Abstract
Summary: bioDBnet is an online web resource that provides interconnected access to many types of biological databases. It has integrated many of the most commonly used biological databases and in its current state has 153 database identifiers (nodes) covering all aspects of biology including genes, proteins, pathways and other biological concepts. bioDBnet offers various ways to work with these databases including conversions, extensive database reports, custom navigation and has various tools to enhance the quality of the results. Importantly, the access to bioDBnet is updated regularly, providing access to the most recent releases of each individual database.
Availability: http://biodbnet.abcc.ncifcrf.gov
Contact: stephensr@mail.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online
1 INTRODUCTION
Deriving maximal biological insights from diverse platforms of high-throughput data frequently requires the data to be converted amongst various database identifiers. There are many online resources which offer cross-references to various external databases but the type and coverage of those varies depending on the resource (http://www.ensembl.org, http://www.genome.jp/kegg, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene, http://www.uniprot.org). Also, the input identifiers for these resources are limited to a narrow subset of potential database types. Therefore, one has to visit many web resources before getting the required conversions as the input data vary widely from one experimental platform to another and would also depend on other factors such as the species.
bioDBnet offers a convenient solution for such database-related queries by integrating many biological databases. With bioDBnet extensive biological reports can be obtained for many types of biological identifiers and data can be converted between and within various biological identifiers. At the same time it does not require learning new technologies or terminologies and also has extensive background information on the technique and the coverage of the databases so as to allow users to customize and use the resource in the most productive way.
2 FUNCTIONALITY
The Advanced Biomedical Computing Center maintains local copies of many widely used biological databases and bioDBnet was created with the intention of integrating all of these databases (http://biodbnet.abcc.ncifcrf.gov/dbInfo/faq.php#net2). It is built by a data warehouse-based integration where the connections are formed by exploiting the existing cross-references in the local copies of various public data sources, mainly Ensembl, UniProt and EntrezGene. The current release of bioDBnet is built by integrating 20 biological databases and recognizes more than 100 different types of database types from the molecular biology database collection (http://www.oxfordjournals.org/nar/database/subcat/3/8). It has 153 database identifiers (nodes) connected by 554 cross-references (edges) (http://biodbnet.abcc.ncifcrf.gov/dbInfo/netGraph.php). It includes gene centric database identifiers like EntrezGene Gene ID, Ensembl Gene ID; protein identifiers like UniProt Accession, Ensembl Protein ID; annotations like GO, InterPro; microarray identifiers from Affymetrix, Agilent; Sequence identifiers from GenBank, RefSeq; and Pathway identifiers—from Biocarta and KEGG.
Various options within bioDBnet offer a variety of functionalities to suit different user needs. All of these tools support batch queries and the results are downloadable as both excel and text files. In addition, the identifiers in the results are linked to external resources wherever applicable.
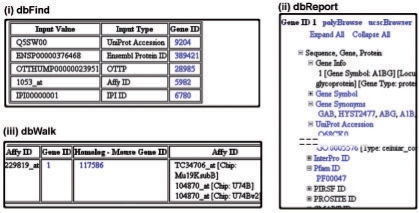
Brief descriptions of the main menu options: ‘db2db’ is a conversion tool that lets users convert from one type of biological database identifier to another. ‘dbFind’ allows users to convert from one identifier to any of the standard identifiers in bioDBnet without specifying the actual type of input. It can be used when the exact type of input is not known or with a mixture of database identifiers (Fig. 1, i). ‘dbReport’ generates an all inclusive report with every possible annotation for a given type of input (Fig. 1, ii). Wherever applicable the reports have links to polyBrowse (http://pbrowse2.abcc.ncifcrf.gov), a gbrowse-based browser (Stein et al., 2002), the UCSC genome browser (http://genome.ucsc.edu, Kent et al., 2002) for visualizing data on the chromosomes and to DAVID (http://david.abcc.ncifcrf.gov, Huang da et al., 2007) for functional annotation clustering. ‘dbWalk’ is a customizable database conversion tool giving the users total control of the type of conversion and the intermediate databases (Fig. 1, iii). This allows a user to incorporate preferences into the path followed, based on the data coverage (http://boidbnet.abcc.ncifcrf.gov/dbInfo/netGraphTbl.php) or the user's confidence in the data quality from a particular database.
Fig. 1.
Partial screen shots of the results from bioDBnet. (i) dbFind to get the types of a mixture of identifiers and converting them to a single type, in this case Gene ID's. (ii) Partial report for Entrezgene Identifier ‘1’ from dbReport. (iii) dbWalk results page displaying the path to get the mouse homologs for human Affymetrix Identifiers.
bioDBnet also offers various supporting tools to enhance connectivity of biological knowledge and annotations. ‘bioText’ can be used to text mine Gene, UniProt or GO (Gene Ontology) annotations. ‘goTree’ displays the GO hierarchy for any GO accession in a top-down manner starting with the input accession to all its parents. Given any type of database identifiers the ‘chrView’ tool tries to find their chromosomal location and displays the results in a movable and zoomable SVG image. This provides for a whole-genome view with an ability to detect clusters. ‘orgTaxon’ provides an easy-to-use search interface to find the taxon ID of any organism.
3 ADVANTAGES
Compared with other integration approaches, bioDBnet is not developed by a federated architecture for integration nor is it a text index-based retrieval system like SRS (Etzold et al., 1996). The data warehouse-based integration of bioDBnet allows for batch queries of database identifiers using SQL and at the same time does not preclude linking over the web once the internal network is constructed.
Unlike other data warehouse-based integrations like Atlas (Shah et al., 2005) and BioWarehouse (Lee et al., 2006), bioDBnet does not have a common integration schema but incorporates the semantics of the data in a separate layer. This approach keeps the database layer independent of the integration layer, which in turn offers greater flexibility and also allows easy updates of the underlying databases. At any given point the databases in bioDBnet are at the most a week apart from the current version of their publicly available database counterpart (http://biodbdev.abcc.ncifcrf.gov/dbInfo/faq.php#data3).
The network-based approach of bioDBnet for linking disparate data sources is similar in part to BIOZON (Birkland and Yona, 2006) and Genmapper (Do and Rahm, 2004), but bioDBnet, compared with the current versions of both these integration tools (http://www.biozon.org/, http://ducati.izbi.unileipzig.de:8080/GenMapper/), has a far wider coverage, is more flexible and has an easy-to-use web interface.
None of the above mentioned integration tools, other than bioDBnet, offer a way to unify multiple types of identifiers. For example, database cross-references from GO to human protein identifiers contain a mixture of RefSeq, Ensembl, UniProt, H-Inv (Human-Invitational Database) and Vega protein identifiers (http://www.geneontology.org/GO.current.annotations.shtml). It would be highly beneficial for converting these into a unified identifier so as to use them in microarray or pathway analysis software. bioDBnet handles such conversions with ease through dbFind (Fig. 1, i). bioDBnet also allows for custom queries, not performed by any other web integration tool, like retrieving homologs, obtained by the integration of the HomoloGene database, for any type of identifiers, (Fig. 1, iii) not just Gene and Protein identifiers. (Refer to Supplementary Material for comparisons between bioDBnet and other database integration tools).
As per our knowledge, compared with any other integration tool, bioDBnet has the most coverage of biological databases (http://biodbnet.abcc.ncifcrf.gov/dbInfo/netNodes.php). This allows for more than 1000 types of conversions of biological annotations and identifiers through db2db and extensive reports for more than 40 different types of identifiers through dbReport.
4 CONCLUSIONS
We think that bioDBnet is going to be very useful for any kind of biological data analysis, as it offers a portal where biological summaries with identifiers for sequence, annotation and feature information can be obtained for multiple, both in terms of number and variety, biological identifiers.
bioDBnet is easily extendable to include additional databases and depending on user interest, many other databases will be added to the network. At this point bioDBnet can handle a few but not all obsolete identifiers. In the next release of bioDBnet we intend to have maximum possible coverage of these identifiers along with literature mining and the ability to extend user data files with additional annotations so as to provide for a complete biological database network.
Supplementary Material
ACKNOWLEDGEMENTS
The authors like to thank all the members in the Bioinformatics Support Group at ABCC especially Gary Smythers, David Liu and Natalia Volfovsky for their help with bioDBnet.
Funding: National Cancer Institute; National Institutes of Health (contract N01-CO-12400).
Conflict of Interest: none declared.
REFERENCES
- Birkland A, Yona G. BIOZON: a hub of heterogenous biological data. Nucleic Acids Res. 2006;34:D235–D242. doi: 10.1093/nar/gkj153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do HH, Rahm E. Proceedings of Advances in Database Technology, Heraklion, Greece. Springer Berlin/Heidelberg; 2004. Flexible integration of Molecular-biological Annotation Data: The GenMapper Approach. [Google Scholar]
- Etzold T, et al. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Huang da W, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, et al. BioWarehouse: a bioinformatics database warehouse toolkit. BMC Bioinformatics. 2006;7:170. doi: 10.1186/1471-2105-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, et al. Atlas - a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.