SUMMARY
We have assembled, annotated, and analyzed a database of over 1700 breakpoints from the most common chromosomal rearrangements in human leukemias and lymphomas. Using this database, we show that although the CpG dinucleotide constitutes only 1% of the human genome, it accounts for 40–70% of breakpoints at proB/pre-B stage translocation regions – specifically, those near the bcl-2, bcl-1, and E2A genes. We do not observe CpG hotspots in rearrangements involving lymphoid-myeloid progenitors, mature B cells, or T cells. The stage-specificity, lineage-specificity, CpG targeting, and unique breakpoint distributions at these cluster regions may be explained by a lesion-specific double-strand breakage mechanism involving the RAG complex acting at AID-deaminated methyl-CpGs.
INTRODUCTION
Chromosomal rearrangements arise when DNA double-strand breaks (DSB) occur at two separate regions of the genome and the resultant DNA ends are aberrantly joined into a new configuration. Pathologic rearrangements are especially frequent in human lymphomas and leukemias, and the most common are the bcl-2 and bcl-1 translocations. However, the molecular mechanisms underlying the DSBs at the bcl-2 and bcl-1 regions are unknown.
Three main types of DSB mechanisms have been identified in human lymphoid malignancies: (1) V(D)J-type, in which the RAG endonuclease creates a site-specific DSB 5′ of a recombination signal sequence (RSS) with a consensus of CACAGTG (12 or 23 spacer) ACAAAAACC in early B or T cells (Bassing et al., 2002; Dudley et al., 2005; Schatz, 2004; Schlissel et al., 2006; Tonegawa, 1983); (2) class-switch recombination (CSR) type, in which activation-induced cytidine deaminase (AICDA or AID) deaminates cytidines on the single-stranded portions of kilobases-long R-loops at immunoglobulin (Ig) switch regions, leading to DSBs in activated B cells (DiNoia and Neuberger, 2007; Ramiro et al., 2004; Unniraman and Schatz, 2006); and (3) random-type breakage, presumed to be due to sequence-independent mechanisms such as reactive oxygen species (ROS) or ionizing radiation which generate DSBs randomly throughout the genome (Greaves and Wiemels, 2003; Lieber et al., 2006).
As we and others have shown, DSBs at the bcl-2 and bcl-1 do not fit into any of these three mechanisms (Jaeger et al., 2000; Marculescu et al., 2002; Raghavan et al., 2004a; Welzel et al., 2001). Rather, breakpoint sequence analysis reveals that DSBs at the bcl-2 and bcl-1 regions, and additionally a third region, E2A, fall into a fourth type of DSB mechanism, CpG-type, marked by a high degree of focusing to the dinucleotide sequence, CpG, and occurrence only in the pro-B/pre-B stage. We propose a novel mechanism of translocations at methyl CpG sequences in early B cells due to the sequential action of AID and the RAG complex.
RESULTS
Overview of bcl-2, bcl-1, and E2A Chromosomal Breakpoint Distributions
Human lymphoid malignancies are predominantly B cell in origin, and many involve chromosomal translocations between one site and the Ig loci, or between two non-Ig loci (Fig. 1). Though the DSBs at the Ig loci are well-characterized, the cause of the DSBs at non-Ig loci, such as bcl-2, bcl-1, and E2A, are poorly understood.
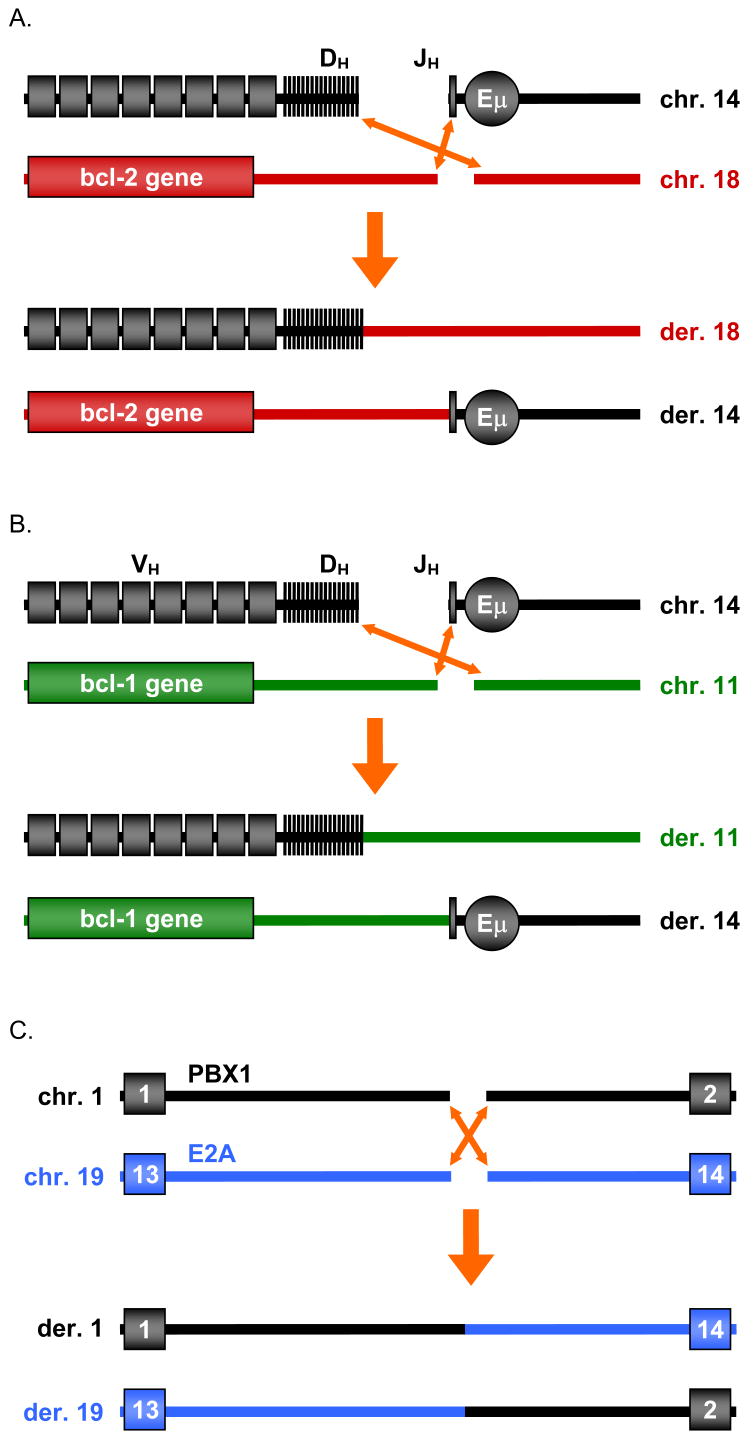
Figure 1. Diagrams of the bcl-2, bcl-1 and E2A Translocations.
(A) In the bcl-2 translocation, a break arises downstream of the coding portion of the bcl-2 gene on chromosome 18. The side of the break closest to the bcl-2 gene (telomeric side) joins to JH segments of the IgH locus on chromosome 14. The centromeric side of the bcl-2 locus usually joins to a DH segment. See Fig. 2 & Suppl. Fig. 1 for description of the MBR, mcr, and icr breakpoint cluster regions downstream of the bcl-2 coding region.
(B) In the bcl-1 translocation, a break at the MTC arises 110 kb centromeric of the bcl-1 gene (cyclin D1). The telomeric portion of the break joins to one of the JH segments at IgH. The centromeric portion usually joins to a DH segment.
(C) In the t(1;19) translocation, the indicated portions are exchanged. In all diagrams, the centromere is located on the same side as the chromosome label.
The first clue for determining how a DSB occurred is its precise location. Free DNA ends are often degraded prior to joining, thus the position of the initial DSB, before degradation, can be difficult to determine. The closest approximation is the breakpoint, or exact position where the rearranged allele ceases to match the normal alleles. Analysis of 695 human translocation breakpoints on bcl-2, bcl-1, and E2A revealed a distinctive pattern of clustering around CpGs (Table 1, Fig. 2, Suppl. Table 1, Suppl. Figs. 1–3). Breakpoints at CpGs account for 43% of events at bcl-2, 35% of events at bcl-1, and 53% of events at E2A (Fig. 2, Suppl. Fig. 3). These frequencies are dramatically higher than the 6% frequency one would expect if breakpoints occurred randomly throughout the breakpoint region (Suppl. Tab. 1, Suppl. Fig. 3) and may be underestimates of the true frequency occurring at CpGs. Due to DNA end degradation prior to joining, breakpoints close to, but not directly at, CpGs may have originated from CpG positions (Suppl. Fig. 3).
Table 1.
Summary of translocation breakpoint features.
| Name | Clinical Entity | Rearrangement | Breakpoints | Stage of Rearrangement | Breakage Types |
|---|---|---|---|---|---|
| bcl-2 | FL and DLBL | t(14;18)(q32;q21) | 551 | pro-B/pre-B | CpG, V(D)J |
| bcl-1 | MCL | t(11;14)(q13;q32) | 114 | pro-B/pre-B | CpG, V(D)J |
| E2A-PBX1 | pre-B/B ALL | t(1;19)(q23;p13) | 30 | pro-B/pre-B | CpG, random |
| TEL-AML1 | pre-B/B ALL | t(12;21)(p12;q22) | 53 | LM HSC | random |
| MLL | primary ALL | t(4;11)(q21;q23)
t(9;11)(p22;q23) other |
254
17 20 |
LM HSC | random |
| MLL | primary AML | t(4;11)(q21;q23)
t(9;11)(p22;q23) other |
11
6 7 |
LM HSC | random |
| MLL | therapy-related ALL and AML | t(4;11)(q21;q23)
t(9;11)(p22;q23) t(11;16)(q23;p13) other |
5
12 11 7 |
LM HSC | random, topoisomerase II-related |
| AML1-ETO | AML | t(8;21)(q22;q22) | 133 | LM HSC | random |
| BCR-ABL | CML | t(9;22)(q34;q11) | 35 | LM HSC | random |
| c-myc | sporadic BL | t(8;14)(q24;q32) | 125 | activated B | CSR, random |
| bcl-6 | mature B lymphomas | t(3;14)(q27;q32) | 38 | activated B | CSR, random |
| SCL-SIL | pre-T/T ALL | del(1)(p32) | 209 | pro-T/pre-T | V(D)J |
| p16 deletion | pre-T/T ALL
T-CLL pre-B/B ALL |
del(9)(p21) | 26
2 13 |
pro-T/pre-T | V(D)J |
| SCL | pre-T/T ALL
pre-B/B ALL |
t(1;14)(p32;q11)
t(1;7)(p32;q34) t(1;3)(p32;q21) t(1;14)(p32;q11) |
14
2 2 1 |
pro-T/pre-T | V(D)J, random |
| LMO2 | pre-T/T ALL | t(11;14)(p13;q11)
t(7;11)(q34;p13) del(11)(p12p13) |
27
2 2 |
pro-T/pre-T | V(D)J, random |
| HOX11 | pre-T/T ALL | t(10;14)(q24;q11) | 13 | pro-T/pre-T | V(D)J |
| TTG1 | pre-T/T ALL | t(11;14)(p15;q11) | 6 | pro-T/pre-T | V(D)J |
| total | 1748 |
Numbers of breakpoints are listed for rearrangements and clinical entities analyzed in the text, followed by estimated stage during which the rearrangement occurs and the apparent types of breakage involved at one or both chromosomes. For example, bcl-2 translocations tend to involve a CpG-type double-strand break (DSB) on chromosome 18 and a V(D)J-type DSB on chromosome 14. “Random”-type breakage does not appear attached to a specific motif, is more regional in nature, and is presumed to be due to ionizing radiation, reactive oxygen species, or some other relatively sequence-independent mechanism. Abbreviations are as follows: follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBL), mantle cell lymphoma (MCL), acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myelogenous leukemia (CML), Burkitt lymphoma (BL), chronic lymphocytic leukemia (CLL), lymphoid-myeloid hematopoietic stem cell (LM HSC), class switch recombination (CSR).
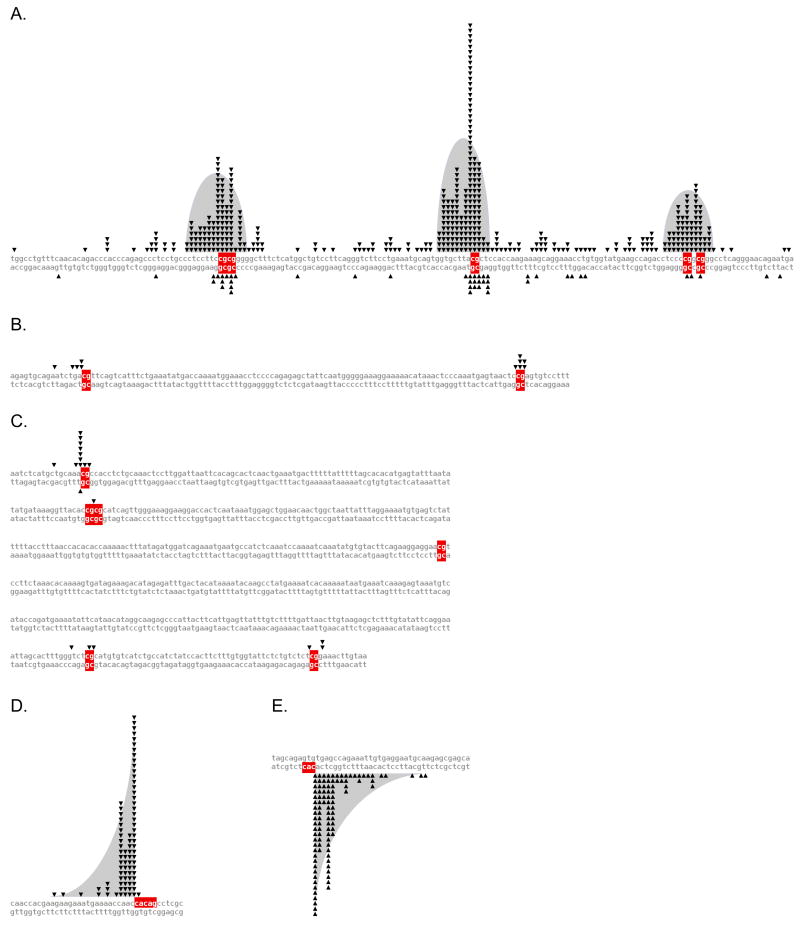
Figure 2. Sequence-Level Breakpoint Diagrams for bcl-2 Clusters and SCL-SIL.
Breakpoint distributions are plotted for the (A) bcl-2 MBR, (B) bcl-2 icr, and (C) bcl-2 mcr. Each breakpoint is represented as a triangle adjoining the breakpoint site, with the top strand sequences running telomeric to centromeric, der(14) breakpoints above, and der(18) breakpoints below. Breakpoint distributions for the (D) main SCL, and (E) SIL breakpoint clusters are shown, with the top strand sequences running telomeric to centromeric, telomeric breakpoints above, and centromeric breakpoints below. Note the different shapes of the distributions in panel A versus panels D and E. Breakpoints in the bcl-2 MBR fall into bell-shaped distributions, with peaks at CpGs and a decline of breakpoints on both sides. Breakpoints at the main SCL and SIL peaks are spiked at CAC, and tail off only in the direction 5′ of the CAC. This implies that different mechanism(s) operate at the MBR versus SCL and SIL.
CpGs in the bcl-2 MBR, icr, and mcr are Hotspots for Translocation Breakpoints
Within the bcl-2 region, breakpoints are highly concentrated at three small clusters. Fifty percent occur in the major breakpoint region (MBR); 13% in the intermediate cluster region (icr) about 18 kb centromeric to the MBR; and 5% in the minor cluster region (mcr) about 11 kb centromeric to the icr (Suppl. Fig. 1A) (Weinberg et al., 2007).
Of the bcl-2 translocations sequenced, 88% occur in the 175 bp MBR, which itself contains three breakpoint peaks (Fig. 2A). Strikingly, CpGs are found at the highest spikes of all three peaks. Of 487 breakpoints in the MBR, 208 (43%) occur precisely at one of the 5 CpG sites in the region (p<10−95). The average MBR breakpoint is 4.4 bp from the nearest CpG, compared to 11.2 bp if the breakpoints were randomly distributed (p<10−41) (Suppl. Table 1).
At the 105 bp icr, eight of the 11 sequenced breakpoints occur at one of the two CpGs (p<10−7), and the remainder are less than 6 bp away from a CpG (Fig. 2B). In contrast, none are observed in the 75 bp region between the two CpGs. Overall, the average distance of these 11 breakpoints from a CpG site is 0.8 bp, versus 22 bp if they were randomly distributed (p<10−6) (Suppl. Table 1).
The 561 bp mcr contains six CpGs, spaced widely apart (Fig. 2C). Again, breakpoints are located very close to CpGs, while intervening regions are devoid of breakpoints (Fig. 2C). Of the 19 sequenced breakpoints, 14 are located directly at CpGs (p<10−17). The average distance between a breakpoint and a CpG site is 0.6 bp, in contrast to 40 bp if these breakpoints were randomly distributed (p<10−12) (Suppl. Table 1).
In addition to the breakpoints located in the three cluster regions described above, 27 sequenced bcl-2 breakpoints are located between these three cluster regions. Of these, 13 (48%) are located less than 10 bp away from a CpG, including 5 (19%) directly at CpGs (p=0.002) (Suppl. Table 1).
These findings strongly suggest that CpG is a DSB hotspot in the MBR, icr, and mcr clusters of bcl-2. Statistical analysis using the binomial distribution, Student’s t-test, and Mann-Whitney U-test indicates that the proximity of these breakpoints to CpGs is highly significant (Suppl. Table 1).
CpGs in the bcl-1 MTC are Hotspots for Translocation Breakpoints
In mantle cell lymphomas, 30% of translocations occur within the 150 bp major translocation cluster (MTC) (Bertoni et al., 2004) which contains 7 CpGs (Suppl. Figs. 1B and 2). The MTC contains 104 of the 114 bcl-1 translocation breakpoints sequenced, 38 (37%) of which occur at CpGs (p<10−8). MTC breakpoints average 2.5 bp from CpGs, compared to 7.8 bp if breakpoints were distributed randomly over the 150 bp region (p<10−11). Notably, the ten breakpoints outside the MTC average 8.8 bp from CpGs, in contrast to 45.3 bp if randomly distributed (p=0.0009) (Suppl. Table 1). Hence, CpGs appear to be DSB hotspots in the MTC as well.
CpGs in the E2A Cluster are Hotspots for Translocation Breakpoints
Most of the breakpoints at E2A occur within a 23 bp zone containing two CpGs (Wiemels et al., 2002) (Suppl. Fig. 1). Fifteen of the 24 sequenced breakpoints occur at CpGs (p=0.0001), averaging 1.1 bp to the closest CpG, versus 3.3 bp if they were randomly distributed in the 23 bp region (p=0.0005). Interestingly, the corresponding breakpoints on PBX1 do not have a tendency to occur at or close to CpGs (p>0.9) and have only a slight tendency towards CAC (p=0.05) (the minimal V(D)J recombination motif), though not directly at CAC (p>0.1) (Suppl. Table 1). Therefore, CpGs appear to be DSB hotspots in the E2A cluster but not in PBX1.
CpG is the Most Significant Dinucleotide Motif at bcl-2, bcl-1, and E2A, and is More Significant than CAC
To confirm our observation, we examined breakpoint proximity to all the other dinucleotide motifs, to A, C, G, T, W, and S, and to ▾CAC/GTG▾ (Suppl. Table 1 and Supplementary text). While certain motifs have lower average distances than CpG (e.g., the mononucleotide motif S (C or G, of which CpG is a subset)), they are not statistically significant. Certain motifs score significantly because they happen to occur next to CpGs but not nearly to the same level as CpG, and these never extend to other translocations or clusters. Among the bcl-2 MBR, bcl-2 icr, bcl-2 mcr, bcl-1 MTC, and E2A breakpoint clusters, and to a lesser degree the unclustered breakpoints (the ones at the bcl-2, bcl-1 or E2A gene loci, but not within the preferred breakpoint zones), CpG is consistently the most significant motif in three different statistical tests.
Initial Chromosomal Breakage Likely Occurs Directly at or Near CpGs
Examination of both derivatives of a reciprocal translocation helps localize, more precisely, the position of the originating DSB (Suppl. Fig. 4A-B). This is because the two breakpoints define a window within which initial breakage likely occurred. This information is only available for a subset of the translocations analyzed above: 46 cases from bcl-2, 23 from bcl-1, and 6 from E2A (Suppl. Fig. 2). Of these, 32 (70%) from bcl-2, 15 (65%) from bcl-1, and 4 (67%) from E2A have one or both breakpoints at CpGs. Significantly, 1 case from bcl-1 and 9 cases from bcl-2, including one case from the bcl-2 mcr, exhibit a breakage window of zero. This strongly suggests that the precise position of the initial DSB was at CpG in these latter 10 cases.
Staging of Translocations from Lymphomas and Leukemias
In order to elucidate the mechanism behind CpG-type translocations, we sought to determine the developmental stage(s) in which they occur. An overview of the major recurrent rearrangements in myeloid and lymphoid tumors is provided in the supplemental text, and the results of our and others’ analyses are shown in Fig. 4.
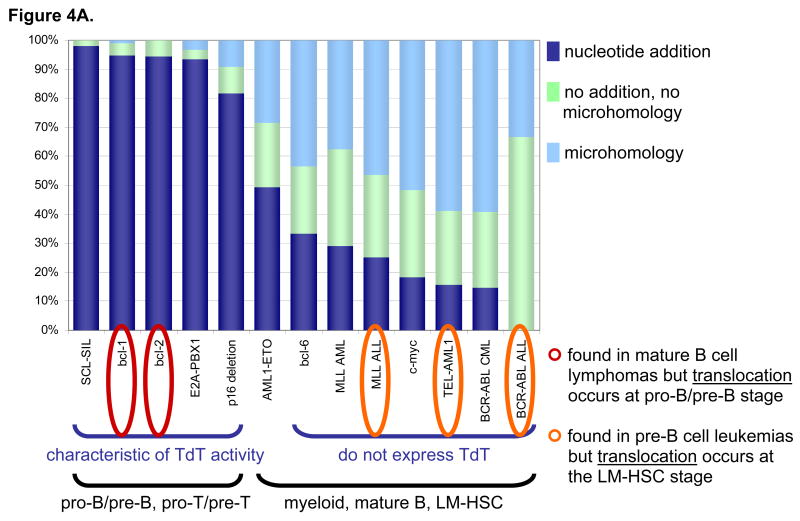
Figure 4. Junctional Additions Help Reveal the Stage of Rearrangement.
(A) Stacked proportions of junctions with nucleotide additions, microhomology, or neither are plotted in dark blue, light blue, and light green, respectively, for various translocations. LM-HSC is lymphoid-myeloid hematopoietic stem cell. Junctions from pro-B/pre-B and pro-T/pre-T rearrangements (i.e. bcl-1, bcl-2, E2A-PBX1, SCL-SIL, and p16 deletions) have a high frequency of nucleotide additions due to strong expression of TdT. In contrast, junctions from rearrangements in other cell stages or types tend to use microhomology because the cells in which they occur do not express TdT. Interestingly, some AMLs with AML1-ETO translocations aberrantly express low levels of TdT, which may account for their intermediate rate of nucleotide additions. This is significant because MLL, TEL-AML1, and BCR-ABL translocations are seen in proB/pre-B stage ALLs but actually occur at the LM-HSC stage. bcl-1 and bcl-2 translocations are found in mature B lymphomas but actually occur at the pro-B/pre-B stage. E2A translocations are found in pro-B/pre-B stage ALLs and occur at that stage. These conclusions are diagrammed in panels B and C.
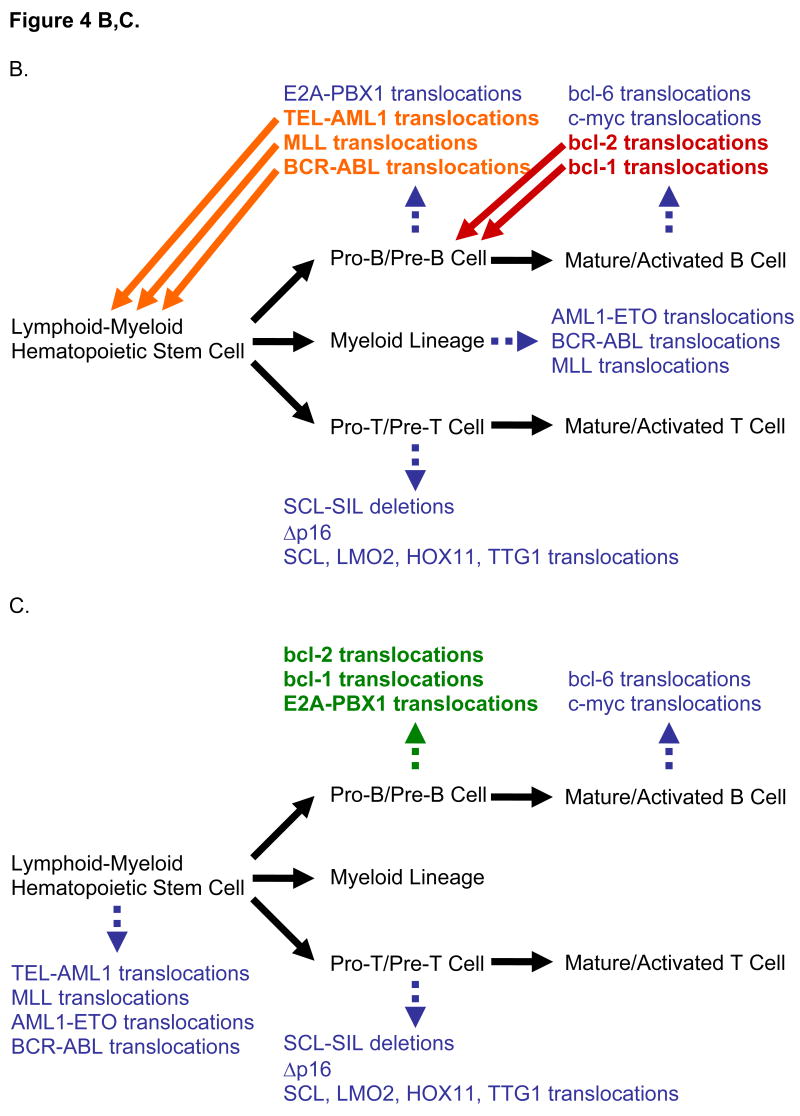
(B) When rearrangements are organized by the stage of the neoplasm in which they are found (i.e., the tumor’s cell of origin), CpG-type hotspots do not appear to follow any pattern. Stages of differentiation are written in black type while rearrangements are written in colored type. (C) Organizing by the stage of development during which the rearrangement occurs reveals that CpG-type hotspots, which occur in the translocations in green type, only occur in the pro-B/pre-B stage.
Staging translocations is not obvious because tumors can be at one stage of development but contain translocations from prior stages (Fig. 4B–C). For instance, bcl-2, bcl-1, c-myc, and bcl-6 translocations are all found in mature B-cell lymphomas. However, we know that bcl-2 and bcl-1 translocations occur at the preceding pro-B stage because they join to IgH RSSs, characteristic of V(D)J recombination which occurs during the pro-B stage. On the other hand, c-myc and bcl-6 translocations occur at the mature/activated B-cell stage, as they involve the IgH switch regions, characteristic of CSR, which occurs during the activated B-cell stage. Similarly, LMO2, HOX11, TTG1, and SCL translocate with TCR RSSs during the pro-T or pre-T stage. But for translocations where neither of the DSBs can be assigned to a stage of development, additional information is required.
Junctional additions at translocation junctions provide such a clue. The presence of apparently random nucleotides between the two breakpoints at a translocation junction is characteristic for the activity of terminal deoxynucleotidyl transferase (TdT), which is normally expressed only in pro-B/pre-B or pro-T/pre-T cells, and aberrantly in some AMLs. As expected, bcl-2, bcl-1, LMO2, HOX11, TTG1, and SCL translocations contain high proportions of junctional additions, while bcl-6 and c-myc translocations contain relatively low proportions (Fig. 4A).
The high proportion of junctional additions in E2A-PBX1 translocations indicates that they occur at the pro-B/pre-B stage, as has been described previously (Wiemels et al., 2002). Additionally, TEL-AML1 and MLL translocations likely occur before the pro-B stage, prior to TdT and RAG expression, as previously inferred biologically (Castor et al., 2005; Hong et al., 2008; Hotfilder et al., 2005; Jansen et al., 2007; Pine et al., 2003) (Fig. 4). BCR-ABL translocations in primary ALLs (not ALLs resulting from CML blast crisis) have been proposed to originate in B-cell progenitors, as opposed to the lymphoid-myeloid hematopoietic stem cell (LM-HSC) origin of BCR-ABL translocations of CMLs (Castor et al., 2005). Nevertheless, BCR-ABL translocations of primary ALLs appear to occur prior to expression of TdT and RAGs, as none of the nine junctions obtained contained nucleotide additions. Based on these analyses, we stage the lymphoid and myeloid translocations as in Fig. 4C.
CpG Hotspots are Specific to Pro-B/Pre-B Stage Translocations
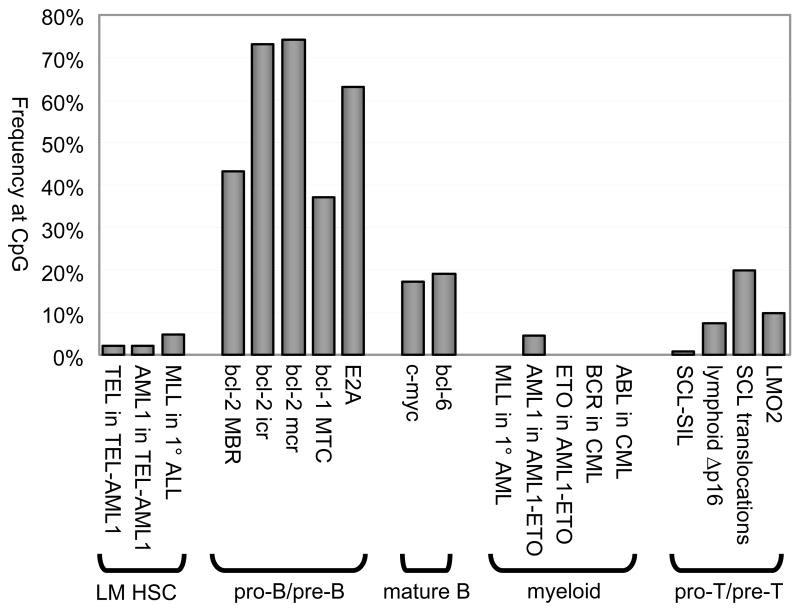
While CpG-type DSBs are prominent in the pro-B/pre-B stage translocations (i.e. bcl-2, bcl-1, and E2A), no significant CpG proximity is found in rearrangements at the LM-HSC stage. In TEL-AML1 translocations, there is no significant CpG proximity among 53 breakpoints spread across 13 kb of TEL or their partners scattered over 165 kb of AML1. The same is true for breakpoints involving the 8.6 kb MLL region in 291 cases of primary ALL, 24 cases of primary AML, and 35 leukemias secondary to topoisomerase II-inhibitor treatment. AML1-ETO translocations show no significant CpG proximity among 67 breakpoints strewn over 24 kb of AML1 or 66 breakpoints dispersed across 57 kb of ETO. Philadelphia chromosome t(9;22)(q34;q11) translocations from CMLs are no different, at either 35 breakpoints found in the 2.8 kb major breakpoint cluster region (M-BCR), or the 25 locatable breakpoints stretched over 125 kb of ABL.
No significant CpG proximity is found in mature/activated B-cell translocations. Neither 125 breakpoints on c-myc, nor 37 breakpoints on bcl-6 spanning 2 kb are significantly close to CpG motifs. Interestingly, c-myc and bcl-6 have higher CpG densities than the bcl-2 MBR and bcl-1 MTC, yet only 17% of breakpoints occur at CpGs in c-myc and only 19% in bcl-6, compared to 43% for the MBR and 37% for the MTC.
T-cell and some pre-B cell rearrangements tend to use a V(D)J-type rather than a CpG-type mechanism, occurring very nonrandomly next to CACs (the minimal heptamer sequence of the RSS) rather than CpGs (Suppl. Table 1, Fig. 3, Suppl. Fig. 3). 208 of 209 SCL-SIL breakpoints spanning 89 kb, 35 of 41 lymphoid Δp16 breakpoints spanning 231 kb, all 13 HOX11 breakpoints, and all 6 TTG-1 breakpoints are compatible with a typical V(D)J-type breakage mechanism. Δp16s from B-ALLs behave similarly (Kitagawa et al., 2002). CpGs are often found in the vicinity of the applicable CACs, but are not used as the site of the breakage.
The 19 SCL breakpoints from SCL-TCR translocations cannot be confidently assigned to a single breakage type, and could be a mixture which includes random-type breakage. Statistical analysis is ambiguous, being significant for both CpG and CAC. CAC is more dominant in 31 LMO2 breakpoints, of which 21 are compatible with V(D)J recombination; however, the remainder could be other types (CpG plus random breaks).
In summary, CpG-type hotspots do not appear to occur outside the pro-B/pre-B stage (Suppl. Table 1, Fig. 3, & Suppl. Fig. 3).
The Widths and Shapes of CpG-type Breakpoint Hotspots Implicate a Novel, Structure-Specific Breakage Mechanism
CpG-type breakpoint clusters differ in width and shape from V(D)J-type, CSR-type, and random-type DSBs. In rearrangements such as BCR-ABL and TEL-AML1, breakpoints extend over tens or hundreds of kilobases and different leukemias rarely share the same breakpoint. Presumably, the breakage mechanism is sequence-independent and, thus, any clustering of breakpoints is due to random chance, selective growth advantage, and/or chromatin accessibility (Greaves and Wiemels, 2003). Such broad clustering effects, however, do not explain how V(D)J-type and CpG-type rearrangements can occur over equally large or larger regions but strongly prefer small windows and even specific nucleotide positions.
The distribution of V(D)J-type and CpG-type breakpoint clusters are also very different from one another, suggesting that they operate by very different mechanisms. V(D)J-type clusters have a “spike-and-single-sided tail” distribution, because a site-specific break first occurs 5′ to the CAC of the RSS, followed by limited recessing only into the pseudo-coding end side of the break (Fig. 2D and E, Suppl. Fig. 5). However, CpG-type breakpoint hotspots are more bell-shaped, with breakpoints on either side of the highest CpG spike; therefore, initial breakage likely occurs around CpG sites with less precision than the V(D)J-type (Fig. 2A, Suppl. Fig. 5). Heterogeneity in nick sites is not typical of sequence-specific processes such as V(D)J recombination, or sequence-specific nucleases such as restriction enzymes; it is, however, consistent with structure-specific nucleases.
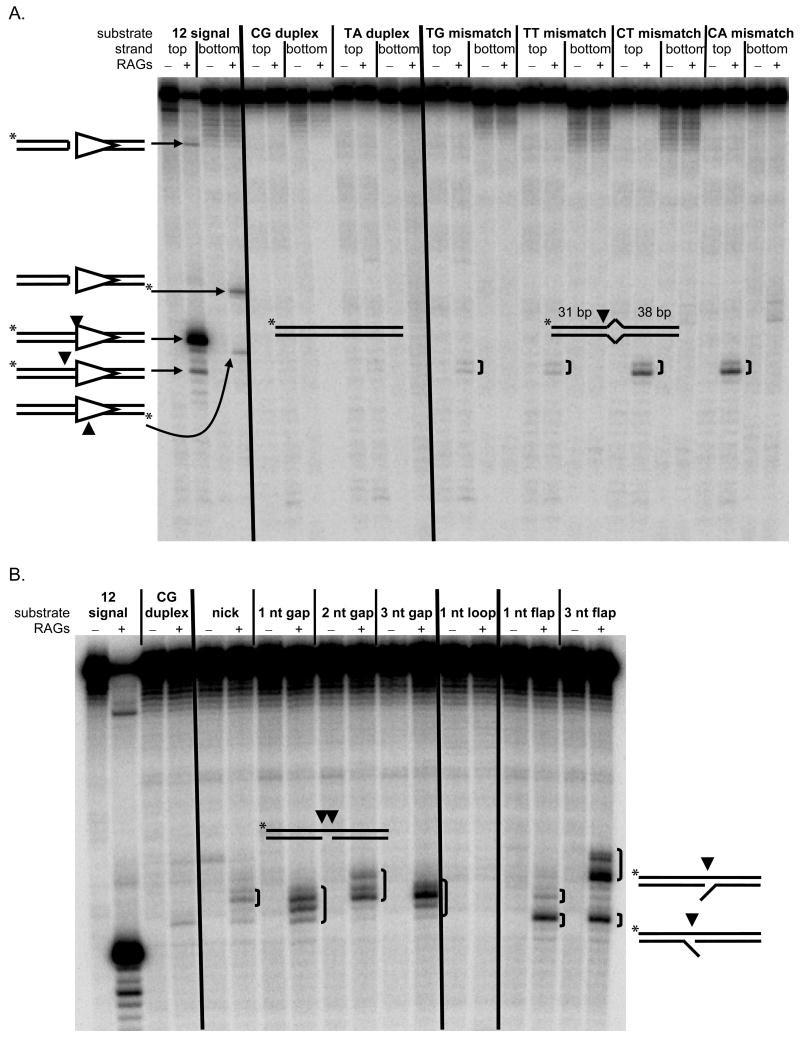
The RAG Endonuclease is Capable of Nicking One-Base Mismatches and Processing Nicks, Gaps, and Flaps into Double-Strand Breaks
In addition to cutting sequence-specifically at an RSS, the RAG complex is also a structure-specific endonuclease (Raghavan et al., 2007; Raghavan et al., 2005a; Santagata et al., 1999). Moreover, RAGs are expressed during the pro-B/pre-B stage when CpG-type DSBs occur, and not expressed during the LM-HSC and mature/activated B-cell stages when CpG-type DSBs do not occur. Thus, the RAG complex is a natural candidate for involvement in CpG-type DSBs. We sought to determine the flexibility of the structure-specific nuclease activity of the RAG complex as it could relate to CpGs.
In accord with its structure-specific endonuclease activity, we have previously shown that the RAG complex can nick large structural deviations such as three to ten-base bubbles and heteroduplexes, but not one-base bubbles (Raghavan et al., 2007; Raghavan et al., 2005a). However, after lengthening the oligonucleotide substrates, we find nicking can occur at one-base bubbles (mismatches) and nicking efficiency appears to correlate with structural instability (Fig. 5A, Suppl. Fig. 6) (Peyret et al., 1999). The most unstable mismatches, C:T and C:A, are nicked at about 6% the efficiency of a very strong RSS. The more stable mismatches T:T and T:G are nicked about six-fold less efficiently, while control C:G and T:A duplexes were not detectably nicked. Although nicking of the T:G mismatch is less efficient than that at C:T and C:A, it is still quite substantial and greater than at certain known sites of V(D)J recombination such as SCL and SIL (Zhang and Swanson, 2008).
Figure 5. The Structure-specific Endonuclease Activity of the RAG Complex at Mismatches.
(A) The structure-specific endonuclease activity of the RAG complex at one-base mismatches is shown. RAGs nick one-base bubbles having arms of 31 bp and 38 bp and lacking CAC at a low level. Fully duplex DNA is not visibly nicked, and a good 12 signal is nicked efficiently. Lanes without RAGs were incubated with HMG1 alone and do not show nicking. The diagrammed structure on the left bottom margin of the gel depicts (expected) RAG nicking on the wrong side of the heptamer adjacent to the spacer. (B) The structure-specific endonuclease activity of the RAG complex is shown at nicks, gaps, and flaps, which are cut at a similar level or higher (relative to the mismatches in panel A) and at heterogeneous positions around the lesion. The 12 signal and duplex DNA controls are similar to those in (A). The diagrammed structures on the right margin depict the two alternative flap configurations that result after annealing the flap oligonucleotides.
The RAG complex also cuts on the anti-parallel strand across from nicks, gaps, and flaps at a rate similar to or higher than that for unstable mismatches in vitro (Fig. 5B). Such activity results in DSBs. Importantly, the nick, gap, and flap substrates display a heterogeneous nicking pattern around the lesion, similar to the pattern of breakpoints seen around CpG at the bcl-2 MBR.
Mechanistically, the RAG complex is capable of generating DSBs in a manner consistent with CpG hotspots.
DISCUSSION AND THEORY
Mechanistic and Developmental Model for Classifying Chromosomal Rearrangements and the Role of the CpG Motif
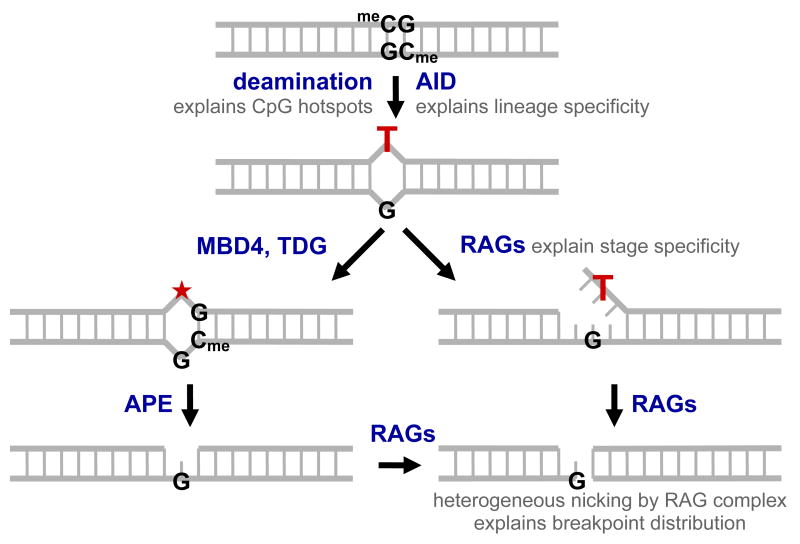
In vertebrates, CG is a very special dinucleotide due to methylation of the C5 position of cytosines in the CG context. Spontaneous deamination of 5-methylcytosine results in thymine, and thus a T:G mismatch. Unmethylated cytosines also deaminate, but result in U:G mismatches (Suppl. Fig. 7). Normally, both types of lesions are corrected by base-excision repair (BER). A glycosylase first removes the damaged base, creating an abasic site. APE1 or spontaneous β-elimination then creates a single-strand gap, which is filled-in by polymerase β, and the resulting nick is sealed by ligase III:XRCC1 (Friedberg et al., 2006; Lindahl et al., 1997; Nash et al., 1997). But when DNA replication occurs before BER can be completed, a polymerase places adenine across from the deaminated base, resulting in a stable point mutation.
T:G mismatches, however, are thought to be more persistent than U:G mismatches. T:G mismatch processing glycosylases, thymine DNA glycosylase (TDG) and methyl-CpG binding domain protein 4 (MBD4), are >2500-fold less efficient in vitro than uracil DNA glycosylase (UDG), which processes U:G (Schmutte et al., 1995; Walsh and Xu, 2006). As a result, CpG to TpG transitions account for a disproportionate number of mutations in p53, BRCA1, BRCA2, p16, and Rb across a wide range of neoplasms (Pfeifer, 2006). Moreover, over evolutionary time, CG has become statistically underrepresented in vertebrate genomes, but is preserved in small and functionally important regions around promoters termed CpG islands (Antequera et al., 1990; Bestor, 2003; Bird, 1992; Takai and Jones, 2003; Yang et al., 1996).
Persistent T:G mismatches, per se, do not lead to DSBs. However, as demonstrated above, the RAG endonuclease could act at the T:G mismatch and/or the vulnerable nick and gap intermediates during the BER process. Direct nicking of the T:G mismatch would result in flap-mismatch lesions, which unlike normal V(D)J-type nicks at CACs, would not be religated readily by ligase I due to the mismatch at the point of ligation (Tomkinson et al., 2006). Alternatively, a one-base gap could first be generated from normal processing by TDG or MBD4 and APE. A subsequent RAG nick across from such a nick or gap would then result in a DSB. We observe that these nick sites are heterogeneous rather than site-specific in vitro, consistent with the heterogeneity in patient breakpoints at CpG hotspots. Involvement of the RAG complex would therefore explain two key aspects of CpG hotspots: (1) why CpG-type rearrangements only occur at the pro-B/pre-B stage during which the RAG complex is expressed and, (2) why the distribution of CpG-type hotspots is heterogeneous around CpGs (Figs. 3, 6 & Suppl. Figs. 5).
Figure 3. Frequencies of Breakpoints at CpG in Various Chromosomal Rearrangements.
Percentages of breakpoints at CpGs are plotted for various chromosomal rearrangements, organized by cell lineage and stage of development when the rearrangement occurs. Frequencies for pro-B/pre-B rearrangements far exceed those from other lineages and stages. Frequencies at various distance intervals from CpG and CAC, along with comparison to random breakpoint distributions, are shown in the supplementary materials (Suppl. Fig. 3). Statistical analysis of this phenomenon is described in the text and supplementary text.
Figure 6. Theoretical Mechanism for CpG-type Double-strand Breakage.
Deamination at a methylcytosine within a CpG creates a T:G mismatch which persists due to catalytic inefficiency of methyl-CpG binding domain protein 4 (MBD4) and thymine DNA glycosylase (TDG) in cleaving the thymine glycosidic bond, leaving an abasic site (★). Single-strand breaks are generated either by the normal base excision repair pathway of glycosylase and AP endonuclease (APE) activity, or by RAG nicking the mismatch directly, resulting in a poorly-ligatable flap. Heterogeneous RAG nicking of the remaining strand creates a double-strand break close to the original site of the T:G mismatch. Nicking by the RAG complex at one-base mismatches and across from nicks, gaps, and flaps is experimentally shown in Fig. 5. Nucleolytic resection of the ends before aberrant joining results in a breakpoint distribution similar to that seen in Fig. 2. We observe the following features at CpG-type translocation hotspots: (1) an extremely high degree of focusing to CpGs, (2) a breakpoint distribution consistent with a structure-specific endonuclease, (3) specificity to the pro-B/pre-B stage, and (4) specificity to the B-cell lineage. Involvement of the RAG complex explains points 2 and 3, while involvement of AID explains point 4 and is strongly suggestive for point 1.
However, T cells also express RAGs but do not appear to have CpG-type rearrangements. One way to account for this may be the recent finding of activation-induced deaminase (AID) expression in early B cells (Crouch et al., 2007; Feldhahn et al., 2007; Mao et al., 2004), in addition to its established pattern of expression in mature activated B cells (and its known absence from T cells). AID in early B cells is consistent with previous work showing that pre-B cell lines can undergo class switch recombination (Alt et al., 1982; Burrows et al., 1983). AID deaminates cytosines in single-stranded DNA in mature B cells during class switch recombination (at R-loops) and somatic hypermutation (DiNoia and Neuberger, 2007). Lower but still significant rates of deamination exist for genomic regions which are outside of the Ig loci, such as c-myc and bcl-6 (Liu et al., 2008). Importantly, AID can act on methylated cytosines (Bransteitter et al., 2003; Morgan et al., 2004). Therefore, involvement of AID would explain why CpG-type rearrangements only occur in the B-cell lineage, and its cytosine deamination activity is exactly the same as the basis for enrichment of point mutations at CpGs in cancers and CpG suppression.
Based on this evidence, we propose the lesion-specific DSB mechanism diagrammed in Fig. 6. A low level of AID expression in early B cells leads to an increased rate of cytosine deaminations, which are adequately repaired at unmethylated cytosines but persist at methylated cytosines as T:G mismatches. These sites are then converted to DSBs by RAGs, before or during attempted repair by T:G mismatch repair enzymes. The resulting DNA ends join to ends from a V(D)J recombination or other double-strand break event and lead to the chromosomal translocations observed so frequently in human B-cell lymphomas.
Mice as Models for Human Lymphomas
For the most part, mice do not develop lymphomas and leukemias in the same way as humans. In the United States, human lymphomas are overwhelmingly of B-cell origin, tend to occur in older individuals, and most contain clonal translocations (Jaffe, 2001; Wada et al., 1993). By contrast, lymphomas in mice, including p53−/− and p53+/− mice, are mostly of T-cell origin and lack clonal translocations (Bassing et al., 2008; Liao et al., 1998). The one exception is mouse plasmacytomas with t(12;15) c-myc-IgH translocations induced by pristane and IL-6 (DiNoia and Neuberger, 2007; Muller et al., 1995). The equivalent in humans is the t(8;14) described previously, which occurs at the mature/activated B-cell stage and not the pro-B/pre-B stage of interest.
The observed incongruency between mouse and human lymphomagenesis may be explained in several ways. First, point mutations of critical genes are sufficient to drive T-cell lymphomagenesis at early ages in mice, masking slower-developing tumors. Experimental demonstration for this comes from a study where mice constitutively expressing c-myc developed only T-cell lymphomas, while RAG1-knockouts constitutively expressing c-myc developed a range of hematological malignancies, including B-LBLs, pre-T cell lymphomas, myeloid progenitor tumors, and cutaneous macrophage tumors (Smith et al., 2005).
Second, translocations and even oncogene overexpression often have no effect on mouse lymphomagenesis. Mice with genome-wide instability, such as H2AX−/−, develop many different translocations, but do not develop tumors (Bassing et al., 2008). Overexpression of the bcl-2 or bcl-1 oncogenes in mice can cause hyperplasia but does not lead to lymphoma (Lovec et al., 1994; McDonnell et al., 1989). Even asymptomatic humans can harbor B cells with t(14;18) translocations (Liu et al., 1994), suggesting additional mutations may be required to drive these translocation-bearing B cells to malignancy.
Third, few mouse models are checked past one year. It may be of some value to follow mice into older ages in an attempt to detect slower-developing tumors. However, even calorie-restricted mice do not live past three years, which may still be simply too short in comparison to human lymphomas which develop in the 5th or 6th decades of life.
Mice as Models for CpG-Type Translocations
According to the proposed mechanism, AID-overexpressing mice should have increased rates of translocation in both the B and T lineages. Similarly, RAG-overexpressing, MBD4-knockout, UNG-knockout, and MSH2-knockout mice should develop more translocations in germinal center B-cells.
AID-overexpressors developed only T-cell lymphomas with no clonal translocations (Okazaki et al., 2003). As described above, point mutations may be driving lymphomas in these mice and masking latent tumors.
Mouse B-cell follicular lymphomas arose in UNG−/− mice after 12 months, but these were not characterized for translocations (Nilsen et al., 2003). MSH2−/− mice developed B and T lymphoblastic lymphomas, and these were not examined for translocations either (Reitmair et al., 1995). It may be valuable to assess them for translocations, though it would not be surprising if AID-induced p53 mutations or c-myc-IgH translocation events were driving these lymphomas. MBD4 knockout mice are not especially susceptible to tumors (Wong et al., 2002). RAG1/2-overexpressing mice are small and die early (Barreto et al., 2001).
Thus CpG-type translocations, as they occur in humans, are not observed in existing mouse models. Part of the problem may be that most studies focus on lymphomagenesis and, with the exception of c-myc, translocations in mice typically do not cause lymphomas. More sensitive assays, such as high-throughput methods, may be required to detect any increases in low-frequency translocations events.
Implications and Testable Predictions
If the theory is correct, it has some intriguing implications. First, individuals whose pro-B/pre-B cells have higher AID activity would be more susceptible to these rearrangements. Second, individuals with SNPs at CpGs within the MBR and MTC, as well as individuals whose pro-B/pre-B cells are unmethylated at those CpGs, would be less susceptible to these rearrangements than individuals in which those CpGs are methylated. However, it is important to note that removal of any single CpG would result in only a small drop in DNA breakage in the MBR and MTC. The model predicts that individuals with SNPs at specific CpGs would not have chromosomal translocations at those SNP sites. The emerging affordability of high-throughput sequencing may allow one to investigate whether the normal pro-B/pre-B cells in patients with such lymphomas have higher levels of AID activity and/or DNA methylation at the breakpoint cluster regions.
There are a multitude of approaches one may take to test the theory by conventional experimental means. The main problem seems to be the exceedingly low frequency of deamination events and rearrangement events. Without accounting for repair, the frequency of spontaneous hydrolytic deamination of cytidines in duplex DNA is on the order of 10−6 per cytosine over 14 days (Frederico et al., 1990). In mice, the highest reported rate of AID-catalyzed mutation at non-V(D)J loci was about 0.1% per base in mature/activated B-cells (Liu et al., 2008; Mao et al., 2004). However, with high-throughput sequencing, AID-induced increases in C to T mutations at CpG sites may be detectable.
As mentioned above, B cell lymphomas arising in UNG−/− or MSH2−/− mice may be worth analyzing for chromosomal translocations (Nilsen et al., 2003; Reitmair et al., 1995). Using high-throughput sequencing, mutated CpG sites could be analyzed for evidence of double-strand breaks in the form of small deletions and additions, indicative of DSBs that were repaired by nonhomologous DNA end joining.
Basis for Translocation Clustering and the Relationship to Translocation Hotspots
The proposed mechanism also has implications for a long-standing mystery —why the translocations cluster in the first place; that is, why they occur so frequently at CpGs within the MBR and MTC, as opposed to distributing across all the other CpGs in the surrounding tens or hundreds of kilobases. The very same problem is seen for CACs in V(D)J-type rearrangement clusters, where breakpoints occur at only a fraction of the available CACs, and not the ones predicted by experimental systems (Marculescu et al., 2002; Raghavan et al., 2001; Zhang and Swanson, 2008). As far as clustering with CpG-type hotspots, possible explanations can be grouped into those which promote breakage and recombination (such as increased methylation, selective AID or targeting of the RAG complex, slow repair rate, chromosomal positioning, and failure to sequester free ends at certain loci) and those which prevent recombination events from appearing in cancers (such as lack of a growth advantage). As the other possibilities are discussed elsewhere, only the first two will be discussed below.
Though unlikely, the most obvious explanation would be selective hypermethylation within the MBR and MTC. Methylation analysis of the MBR and MTC showed considerable variation within and between FACS-sorted pre-B cells from five healthy individuals of similar age. CpGs outside the clusters were methylated at the same or higher rate (Suppl. Fig. 8). Hence, while there are plenty of methylated CpG targets for deamination-based translocations, selective hypermethylation does not appear to explain targeting to the MBR or MTC – at least to the extent that our samples are similar to those which develop into lymphoma (Fig. 4A).
Hydrolytic deamination of methylcytosine is extremely slow in duplex DNA, but is accelerated 500 to 1000-fold when the base is unpaired or single-stranded (Frederico et al., 1990). Moreover, AID is much more active on unpaired cytosines than on base-paired cytosines (Bransteitter et al., 2003; DiNoia and Neuberger, 2007; Pham et al., 2003; Yu et al., 2004). Therefore, one would expect CpGs in single-stranded regions to be more susceptible to deamination. We have previously shown that the bcl-2 MBR reacts with the single-strand-specific chemical probe bisulfite, suggesting some degree of single-stranded character inherent to the DNA in this region (Raghavan et al., 2004a). The centromeric end of the 150 bp bcl-1 MTC also shows a high degree of bisulfite reactivity (AT & MRL, unpublished). This naturally leads to the hypothesis that AID may target cytosines within these regions due to their increased propensity for single-strandedness.
Single-stranded character may also be caused by slippage at repeat sequences, induced by a passing RNA or DNA polymerase. Such a mechanism has been suggested based on sequence data from a yeast LYS2 reporter system (Kim et al., 2007). AID is more active on small bubble structures (such as 3 bp to 7 bp) than on larger bubble structures, and more active on supercoiled DNA than nicked circular DNA (Larijani and Martin, 2007; Shen and Storb, 2004). Therefore, even slight perturbations in the DNA can significantly enhance AID action. At the bcl-2 mcr, there is an eight base pair direct repeat on each side of one CpG hotspot. A slippage event between the top and bottom strand would place the CpG within a loop, thereby making it vulnerable to hydrolytic deamination or to AID.
In summary, we have analyzed over 1700 human chromosomal rearrangement breakpoints grouped according tissue type, lineage, and developmental stage. Based on this, we have identified a new type of double-strand breakage motif, which appears to shape the breakpoint distributions within the bcl-2 MBR, bcl-2 icr, bcl-2 mcr, bcl-1 MTC, and E2A cluster regions. This CpG-type breakage is prominent in pro-B/pre-B cells and apparently absent or heavily diminished in other cell types. We propose that deaminated methyl-CpGs within cluster regions are intercepted by RAGs before repair can be completed, generating the requisite double-strand breaks in several of the most common translocations in human lymphoma.
EXPERIMENTAL PROCEDURES
Statistical Analysis
Junctions and breakpoints were obtained from literature and GenBank searches and aligned with hg18 from the UCSC genome browser (Kent et al., 2002) using BLAT (Kent, 2002) and verified by eye. A breakpoint “at” a CpG is defined as being either 5′ to the C, between the C and G, or 3′ to the G, i.e. ▾CG, C▾G, or CG▾. For binomial probabilities, the random probability p is calculated as the number of sites at motifs divided by the total number of sites from the most 5′ to the most 3′ breakpoint. “Distance” to a motif is calculated as the number of nucleotides between the breakpoint and the closest motif site. Distance distributions for uniformly or “randomly” distributed breakpoints were generated by traversing the region from the most 5′ to the most 3′ breakpoint, treating each strand independently, and calculating the distance to the closest motif for each site along the way. These distributions were used for a Student’s t-test with a standard Box-Cox log (x+1) transformation to correct for non-normality, and for a Mann-Whitney U-test. t and z-values were converted to p-values in Excel. Details of this analysis are discussed in the supplementary text. The database of annotated breakpoints and junctions will be available online in computer-readable format.
RAG Endonuclease Assays
Substrates were prepared by boiling PAGE-purified, 5′-labeled oligonucleotide with 1.5-fold excess PAGE-purified cold oligonucleotide(s) and slow cooling. Protein purification and assay were as described previously (Raghavan et al., 2007). (See supplement.)
Supplementary Material
Acknowledgments
We thank Dr. Susan Groshen and the USC Norris Cancer Center Biostatistics Core Shared Resources; Drs. Darryl Shibata and Allen Yang for comments on the manuscripts; and members of the Lieber lab.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt FW, Rosenberg N, Casanova RJ, Thomas E, Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. 1982;296:325–331. doi: 10.1038/296325a0. [DOI] [PubMed] [Google Scholar]
- Antequera F, Boyes J, Bird AP. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Barreto V, Marques R, Demengeot J. Early death and severe lymphopenia caused by ubiquitous expression of the Rag1 and Rag2 genes in mice. Eur J Immunol. 2001;31:3763–3772. doi: 10.1002/1521-4141(200112)31:12<3763::aid-immu3763>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Ranganath S, Murphy M, Savic V, Gleason M, Alt FW. Aberrant V(D)J recombination is not required for rapid development of H2ax/p53-deficient thymic lymphomas with clonal translocations. Blood. 2008;111:2163–2169. doi: 10.1182/blood-2007-08-104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Bertoni F, Zucca E, Cotter FE. Molecular basis of mantle cell lymphoma. Br J Haematol. 2004;124:130–140. doi: 10.1046/j.1365-2141.2003.04761.x. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Unanswered questions about the role of promoter methylation in carcinogenesis. Ann N Y Acad Sci. 2003;983:22–27. doi: 10.1111/j.1749-6632.2003.tb05959.x. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PD, Beck-Engeser GB, Wabl MR. Immunoglubulin Heavy-Chain Class Switching in a pre-B Cell Line is Accompanied by DNA Rearrangement. Nature. 1983;306:243–246. doi: 10.1038/306243a0. [DOI] [PubMed] [Google Scholar]
- Castor A, Nilsson L, Astrand-Grundstrom I, Buitenhuis M, Ramirez C, Anderson K, Strombeck B, Garwicz S, Bekassy AN, Schmiegelow K, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNoia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Ann Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Feldhahn N, Henke N, Melchior K, Duy C, Soh BN, Klein F, von Levetzow G, Giebel B, Li A, Hofmann WK, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med. 2007;204:1157–1166. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington D.C: ASM Press; 2006. [Google Scholar]
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, Green J, Colman S, Piacibello W, Buckle V, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- Hotfilder M, Rottgers S, Rosemann A, Schrauder A, Schrappe M, Pieters R, Jurgens H, Harbott J, Vormoor J. Leukemic stem cells in childhood high-risk ALL/t(9;22) and t(4;11) are present in primitive lymphoid-restricted CD34+CD19-cells. Cancer Res. 2005;65:1442–1449. doi: 10.1158/0008-5472.CAN-04-1356. [DOI] [PubMed] [Google Scholar]
- Jaeger U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba A, Mannhalter C, Nadel B. Follicular lymphomas BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- Jaffe ES. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon; Washington, D.C: IARC Press; 2001. [Google Scholar]
- Jansen MW, Corral L, van der Velden VH, Panzer-Grumayer R, Schrappe M, Schrauder A, Marschalek R, Meyer C, den Boer ML, Hop WJ, et al. Immunobiological diversity in infant acute lymphoblastic leukemia is related to the occurrence and type of MLL gene rearrangement. Leukemia. 2007;21:633–641. doi: 10.1038/sj.leu.2404578. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y, Inoue K, Sasaki S, Hayashi Y, Matsuo Y, Lieber MR, Mizoguchi H, Yokota J, Kohno T. Prevalent involvement of illegitimate V(D)J recombination in chromosome 9p21 deletions in lymphoid leukemia. J Biol Chem. 2002;277:46289–46297. doi: 10.1074/jbc.M208353200. [DOI] [PubMed] [Google Scholar]
- Larijani M, Martin A. Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol Cell Biol. 2007;27:8038–8048. doi: 10.1128/MCB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MJ, Zhang XX, Hill R, Gao J, Qumsiyeh MB, Nichols W, Van Dyke T. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol Cell Biol. 1998;18:3495–3501. doi: 10.1128/mcb.18.6.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Karran P, Wood RD. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hernandez A, Shibata D, Cortopassi GA. Bcl2 Translocation Frequency Rises with Age in Humans. Proc Natl Acad Sci. 1994;91:8910–8914. doi: 10.1073/pnas.91.19.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. Embo J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Jiang L, Melo-Jorge M, Puthenveetil M, Zhang X, Carroll MC, Imanishi-Kari T. T cell-independent somatic hypermutation in murine B cells with an immature phenotype. Immunity. 2004;20:133–144. doi: 10.1016/s1074-7613(04)00019-6. [DOI] [PubMed] [Google Scholar]
- Marculescu R, Le T, Simon P, Jaeger U, Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J Exp Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, Korsmeyer SJ. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muller JR, Janz S, Potter M. Differences between Burkitt’s lymphoma and mouse plasmacytoma in the immunoglobulin heavy chain/c-myc recombinations that occur in their chromosomal translocations. Cancer Res. 1995;55:5012–5018. [PubMed] [Google Scholar]
- Nash RA, Caldecott KW, Barnes DE, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Stamp G, Andersen S, Hrivnak G, Krokan HE, Lindahl T, Barnes DE. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalyzed cytosine deamination on single-stranded DNA stimulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Pine SR, Wiemels JL, Jayabose S, Sandoval C. TEL-AML1 fusion precedes differentiation to pre-B cells in childhood acute lymphoblastic leukemia. Leuk Res. 2003;27:155–164. doi: 10.1016/s0145-2126(02)00183-2. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Gu J, Swanson PC, Lieber MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair (Amst) 2007;6:751–759. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Kirsch IR, Lieber MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J Biol Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 Mbr and at other non-B DNA structures in vitro. Mol Cell Biol. 2005a;25:5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the bcl-2 major break point region is cleaved by the RAG complex. Nature. 2004a;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosomal translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Reitmair AH, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker HW, Wakeham A, Liu B, et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- Santagata S, Besmer E, Villa A, Bozzi F, Allingham JS, Sobacchi C, Haniford DB, Vezzoni P, Nussenzweig MC, Pan ZQ, Cortes P. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Kaffer CR, Curry JD. Leukemia and lymphoma: a cost of doing business for adaptive immunity. Genes Dev. 2006;20:1539–1544. doi: 10.1101/gad.1446506. [DOI] [PubMed] [Google Scholar]
- Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U:G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Harris AW, Cory S. T-cell lymphomas mask slower developing B-lymphoid and myeloid tumours in transgenic mice with broad haemopoietic expression of MYC. Oncogene. 2005;24:3544–3553. doi: 10.1038/sj.onc.1208399. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair (Amst) 2006;5:1259–1264. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Wada M, Bartram CR, Nakamura H, Hachiya M, Chen DL, Borenstein J, Miller CW, Ludwig L, Hansen-Hagge TE, Ludwig WD, et al. Analysis of p53 mutations in a large series of lymphoid hematologic malignancies of childhood. Blood. 1993;82:3163–3169. [PubMed] [Google Scholar]
- Walsh CP, Xu GL. Cytosine methylation and DNA repair. Curr Top Microbiol Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- Weinberg OK, Ai WZ, Mariappan MR, Shum C, Levy R, Arber DA. “Minor” BCL2 breakpoints in follicular lymphoma: frequency and correlation with grade and disease presentation in 236 cases. J Mol Diagn. 2007;9:530–537. doi: 10.2353/jmoldx.2007.070038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel NTTL, Marculescu R, Mitterbauer G, Chott A, Pott C, Kneba M, Du MQ, Kusec R, Drach J, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001:1629–1636. [PubMed] [Google Scholar]
- Wiemels JL, Leonard BC, Wang Y, Segal MR, Hunger SP, Smith MT, Crouse V, Ma X, Buffler PA, Pine SR. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2002;99:15101–15106. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci U S A. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Gonzalgo ML, Zingg JM, Millar RP, Buckley JD, Jones PA. The rate of CpG mutation in Alu Repetitve elements within the p53 tumor suppressor gene in the primate germline. J Mol Biol. 1996;258:240–250. doi: 10.1006/jmbi.1996.0246. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- Zhang M, Swanson PC. V(D)J recombinase binding and cleavage of cryptic recombination signal sequences identified from lymphoid malignancies. J Biol Chem. 2008;283:6717–6727. doi: 10.1074/jbc.M710301200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.