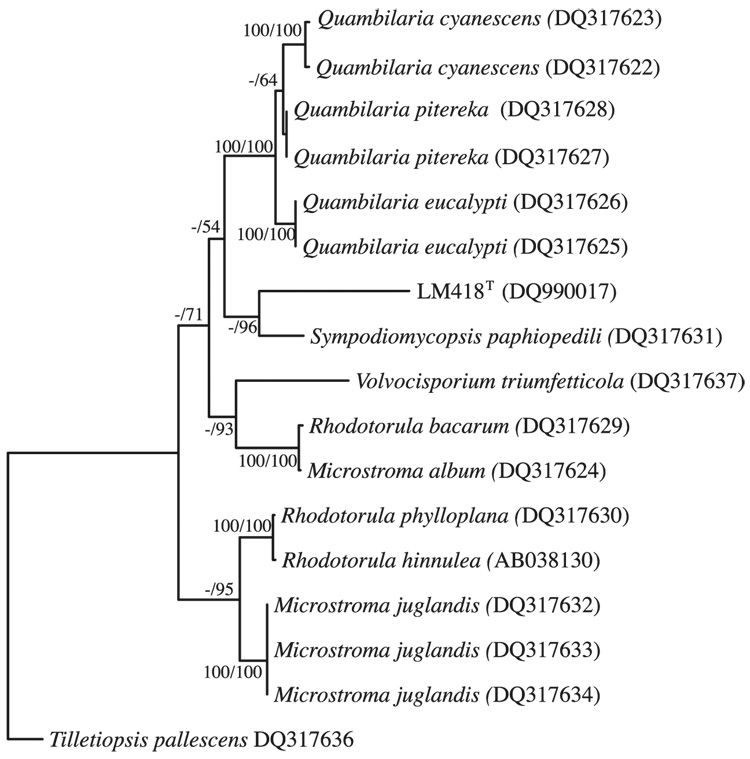
Our initial 18S rRNA gene phylogenetic analysis was subsequently supported by Bayesian analysis of a D1/D2 region of the LSU rRNA gene nucleotide sequence (Fig. 1). However, this analysis did not conclusively identify within-clade affiliations of the Microstromatales, as evidenced by the branching polytomy (Fig. 1). Similarly there was no consensus of placement or bootstrap support using neighbour-joining, parsimony or maximum-likelihood methods (data not shown). Within-clade analysis using ITS sequences has LM418T branching with S. paphiopedili (Fig. 2), a position maintained in maximum-likelihood, neighbour-joining and parsimony analysis (data not shown), but with little bootstrap support. Both D1/D2 and ITS analyses do support an incertae sedis position for S. paphiopedili and LM418T within the Microstromatales as per de Beer et al. (2006).
Fig. 1.
Consensus tree of Bayesian analysis of c. 522 bp D1/D2 region of the LSU rRNA gene of LM418T and related species. Volvocisporium triumfetticola was removed from the analysis to eliminate the effects of long-branch attraction. Bayesian analysis was performed with mrbayes 3.1.2 for 500 000 generations using four independent chains and the GTR+I+Γ model of nucleotide substitution. Numbers at nodes refer to posterior probabilities determined using the sumt command of mrbayes. Values smaller than 50% are not shown.
Fig. 2.
Phylogram obtained by maximum-likelihood analysis of 509 bp alignment of the ITS 1, 5.8S rRNA gene and ITS 2 region LM418T of related species in the Microstromatales. Numbers at nodes refer to percentage bootstrap values of 1000 replicates of neighbour-joining analysis, and to clade posterior probabilities as determined using the sumt command of mrbayes, where tree topology was conserved. Values smaller than 50% are not shown.
Sympodiomycopsis paphiopedili and LM418T share 93.8% sequence identity in the variable D1/D2, and 78% sequence identity across 457 base pairs of the ITS variable region. Both species produce true hyphae and assimilate glucose, maltose, raffinose, d-mannitol, d-glucitol and sodium succinate, and do not assimilate l-rhamnose, hexadecane or produce starch. Neither species ferments glucose. The two species can be separated on their abilities to utilize sucrose, cellobiose, trehalose, melezitose, xylose, ribose and erythritol (Table 1). Both species develop hyphae, however S. paphiopedili has sympodial formation of conidia, which is not observed in LM418T.
Table 1.
Physiological properties of Sympodiomycopsis lanaiensis LM418T and related taxa*
| Assimilation | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| l-Sorbose | + | + | − | v | − |
| Sucrose | − | + | + | + | + |
| Cellobiose | − | + | + | s | − |
| Trehalose | − | + | + | + | − |
| Meliboise | + | + | − | − | − |
| Melezitose | − | + | + | + | + |
| Soluble starch | + | w | − | + | − |
| d-Xylose | − | + | − | + | − |
| l-Arabinose | − | + | + | + | + |
| d-Arabinose | − | + | − | + | − |
| d-Ribose | − | + | − | + | − |
| Ethanol | − | + | v | − | + |
| Erythritol | − | + | + | + | + |
| 2-Keto d-gluconate | − | + | − | − | − |
| Vitamin free | + | + | + | − | + |
| dl-Lactic acid | w | w | − | − | − |
| Sodium citrate | + | − | w | s | − |
| Inositol | − | + | + | − | s |
| True or pseudohyphae | + | + | − | w+ | − |
1, LM418T; 2, Sympodiomycopsis paphiopedili; 3, Rhodotorula hinnulea; 4, Rhodotorula bacarum; 5, Rhodotorula phylloplana
w, weak; n, not done; s, slow; v, variable. Only S. paphiopedili assimilates lactose and inulin, and in both cases it is weak. All strains assimilate d-mannitol, but it is weak in R. bacarum. Data from Kurtzman & Fell (1998) and the CBS website.
The generic designation of the Rhodotorula species in the Microstromatales was based on traditional phenotypic characteristics. For example, while many of the species are lightly to strongly red-pigmented, molecular systematics demonstrated the majority of the Rhodotorula species belong to the Pucciniomycetes (Fell et al., 2000; Scorzetti et al., 2002). In contrast, Rhodotorula species in the Microstromatales are cream to buff colored, as observed with Sympodiomycopsis. The molecular analyses strongly suggest that a new generic designation for these Rhodotorula species should be proposed. However, the lack of a teleomorphic connection is a limiting factor for such a proposal. The Rhodotorula and Sympodiomycopsis species have considerable variability in their abilities to utilize carbon compounds with a single uniform difference between the genera, which is melibiose assimilation (Table 1).
The placement of LM418T in Sympodiomycopsis is not unequivocal. The ITS data (Fig. 2) indicates a closer relationship of LM418T to Sympodiomycopsis than to R. bacarum, Rhodotorula phylloplana and Rhodotorula hinnulea. Importantly, our knowledge of yeasts is evolving. Although there are perhaps 1200 described species of yeasts, this number may only represent 1% of the species in nature. In particular, research on the extent of species diversity within the Microstromatales has been sparse. Consequently, as the database expands, the relationship of these incertae sedis yeasts should be clarified based on teleomorphic/anamorphic connections and descriptions of new genera and species. With the intent of supporting this needed expansion of information, we propose the species S. lanaiensis.
Latin diagnosis
Sympodiomycopsis lanaiensis Mahdi, Statzell-Tallman, Fell, Brown & Donachie sp. nov.
Ustilaginomycotina, Microstromatales
Reproductio per gemmationem, Cellulae globosae aut ovoideae (1–9 mm × 1–4 mm). Cultura glabra, butyrosa, lucida, cremea aut rosea. Sexualis coniunctio non manifesta. Ballistoconidia non formantur. Mycelium formatur. Glucosum, l-sorbosum, maltosum, raffinosum, melibiosum, α-methyl-d-glucosidum, amylum solubilis, d-mannitolum, d-glucitolum, sodii succinatum nitratumque assimilantur. Lactosum, inulinum, d-l-acidum lacticum exigue assimilantur. Fermentatio nulla. Maxima temperatura crescentiae: 30 °C.
Typus LM418T isolatus ex insula Lanai (Hawai‘i) conser-vatur sub numero DSM 18755 in collectione microorganis-morum alemanna (German Collection of Microorganisms and Cell Cultures), sub numero MYA-4092 in collectione zymotica americana (American Type Culture Collection).
Standard description
Sympodiomycopsis lanaiensis Mahdi, Statzell-Tallman, Fell, Brown & Donachie sp. nov. S. lanaiensis sp. nov. (la.nai.ensis’ L. adj., refers to the geographical origin of the species: the island of Lana‘i, Hawai‘i).
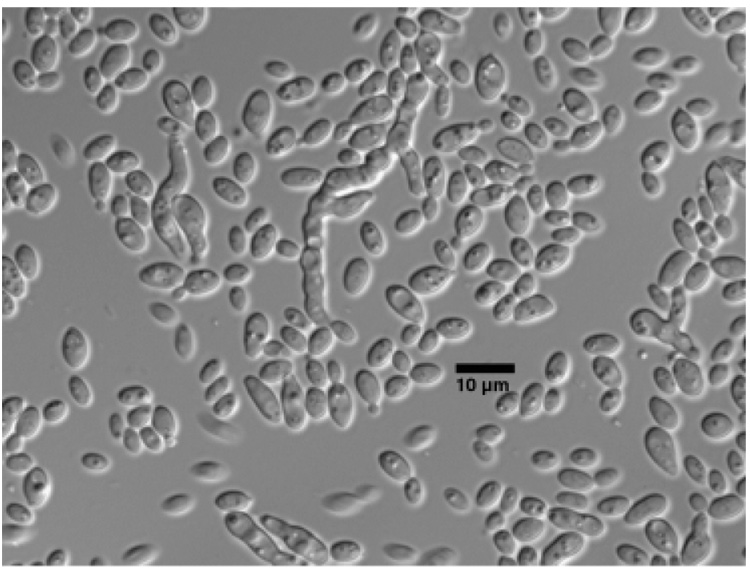
Cells in liquid malt extract (ME) were spheroid to ovoid and ranged from 1.3–9.4 µm long to 1.3–4 µm wide and occurred singly or in pairs. Reproduction was by polar budding (Fig. 3). A sediment was present, but neither a ring nor pellicle were present.
Fig. 3.
Bipolar budding by LM418T in ME broth after 3 days at 25 °C. Scale bar = 10 µm.
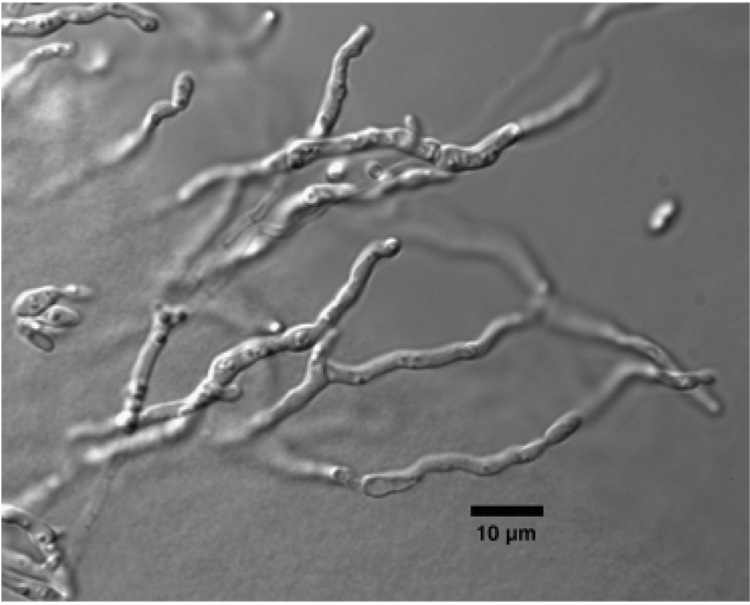
Colonies on MEA [with 2% (w/v) NaCl] at 22 °C were smooth, opaque white to rosy buff (Rayner, 1970), and convex with entire margins. Colony texture was creamy with a sticky consistency. Based on comparative observations, colonies on MEA containing 2% (w/v) NaCl were larger than those grown on MEA without NaCl. Ballistoconidia were not observed on inverted ME plates. Hyphal strands were present with conidial formation at the internodes (Fig. 4). Sexual structures were also not observed after 30 days on cornmeal Dalmau plates. Extensive hyphae were present on PDA.
Fig. 4.
Pseudohyphal formation by LM418T on MEA (with 2% NaCl) after 14 days at 25 °C. Scale bar = 10 µm.
Assimilation (liquid medium): raffinose, melibiose, methyl-α-d-glucoside, soluble starch, l-sorbose, l-mannitol, l-glucitol, succinate, citrate, and nitrate are assimilated; glucose, maltose, glycerol and dl-lactate are assimilated weakly. Inulin, sucrose, galactose, lactose, trehalose, melezitose, cellobiose, salicin, l-rhamnose, xylose, l-arabinose, d-arabinose, d-ribose, methanol, ethanol, erythritol, ribitol, galactitol, inositol, d-glucosamine, K-d-saccharate, creatine, creatinine, 2-keto-d-gluconate, 5-keto-d-gluconate and d-glucuronate are not assimilated. Starch is not produced. Gelatin agar hydrolysis is negative. Growth on both 50% glucose agar and 10% NaCl/5% glucose agar is weak. Vitamins are not required for growth. Urease is present. The diazonium blue B reaction is positive.
Maximum temperature for growth is 30 °C.
Type strain: LM418 (ATCC = MYA-4092, DSMZ = DSM 18755, NRRL Y-48466, CBS 10858) isolated from driftwood in the intertidal zone of ‘Shipwreck Beach’ on the island of Lana‘i, Hawai‘i (20°55′15″N, 156°54″30″W) November 2005.
Sequences submitted to GenBank were assigned the accession nos D1/D2 LSU rRNA = DQ990016, ITS rRNA = DQ990017 and partial 18S rRNA gene = DQ990018.
Acknowledgements
This research was funded by awards to S.P.D. under a Pilot Project program from the University of Hawai‘i Pacific Research Center for Marine Biomedicine, part of the Centers for Oceans and Human Health (COHH) program of the National Institute of Environmental Health Sciences (P50ES012740), National Institutes of Health, and the National Science Foundation (OCE04-32479), PIs Drs Edward Laws and Richard Yanagihara. Funding at Rosenstiel School of Marine and Atmospheric Science was provided in part by NSF & NIEHS Ocean and Human Health Center Program (NSF 0432368 and NIEHS P50 ES12736) and NSF DEB 0206521. Travel costs were supported in part by an American Philosophical Society ‘Lewis and Clark Exploration Award’ to L.E.M. M.V.B. acknowledges support from the NASA Astrobiology Institute under Cooperative Agreement NNA04CC08A at the Institute for Astronomy (University of Hawai‘i at Mānoa).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadon D. Ecology and the evolution of some Hawaiian birds. Evolution. 1947;1:63–68. [Google Scholar]
- Barr ME, Hodges CS., Jr Hawaiian forest fungi. VIII. New species in Gnomoniella and Stigmochora. Mycologia. 1987;79:782–786. [Google Scholar]
- Carr GDR, Robichaux RH, Witter MS, Kyhos DW. Adaptive radiation of the Hawaiian silversword alliance (Compositae–Madiinae): a comparison with Hawaiian picture-winged Drosophila. In: Giddings LV, Kaneshiro KY, Anderson WW, editors. Genetics, Speciation and the Founder Principle. New York: Oxford University Press; 1989. pp. 79–97. [Google Scholar]
- de Beer ZW, Begerow D, Bauer R, Pegg GS, Crous PW, Wingfield MJ. Phylogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Stud Mycol. 2006;55:289–298. doi: 10.3114/sim.55.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie SP, Zdanowski MK. Potential digestive function of bacteria in krill, Euphausia superba stomach. Aquat Microb Ecol. 1998;14:129–136. [Google Scholar]
- Donachie SP, Hou S, Gregory TS, Malahoff A, Alam M. Idiomarina loihiensis, sp. nov., a new halophilic γ-Proteobacterium isolated from the Lō‘ihi submarine volcano, Hawai‘i. Int J Syst Evol Microbiol. 2003;53:1873–1879. doi: 10.1099/ijs.0.02701-0. [DOI] [PubMed] [Google Scholar]
- Donachie SP, Bowman JP, Alam M. Psychroflexus tropicus, sp. nov., a novel, obligately halophilic Cytophaga–Flavobacterium–Bacteroides group bacterium isolated from an Hawaiian hypersaline lake. Int J Syst Evol Microbiol. 2004a;54:935–940. doi: 10.1099/ijs.0.02733-0. [DOI] [PubMed] [Google Scholar]
- Donachie SP, Hou S, Lee K-S, et al. The Hawaiian archipelago: a microbial diversity hotspot. Microb Ecol. 2004b;48:509–520. doi: 10.1007/s00248-004-0217-1. [DOI] [PubMed] [Google Scholar]
- Donachie SP, Bowman JP, On SLW, Alam M. Arcobacter halophilus sp. nov., the first obligately halophilic species in the genus Arcobacter. Int J Syst Microbiol. 2005;55:1271–1277. doi: 10.1099/ijs.0.63581-0. [DOI] [PubMed] [Google Scholar]
- Donachie SP, Bowman JP, Alam M. Nesiotobacter exalbescens gen. nov., sp. nov., a moderately thermophilic alpha-Proteobacterium from an Hawaiian hypersaline lake. Int J Syst Evol Microbiol. 2006;56:563–567. doi: 10.1099/ijs.0.63440-0. [DOI] [PubMed] [Google Scholar]
- Dring DM, Meeker J, Goos R. Clathrus oahuensis, a new species from Hawaii. Mycologia. 1971;63:893–897. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JW. Yeasts in oceanic regions. In: Jones EBG, editor. Recent Advances in Aquatic Mycology. London: Elek Science; 1976. pp. 93–124. [Google Scholar]
- Fell JW. Collection and identification of marine yeasts. Collection and identification of marine yeasts. Marine Microbiology. In: Paul JH, editor. Methods in Microbiology. Vol. 30. San Diego, CA: Academic press; 2001. pp. 347–356. [Google Scholar]
- Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large subunit rD1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Seattle, WA: Distributed by the author, Department of Genetics, University of Washington; 1993. [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeyer J. Marine fungi of Hawaii including the new genus Helicascus. Can J Bot. 1969;47:1469–1487. [Google Scholar]
- Kohlmeyer J. Nimbospora octonae, a new marine ascomycete from Hawaii. Can J Bot. 1985;63:1122–1125. [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B. Hawaiian marine fungi, including two new genera of Ascomycotina. Mycol Res. 1989;92:410–421. [Google Scholar]
- Kurtzman CP, Fell JW. The Yeasts: A Taxonomic Study. 4th edn. Amsterdam: Elsevier; 1998. p. 1055. [Google Scholar]
- Liu Z, Jensen PR, Fenical W. A cyclic carbonate and related polyketides from a marine-derived fungus of the genus Phoma. Phytochemistry. 2003;64:571–574. doi: 10.1016/s0031-9422(03)00272-3. [DOI] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nagahama T, Hamamoto M, Nakase T, Takami H, Horikoshi K. Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie Van Leeuwenhoek. 2001;80:101–110. doi: 10.1023/a:1012270503751. [DOI] [PubMed] [Google Scholar]
- Rayner RW. A Mycological Colour Chart. Kew, Surry: Commonwealth Mycological Institute; 1970. p. 34. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. Systematics of basidiomycetous yeasts: a comparison of large sub-unit D1D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Sugiyama J, Suh S-O. Sympodiomycopsis Sugiyama, Tokuoka, Komagata. In: Kurtzman CP, Fell JW, editors. The Yeasts: A Taxonomic Study. 4th edn. Amsterdam, The Netherlands: Elsevier; 1998. pp. 846–847. [Google Scholar]
- Sugiyama J, Tokuoka K, Suh SO, Hirata A, Komagata K. Sympodiomycopsis: a new yeast like anamorph genus with Basidiomycetous nature from orchid nectar. Antonie Van Leeuwenhoek. 1991;59:95–108. doi: 10.1007/BF00445653. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Miura S, Yamashita Y, Ohta T. Aspermytin A: a new neurotrophic polyketide isolated from a marine-derived fungus of the genus Aspergillus. Bioorg Med Chem Lett. 2004;14:417–420. doi: 10.1016/j.bmcl.2003.10.053. [DOI] [PubMed] [Google Scholar]
- Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW, editors. The Yeasts: A Taxonomic Study. 4th edn. Amsterdam, The Netherlands: Elsevier; 1998. pp. 77–100. [Google Scholar]