Abstract
Objectives
To define patterns of aberrant DNA methylation, p53 mutation and Her-2/neu overexpression in tissues from benign (N=29), malignant (N=100), and border line malignant ovaries (N=10), as compared to normal (N=68) ovarian tissues. Further, to explore the relationship between the presence of genetic and epigenetic abnormalities in ovarian cancers, and assess the association between epigenetic changes and clinical stage of malignancy at presentation and response to therapy.
Methods
The methylation status of 23 genes that were previously reported associated with various epithelial malignancies was assessed in normal and abnormal ovarian tissues by methylation specific PCR. The presence of p53 mutation (N=82 cases) and Her-2/neu overexpression (N=51 cases) were assessed by DNA sequencing and immunohistochemistry, respectively.
Results
Methylation of four genes (MINT31, HIC1, RASSF1, and CABIN1) was significantly associated with ovarian cancer but not other ovarian pathology. Her-2/neu overexpression was associated with aberrant methylation of three genes (MINT31, RASSF1, and CDH13), although aberrant methylation was not associated with p53 mutations. Methylation of RASSF1 and HIC1 was more frequent in early compared to late stage ovarian cancer, while methylation of CABIN1 and RASSF1 was associated with response to chemotherapy.
Conclusion
DNA methylation of tumor suppressor genes is a frequent event in ovarian cancer, and in some cases is associated with Her-2/neu overexpression. Methylation of CABIN1 and RASSF1 may have the utility to predict response to therapy.
Keywords: hypermethylation, Her-2/neu overexpression, p53, ovarian cancer
Introduction
Early stage ovarian cancer (OC) is highly curable, with a 5-year survival of 90% for women presenting with disease limited to the ovary [1]. Unfortunately, at present most women have advanced stage disease at the time of initial diagnosis, and despite aggressive treatment, have poor 5-year survival rates (10–20%) [1]. Developing strategies for early diagnosis and improved treatment of OC will likely require gaining insights into the molecular pathogenesis of OC which presently remains largely undefined [1]. In general, progression to cancer is associated with loss of function of genes regulating tumor suppression, DNA repair, apoptosis, and gain in function of oncogenes that facilitate development of malignancy. Complete loss of function of a gene generally requires changes in both copies of the gene, either as a result of the acquisition of heritable changes in DNA sequence (genetic mutations or loss/deletion), and/or through acquisition of “epigenetic” changes (heritable changes in gene expression not related to alterations in the primary nucleotide sequence) such as DNA hypermethylation [2]. Both pathways play a role in most sporadic cancers. Previous studies have examined mutations in a large number of OCs and shown that 5–10% of familial OC contain mutations of BRCA1, BRCA2 and mismatch repair genes (hMSH2 or hMLH1), with up to 50% of sporadic OC containing p53 mutations. Further, 10–20% of sporadic OC have been described as having overexpression of Her-2/neu [3]. Such changes have been shown to play a central role in the pathogenesis of OC [1] and to influence responses to therapy [4]. Fewer data are available concerning epigenetic changes associated with OC. Aberrant DNA methylation of a few genes, which are commonly methylated in many other cancers [2], has been reported to be present in OC cell lines and in a small number of primary OCs examined thus far. However, studies of OC associated changes in methylation have been limited by the examination of only a small number of genes, and by the lack of inclusion of adequate numbers of normal ovarian tissue controls [5–11], making it difficult to interpret the significance of reported findings.
Although it is well established that both genetic and epigenetic alterations play important roles in ovarian cancer tumorigenesis, little is known about whether these two pathways interact and collaborate during tumor development. Several recent studies of colon and non-small-cell lung cancers have reported specific associations between genetic and epigenetic changes [12, 13]. For example, methylation of five genes (CDKN2A, MINT1, MINT2, MINT31 and MLH1) has been associated with mutations of BRAF in colon cancer [14], and methylation of CDKN2A has been associated with K-ras mutations in non-small-cell lung cancer [12]. This has not yet been examined in the case of OC.
To gain further insight into the relationship between genetic and epigenetic changes in sporadic OC cancer and response to therapy, we examined stored tissues samples from women with and without primary OC for the presence of p53 mutations, Her-2/neu overexpression, and DNA hypermethylation of 23 genes frequently hypermethylated in other epithelial malignancies [5, 15–27]. All tissues from women with OC were obtained prior to treatment.
Materials and Methods
Collection of clinical specimens
Ovarian tissues and associated clinical information from 207 women, including 68 with normal histology, 29 with benign ovarian disease (including serous cystadenoma, serous cystadenofibroma, mucinous cystadenoma, endometriosis, and simple cyst), 10 with serous carcinoma of low malignant potential (LMP) and 100 with ovarian carcinoma were obtained from the ovarian SPORE tissue bank as approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center (Table 1). These tissues had been originally collected from women undergoing ovarian-related surgery for one of the following reasons: pelvic mass, uterine fibroids, cervical-related diseases, or endometrial-related diseases (with malignant, benign or normal ovaries) and were placed in 1 ml Specimen Transport Media (STM) (Qiagen, Valencia, CA). Of the 100 patients with OC, 91 subsequently received carboplatin-paclitaxel chemotherapy. Patients were followed for 5 years after surgery and survival information was extracted from their medical records. Among patients who received chemotherapy, responsive cases were defined as those who had a progression-free survival of at least 6 months after the completion of chemotherapy. Non-responsive cases were defined as patients who progressed through, had persistent disease at the completion of chemotherapy, or had recurrent disease within 6 months after the chemotherapy. None of the study subjects had a family history of breast cancer and/or OC, or BRCA1 germ-line mutations. Some of the demographic data were missing because earlier questionnaires did not effectively capture those specific data (Table 2).
Table 1.
Clinical features of malignant OCs and nonmalignant benign ovarian tissues
| Tissue diagnosis | No. |
|---|---|
| Normal ovarian tissue | 68 |
| Non malignant pathology (Benign) | 29 |
| Serous cystadenoma | 7 |
| Serous cystadenofibroma | 9 |
| Mucinous cystadenoma | 5 |
| Endometriosis | 3 |
| Simple cyst | 5 |
| Neoplasia of Low Malignant Potential (LMP) – Serous | 10 |
| Malignant Neoplasia | 100 |
| Serous adenocarcinoma | 73 |
| Mucinous adenocarcinoma | 5 |
| Clear cell adenocarcinoma | 6 |
| Endometrioid adenocarcinoma | 4 |
| Undifferentiated cancer | 12 |
| FIGO Stage | |
| I | 19 |
| II | 2 |
| III | 69 |
| IV | 10 |
| Chemotherapeutic responsea | |
| Responsive | 60 |
| Non-responsive | 31 |
Only 91 patients underwent chemotherapy.
Table 2.
Demographics of study population
| Normal (n=68) |
Benign (n=29) |
LMP (n=10) |
Cancer (n=100) |
|
|---|---|---|---|---|
| Age (mean years ± sd)* | 53.1 ± 11.8 | 59.0 ± 14.7 | 39.1 ± 9.1 | 62.1 ± 12.3 |
| <50 | 12/67 (40) | 8/29 (28) | 9/10 (90) | 17/100 (17) |
| 50–59 | 21/67 (31) | 11/29 (38) | 1/10 (10) | 26/100 (26) |
| ≥60 | 19/67 (28) | 10/29 (34) | 0/10 (0) | 57/100 (57) |
| Caucasian** | 34/39 (87) | 5/5 (100) | 4/5 (80) | 66/71 (93) |
| Ever Birth Control*** | 29/39 (74) | 2/5 (40) | 5/6 (83) | 42/74 (57) |
| Ever Pregnant*** | 34/39 (87) | 4/5 (80) | 5/6 (83) | 61/74 (82) |
| Menopause**** | ||||
| Pre Menopause | 8/35 (23) | 0/4 (0) | 3/6 (50) | 3/69 (4) |
| Possible Menopause | 7/35 (20) | 1/4 (25) | 0/6 (0) | 1/69 (1) |
| Natural Menopause | 6/35 (17) | 2/4 (50) | 1/6 (17) | 25/69 (36) |
| Due to HRT | 5/35 (14) | 1/4 (25) | 2/6 (33) | 20/69 (29) |
| Due to Surgery | 8/35 (23) | 0/4 (0) | 0/6 (0) | 18/69 (26) |
| Due to Medical TR | 1/35 (3) | 0/4 (0) | 0/6 (0) | 2/69 (3) |
Missing 1 subject’s age
Missing 87 subject’s race
Missing 83 subject’s birth control and pregnancy status
Missing 93 subject’s menopausal status
DNA isolation from ovarian tissue and examination for methylation by methylation specific PCR (MSP)
Ovarian tissues collected in STM were digested with proteinase K at 37°C overnight. Genomic DNA was isolated from 200 µl of digested tissue using the QIAamp blood DNA mini kit (Qiagen, Valencia, CA). DNA concentration was determined with the Picogreen dsDNA assay (Molecular Probes, Eugene, OR). One microgram of genomic DNA was bisulfite modified using Intergen’s CpGenome DNA modification kit (Chemicon International, Temecula, CA) [28]. Briefly, genomic DNA was modified by sodium bisulfite, desulfonated with NaOH, purified and resuspended in TE (10 mM Tris,0.1 mM EDTA, pH 7.5). Human sperm DNA and in vitro methylated (using SssI CpG methyltransferase) human sperm DNA were converted with sodium bisulfite along with clinical DNA samples, and used as unmethylated (U) and methylated (M) control DNA respectively. Primers specific for methylated DNA were designed for each of the 23 genes examined (MINT31, HIC1, RASSF1, APC, BRCA1, CDH1, TERC, CDH13, CABIN1, RARB,SYK, DAPK1, CDKN2B, ERBB2, MLH1, CDKN2A, GSTP1, PRDM2, CPG15G2, BIRC5, TES, TP73 and ESR1) (Table 3). Hot start PCR was performed using AmpliTag Gold (Applied Biosystems, Foster City, CA) and the following parameters, 95°C for 5 min; 35 cycles of 95°C for 45 sec, Ta°C for 45 sec (Table 3), 72°C for 1 min; 72°C for 10 min. PCR products were analyzed on a 2% agarose gel containing ethidium bromide. Methylation of a specific gene was considered to be present if both the specimen and the M control DNA, but not the U control DNA, were amplified. The methylation index (MI) was calculated by dividing the number of genes methylated in each sample by the number of genes examined.
Table 3.
Methylation specific PCR primers
| Gene symbol | Gene name | Gene function | Sequence | Ta* | Previously reported methylated in cancer | Reference |
|---|---|---|---|---|---|---|
| MINT31 | Methylated in tumor 31 | Unknown | AGGGTAATTAGGGAGACGAC AAAACGCTTACGCCACTACG |
55 | OC | [15] |
| RASSF1 | Ras association (RalGDS/AF-6) domain family 1 |
Cell cycle control | GGGTTTTGCGAGAGCGCG GCTAACAAACGCGAACCG |
55 | Cervical cancer Breast cancer |
[5] |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) |
Adhesion | TTAGGTTAGAGGGTTATCGCGT TAACTAAAAATTCACCTACCGAC |
52 | OC | [16] |
| CABIN1 | Calcineurin binding protein 1 |
Signal transduction | GCGAAAGCGAAAGTCGTTCG CCCAACGCACATAACGAACC |
58 | OC | [15] |
| CDH13 | Cadherin 13, H-cadherin (heart) |
Adhesion | TCGCGGGGTTCGTTTTTCGC GACGTTTTCATTCATACACGCG |
55 | Breast cancer Lung cancer | [17] |
| BRCA1 | Breast cancer 1 | DNA repair Cell cycle control |
GAGTTTCGAGAGACGTTTGG AATCTCAACGAACTCACGCC |
58 | OC | [15] |
| HIC1 | Hypermethylated in cancer 1 | Cell cycle control | TCGGTTTTCGCGTTTTGTTCGT GCGATACCCGCCCTAACGCCG |
60 | OC | [15] |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
Cell cycle control | GCGTTCGTATTTTGCGGTT CGTACAATAACCGAACGACCGA |
60 | Leukemia | [18] |
| APC | Adenomatosis polyposis coli | Cell cycle control Adhesion |
CGTTGGATGCGGATTAGGGC CCACATATCGATCACGTACG |
60 | OC | [20] |
| TERC | Telomerase RNA component | Cellular senescence, Chromosomal repair | GACGTAAAGTTTTTTTCGGACG ACCCGATACGCTACCGAACG |
55 | OC | [15] |
| SYK | Spleen tyrosine kinase | Cell proliferation | CGATTTCGCGGGTTTCGTTC AAAACGAACGCAACGCGAAAC |
55 | Breast cancer | [19] |
| RARB | Retinoic acid receptor, beta | Cell differentiation | TCGAGAACGCGAGCGATTCG GACCAATCCAACCGAAACGA |
60 | OC | [16] |
| DAPK1 | Death-associated protein kinase 1 |
Apoptosis | GGATAGTCGGATCGAGTTAACGTC CCCTCCCAAACGCCGA |
60 | Cervical cancer Head and neck cancer |
[21] |
| ERBB2 | Her-2/neu | Signal transduction | TATTGTAGTACGTAGTCGCGG AACTTCCGATTACTCCGAAAATA |
55 | OC | [22] |
| MLH1 | MutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
DNA repair | ACGTAGACGTTTTATTAGGGTCGC CCTCATCGTAACTACCCGCG |
60 | OC | [15] |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) |
Cell cycle control | TTATTAGAGGGTGGGGCGGATCGC GACCCCGAACCGCGACCGTAA |
60 | Cervical cancer Breast cancer |
[18] |
| GSTP1 | Glutathione S-transferase pi | Cellular detoxification |
TTCGGGGTGTAGCGCTCGTC GCCCCAATACTAAATCACGACG |
55 | Cervical cancer Breast cancer Renal carcinoma |
[23] |
| PRDM2 | PR domain containing 2 | Transcription regulation |
GTGGTGGTTATTGGGCGACGGC GCTATTTCGCCGACCCCGACG |
68 | Breast cancer Liver cancer |
[24] |
| CpG15G2 | Unknown | ATACCCTATAACTCCTTTTAAAACTCGAA CGCGGACGTTTAGTTAAGGTTC |
59 | OC | [25] | |
| BIRC5 | Baculoviral IAP repeat- containing 5 |
Apoptosis Cell cycle control |
AGATTTGAATCGCGGGATTCG CCGCTCCGAAATACAAACGC |
60 | OC | [22] |
| TES | Testis derived transcript | Unknown | GTCGCGGGAGTTTCGTAGG CCAAATCCATATTAACGCGTCC |
60 | OC Cervical cancer |
[26] |
| TP73 | Tumor protein p73 | DNA repair Cell cycle control |
GGACGTAGCGAAATCGGGGTTC ACCCCGAACATCGACGTCCG |
68 | OC | [15] |
| ESR1 | Estrogen receptor 1 | Cell cycle control | ACGAGTTTAACGTCGCGGTC ACCCCCCAAACCGTTAAAAC |
57 | OC | [27] |
p53 mutation analysis
Adequate material was available from 82 of the cases of OC examined above for aberrant methylation to carry out additional analyses for p53 mutations. There were no significant differences in frequency of methylation between OC cases with and without material available for p53 analyses (data not shown). Because mutations in exons 2, 3 and 11 are known to be exceedingly rare in OC [29], we sequenced exons 4–10 of the p53 gene. DNA isolated from OC tissue was amplified in separate PCR reactions for p53 exons 4–10 [29]. PCR products were purified and sequenced with Big Dye Terminator chemistry (Perkin-Elmer, Boston, MA) and run on an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, CA). Sequencing data were analyzed with Sequencher software (Gene Codes Corporation, Ann Arbor, MI). All mutations were confirmed in two separate sequencing reactions.
Immunohistochemical analysis of Her-2/neu overexpression
Material was available from 51 of the cases of OC for immunohistochemical analysis of Her2/neu overexpression. There was no significant difference in the frequency of methylation between OC cases with and without material available for Her2/neu overexpression analyses (data not shown). Four-micron sections of formalin fixed paraffin embedded tissue were cut and placed on Superfrost Plus microscope slides (VWR, San Francisco, CA). The tissue sections were deparaffinized and rehydrated through graded alcohols. Endogenous peroxidase activity was blocked by incubation in 3% H2O2. Antigen retrieval was carried out with 0.01M citrate buffer pH 6.0 and microwave heat induction. Approximately 100 µl of the primary rabbit polyclonal antibody (diluted 1:8,000) (DakoCytomation Carpinteria, CA) or antibody diluent lacking the primary antibody was applied to each slide. The slides were washed, and a biotinylated anti-rabbit antibody (diluted 1:500) (Vector Laboratories Burlingame, CA) was applied. After a second wash, the avidin-biotin-peroxidase complex (Vector Laboratories Burlingame, CA) was applied. Color development was accomplished by incubation in diaminobenzidine with 3% H2O2 as a substrate, and nickel chloride enhancement. The slides were counterstained with methyl green, dehydrated through graded alcohols, cleared in xylene, and coverslipped with permanent mounting media. All cases were reviewed and scored without knowledge of other laboratory or clinical results. A case was scored as positive for Her-2/neu overexpression if it exhibited a staining intensity of 2+ to 3+ with circumscribed membrane staining in more than 10% tumor cells, but not in normal tissues. A case was scored as negative for Her-2/neu overexpression if staining was absent, staining intensity was 1+ or less, or if circumscribed membrane staining was absent regardless of staining intensity. Each batch of slides was run with a known tissue lacking Her-2/neu overexpression as the negative control and a known tissue with Her-2/neu overexpression as the positive control.
Statistical analysis
All statistical analyses were conducted using SAS version 9.1 (SAS Institute Inc., Cary, NC). Pearson’s chi-square tests were used in univariate analyses to compare dichotomous variables and Student’s t-tests were used to compare continuous variables. Multivariable logistic regression, adjusting for age, was used to determine differences in methylation frequency of each gene in women with OC and those with normal or benign histology. In that analysis, p-values and 95% confidence intervals were adjusted to take into account multiple comparisons by setting the false discovery rate (FDR) equal to 0.05, using PROC MULTTEST [30] and subsequently recalculating confidence intervals [31]. In order to assess the potential importance of gene methylation by biologic mechanism, genes shown to be more frequently methylated in cancers compared to controls were grouped into eight non-mutually exclusive biologic pathways, including apoptosis (DAPK1 and HIC1), cell adhesion and invasion/metastasis (CDH13, APC, CDH1), cell cycle control (RASSF1, APC, CDKN2B), cell proliferation/differentiation (RARB, SYK, ERBB2), DNA repair/detoxification (BRCA1, HIC1), calcium and MAPK signal transduction (MINT31), signal transduction and chromatin modification (CABIN1), and cellular senescence (TERC). Logistic regression was additionally used to determine differences in methylation frequency of each gene in OC patients in relation to clinical stage, response to chemotherapy and p53 mutation status or Her-2/neu overexpression, and odds ratios (OR) and 95% confidence intervals were calculated based upon test-based methods. Differences in MI between clinical and pathologic variables were assessed using Quasi-Poisson regression modeling, to take into account overdispersion potentially induced by the clustering of methylation of multiple genes in the same subject [32]. Kappa statistics were used to evaluate gene-specific concordance. A two-sided 0.05 test level determined statistical significance for all analyses.
Results
Subjects with ovarian carcinoma were significantly older (62.1 vs. 54.9, p<0.001; Table 3) and were more likely to be menopausal (94% vs. 59%, p<0.001) compared to those subjects with normal ovarian tissues (i.e. normal pathology) and benign (serous cystadenoma, serous cystadenofibroma, mucinous cystadenoma, endometriosis, and simple cyst) ovarian disease. Most subjects were Caucasian (91%), had at least one pregnancy (84%) and had a history of birth control usage (63%); these rates did not differ between women with ovarian carcinoma and with normal histology or benign disease. No study subjects reported a family history of breast cancer or OC.
Association of gene hypermethylation and OC
Overall, the prevalence of methylation for each of the genes examined varied from 0% to 88.7%. Seven genes (MLH1, CDKN2A, GSTP1, PRDM2, BIRC5, TES and TP73) were rarely methylated (0–1.6%) while two genes (ESR1 and CpG15G2) were frequently methylated (42.7% and 88.7% respectively) in all ovarian tissues regardless of histological diagnosis (data not shown), and were therefore excluded from further analysis. On average, tissues from OCs had a higher MI than did tissues from normal ovaries and ovaries with benign pathology (0.18 vs. 0.07, p<0.001) (LMP tissue samples were excluded from the analysis), with methylation of four genes (MINT31, HIC1, RASSF1, and CABIN1) occurring significantly more often in OC tissues than in normal or benign ovarian tissues, with odd ratios ranging from 4.1 to 20.4 (Table 4). The remaining genes (APC, BRCA1, CDH1, TERC, CDH13, RARB, DAPK1, SYK, CDKN2B, and ERBB2) all had increased rates of methylation in cancer compared to normal/benign tissues, but these differences did not attain statistical significance after adjustment for multiple comparisons. A representative gel electrophoresis of MSP analysis of these genes was presented in Figure 1.
Table 4.
Aberrant gene methylation in normal, benign, borderline and malignant ovarian tissues
| Gene | Histology |
OR (95% CI)* | |||
|---|---|---|---|---|---|
| Normal N=68 |
Benign N=29 |
LMP N=10 |
Cancer N=100 |
||
| MINT31 | 7/68 (10) | 7/29 (24) | 4/10 (40) | 42/100 (42) | 4.3 (1.9–9.8) |
| HIC1 | 10/68 (15) | 1/29 (3) | 2/10 (20) | 34/100 (34) | 4.1 (1.7–9.8) |
| RASSF1 | 2/68 (3) | 0/29 (0) | 0/10 (0) | 34/100 (34) | 20.4 (3.7–111) |
| APC | 6/68 (9) | 4/29 (14) | 0/10 (0) | 22/100 (22) | 2.2 (0.8–6.4) |
| BRCA1 | 7/68 (10) | 3/29 (10) | 0/10 (0) | 21/100 (21) | 2.4 (0.8–7.3) |
| CDH1 | 4/67 (6) | 4/29 (14) | 0/10 (0) | 18/100 (18) | 2.1 (0.7–6.5) |
| TERC | 5/67 (7) | 3/29 (10) | 0/10 (0) | 17/100 (17) | 2.3 (0.7–7.1) |
| CDH13 | 5/68 (7) | 4/29 (14) | 1/10 (10) | 15/100 (15) | 1.6 (0.4–6.1) |
| CABIN1 | 3/67 (4) | 0/29 (0) | 0/10 (0) | 14/100 (14) | 5.2 (1.0–26.4) |
| RARB | 3/68 (4) | 2/29 (7) | 0/10 (0) | 8/100 (8) | 1.6 (0.4–6.1) |
| DAPK1 | 2/68 (3) | 0/29 (0) | 0/10 (0) | 7/100 (7) | 3.1 (0.5–19.9) |
| SYK | 1/67 (1) | 4/29 (14) | 1/10 (10) | 6/97 (6) | 0.9 (0.1–7.6) |
| CDKN2B | 0/68 (0) | 0/29 (0) | 1/10 (10) | 6/100 (6) | 4.7 (0.4–55.0)** |
| ERBB2 | 1/68 (1) | 0/29 (0) | 0/10 (0) | 3/100 (3) | 3.7 (0.3–52.9) |
| Mean MI | 0.06 ± 0.08 | 0.08 ± 0.08 | 0.06 ± 0.06 | 0.18 ± 0.16 | <0.001*** |
Multivariable Logistic Regression – Cancer (n=100) versus normal/benign (n=97), p-values and 95% confidence intervals adjusted for age as well as multiple comparisons
Exact Logistic Regression
Multivariable Quasi-Poisson Regression P-value – Cancer versus normal/benign, adjusted for age
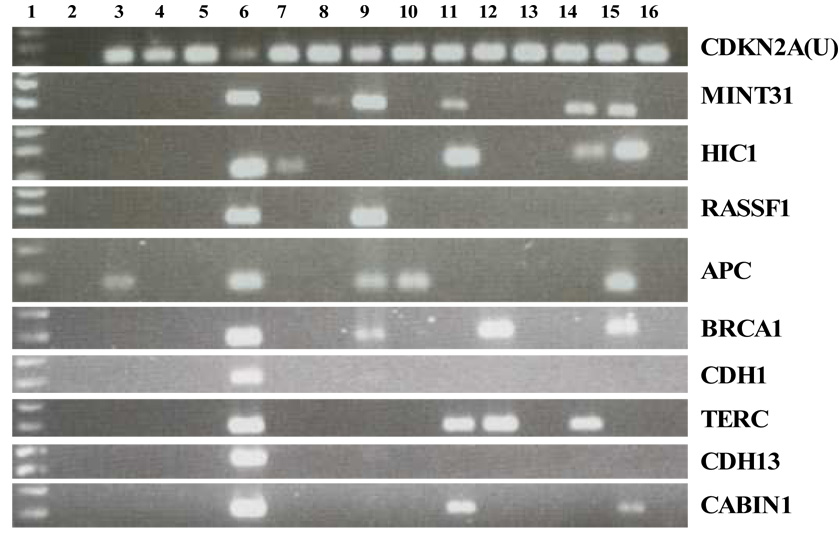
Figure 1.
Representative gel electrophoresis of methylation specific PCR (MSP) on ovarian tissue samples. Lane 1: molecular marker; lane 2: no template control; lane 4: human sperm DNA as unmethylated DNA control; lane 6: in vitro SssI methylated human sperm DNA as methylated DNA control; lane 3, 5, 7–16: clinical sample 1–12. CDKN2A(U) assay detects unmethylated gene copies, while the remaining gene assays detect individual methylated gene copies.
A number of genes (MINT31, APC, CDH1, SYK, CDH13, and RARB) showed increased frequency of methylation in ovarian tissues with benign pathology (n=29) as compared to normal tissues (n=68), but again, these differences were not statistically significant. Although SYK methylation was detected in only 1 (1%) of 67 normal tissues and in 6 (6%) of 97 OC tissues, it was detected in 4 (14%) of 29 benign ovarian tissues (2 serous cystadenoma, 1 serous cystadenofibroma and 1 benign cyst). As regards to LMP tumors (n=10), methylation of MINT31 was present in 40% of LMP tumors, but only in 10% of normal ovarian tissues (OR=5.8, 95% CI=1.3–26). Methylation of the other genes examined did not differ between LMP tumors and normal tissues. All of the genes examined, except MINT31, SYK, and CDKN2B, appeared to be more common in ovarian cancerous tissues as compared to LMP tumors, although this increase was significant only for RASSF1 (OR=7.0, 95% CI=1.1–∞) (data not shown).
Aberrant methylation of multiple genes was noted in 60 of 78 (77%) OC tissues. In order to assess whether, within a cancerous tissue sample, certain genes were usually methylated independently or concordantly, we analyzed the methylation status of all gene pair combinations. A number of genes within a sample tended to be concurrently hypermethylated. For example, these eight gene pairs were frequently methylated in the same cancerous tissues: MINT31 and RASSF1A (kappa=0.45), MINT31 and HIC1 (kappa=0.41), RASSF1A and APC (kappa=0.31), RASSF1A and HIC1 (kappa=0.29), RASSF1A and CABIN1 (kappa=0.27), CDH1 and BRCA1 (kappa=0.27), and CABIN1 and APC (kappa=0.26). In contrast, six gene pairs tended to be discordantly methylated (kappa between 0 and −0.1) and were often not methylated in the same cancerous material, including BRCA1 and CDKN2B, CDH1 and CABIN1, BRCA1 and HIC1, RASSF1A and CDKN2B, CABIN1 and BRCA1, and CABIN1 and CDKN2B.
In order to assess whether downregulation of specific pathways via methylation was especially important in the development of ovarian cancer, we combined various genes by known biologic pathways. The 14 genes which were aberrantly methylated more frequently in OC tissues are part of eight non-mutually exclusive biologic pathways. Genes in all of these eight basic pathways tended to be more frequently methylated in cancerous tissues as compared to normal ovarian tissue, including the apoptosis pathway (methylated in 12% of normal or benign tissue compared to 37% of tumor tissues; OR=4.2, 95% CI=1.9–9.2), cell adhesion and invasion/metastasis pathways (23% vs. 42%; OR=2.3, 95% CI=1.2–4.5), cell cycle control (12% vs. 44%; OR=5.6, 95% CI=2.6–12.3), cell proliferation/differentiation (11% vs. 16%, OR=1.5, 95% CI=0.6–3.7), DNA repair/detoxification (20% vs. 49%; OR=3.7, 95% CI=1.9–7.3), calcium and MAPK signal transduction (14% vs. 42%; OR=4.3, 95% CI=2.1–9.1), signal transduction and chromatin modification (3% vs. 14%; OR=5.1, 95% CI=1.3–23.2), and cellular senescence (8% vs. 17%; OR=2.3, 95% CI=0.9–6.1). Interestingly, within the cell cycle control pathway, RASSF1 tended to be methylated concordantly with APC but discordantly CDKN2B, while within the DNA repair/detoxification pathway, BRCA1 and HIC1 were often discordantly methylated. However, clear patterns of methylation concordance/discordance were not identified between any specific pathways.
Association of gene hypermethylation to cancer stage and response to chemotherapy
Subjects presenting with early, as compared to subjects presenting with late stage disease had a similar mean MI (p=0.34). Similarly, subjects who responded to therapy, as compared to subjects who did not respond to therapy had a similar mean MI (p=0.57) (Table 5). However, methylation of only a few specific genes was significantly associated with stage of disease and response to therapy. Methylation of HIC1 and RASSF1 was more frequent in women presenting with early stage diseases (stage I and II) as compared to late stage diseases (stage III and IV) (OR=0.3, 95% CI=0.1–0.8 and OR=0.4, 95% CI=0.1–1.0, respectively), while methylation of BRCA1 and RARB were somewhat more frequent among women presenting with late stage disease. Women who responded to therapy had significantly higher frequencies of CABIN1 hypermethylation (OR=0.1, 95% CI=0.02–0.97) and had moderately higher frequencies of RASSF1 hypermethylation (OR=0.4, 95% CI=0.2–1.1).
Table 5.
Association of gene hypermethylation with clinical stages and response to therapy in OC tissues
| Gene | Stages |
OR (95% CI)* | Responses to chemotherapy |
OR (95% CI)* | ||
|---|---|---|---|---|---|---|
| I–II (N=21) |
III–IV (N=79) |
Yes (n=60) |
No (N=31) |
|||
| MINT31 | 12 (57) | 30 (38) | 0.5 (0.2–1.2) | 25 (42) | 14 (45) | 1.2 (0.5–2.8) |
| HIC1 | 12 (57) | 22 (28) | 0.3 (0.1–0.8) | 21 (35) | 10 (32) | 0.9 (0.4–2.2) |
| RASSF1 | 11 (52) | 23 (29) | 0.4 (0.1–1.0) | 25 (42) | 7 (23) | 0.4 (0.2–1.1) |
| APC | 5 (24) | 17 (22) | 0.9 (0.3–2.7) | 14 (23) | 6 (19) | 0.8 (0.3–2.3) |
| BRCA1 | 1 (5) | 20 (25) | 6.8 (0.9–54) | 14 (23) | 6 (19) | 0.8 (0.3–2.3) |
| CDH1 | 2 (10) | 16 (20) | 2.4 (0.5–12) | 10 (17) | 7 (23) | 1.5 (0.5–4.3) |
| TERC | 5 (24) | 12 (15) | 0.6 (0.2–1.9) | 11 (18) | 4 (13) | 0.7 (0.2–2.3) |
| CDH13 | 4 (19) | 11 (14) | 0.7 (0.2–2.4) | 10 (17) | 4 (13) | 0.7 (0.2–2.6) |
| CABIN1 | 5 (24) | 9 (11) | 0.4 (0.1–1.4) | 13 (22) | 1 (3) | 0.1 (0.02–0.97) |
| RARB | 0 (0) | 8 (10) | 3.2 (0.5–∞)** | 3 (5) | 5 (16) | 3.7 (0.8–16.5) |
| DAPK1 | 1 (5) | 6 (8) | 1.6 (0.2–15) | 3 (5) | 4 (13) | 2.8 (0.6–13.5) |
| SYK^ | 1 (5) | 5 (7) | 1.4 (0.2–13) | 3 (5) | 3 (10) | 2.0 (0.4–10.8) |
| CDKN2B | 1 (5) | 5 (6) | 1.4 (0.2–12) | 4 (7) | 2 (6) | 1.0 (0.2–5.6) |
| ERBB2 | 1 (5) | 2 (3) | 0.5 (0.1–6.0) | 3 (5) | 0 (0) | 0.5 (0.0–4.7)** |
| Mean MI | 0.21 ± 0.16 | 0.17 ± 0.16 | 0.34*** | 0.19 ± 0.15 | 0.17 ± 0.18 | 0.57*** |
SYK methylation status missing for 3 cases in stage and chemotherapy analyses
Univariable Logistic Regression
Univariable Exact Logistic Regression
Univariable Quasi-Poisson Regression P-value
Association of genetic alterations and gene hypermethylation in OC
Mutations in exons 4–10 of p53 were detected in 50% (41/82) of the OC cases while Her-2/neu overexpression was detected in 23.5% (12/51) cases (Table 6). Thirty-one (66%) of the 47 cancer analyzed for both p53 mutations and Her-2/neu overexpression had at least one of these alterations present, while 5 (11%) had both p53 mutation and Her-2/neu overexpression. Subjects with p53 mutations, as compared to wildtype, had a similar mean MI (p=0.78), and the presence of p53 mutations was not significantly associated with DNA methylation of any specific genes. However, aberrant methylation of BRCA1 and CDKN2B tended to be somewhat more frequent in cancers having a p53 mutation, as compared to tumors with wildtype p53, and methylation of HIC1 was somewhat more frequent in tumors without p53 mutations (OR=0.4, 95% CI=0.2–1.04).
Table 6.
Correlation of gene hypermethylation and genetic alterations in OC tissues
| Gene | p53 |
OR (95% CI)* | Her-2/neu |
OR (95% CI)* | ||
|---|---|---|---|---|---|---|
| Wildtype (n=41) |
Mutant (n=41) |
Normal (n=39) |
Overexpressed (n=12) |
|||
| MINT31 | 18 (44) | 16 (39) | 0.8 (0.3–2.0) | 13 (33) | 9 (75) | 6.0 (1.4–26.0) |
| HIC1 | 17 (41) | 9 (22) | 0.4 (0.2–1.0) | 8 (21) | 4 (33) | 1.9 (0.5–8.1) |
| RASSF1 | 14 (34) | 15 (37) | 1.1 (0.5–2.8) | 8 (21) | 9 (75) | 11.6 (2.5–53.2) |
| APC | 10 (24) | 10 (24) | 1.0 (0.4–2.7) | 11 (28) | 4 (33) | 1.3 (0.3–5.1) |
| BRCA1 | 5 (12) | 12 (29) | 3.0 (0.9–9.4) | 7 (18) | 2 (17) | 0.9 (0.2–5.1) |
| CDH1 | 7 (17) | 9 (22) | 1.4 (0.5–4.1) | 9 (23) | 2 (17) | 0.7 (0.1–3.6) |
| TERC | 6 (15) | 9 (22) | 1.6 (0.5–5.1) | 5 (13) | 3 (25) | 2.3 (0.5–11.3) |
| CDH13 | 5 (12) | 6 (15) | 1.2 (0.4–4.4) | 0 (0) | 4 (33) | 22.2 (2.6–∞)** |
| CABIN1 | 5 (12) | 8 (20) | 1.8 (0.5–5.9) | 6 (15) | 4 (33) | 2.8 (0.6–12.1) |
| RARB | 4 (10) | 4 (10) | 1.0 (0.2–4.3) | 4 (10) | 1 (8) | 0.8 (0.1–7.9) |
| DAPK1 | 4 (10) | 1 (2) | 0.2 (0.03–2.2) | 4 (10) | 0 (0) | 0.6 (0.0–5.0)** |
| SYK^ | 4 (10) | 2 (5) | 0.5 (0.1–2.8) | 1 (3) | 0 (0) | 3.1 (0.0–120.3)** |
| CDKN2B | 1 (2) | 3 (7) | 3.2 (0.3–32) | 2 (5) | 0 (0) | 1.3 (0.0–17.7)** |
| ERBB2 | 0 (0) | 2 (5) | 2.5 (0.2–∞)** | 1 (3) | 2 (17) | 7.6 (0.6–92.5) |
| Mean MI | 0.17 ± 0.17 | 0.19 ± 0.15 | 0.78*** | 0.15 ± 0.15 | 0.26 ± 0.19 | 0.04*** |
SYK methylation status missing for 2 cases in p53 and Her-2/neu analyses
Univariable Logistic Regression
Univariable Exact Logistic Regression
Univariable Quasi-Poisson Regression P-value
On the other hand, subjects with Her-2/neu overexpression had a significantly higher MI than women without Her-2/neu overexpression (p=0.04). Methylation of MINT31, RASSF1 and CDH13 was significantly more frequent in OCs with, as compared to those without Her-2/neu overexpression (OR=6.0, 95% CI=1.4–26, OR=11.6, 95% CI=2.5–53, and OR=22.2, 95% CI=2.6–∞, respectively).
Discussion
We examined normal and abnormal ovarian tissues for the presence of abnormal CpG island promoter methylation of 23 genes, as well as for p53 mutations and overexpression of Her-2/neu. Four genes (MINT31, HIC1, RASSF1, and CABIN1) were significantly more frequently methylated in OC tissue than in tissue from normal ovaries or ovaries with benign disease while a number of other genes, including APC, BRCA1, CDH1, and CDKN2B, were somewhat more likely to be methylated in OC tissue, although these differences did not reach statistical significance after adjustment for multiple comparisons. The frequency of hypermethylation of five of these genes in OC tissues (MINT31, 42%; RASSF1, 34%; CDH1, 18%; HIC1, 34%; APC, 22%) was similar to frequencies reported in previous studies. Methylation of MINT31 has been reported in 54% [15], RASSF1 in 10%–50% [5, 20, 33–36], and CDH1 in 26%–42% [20, 35, 37], HIC1 in 16–35% [15, 20, 36], and APC in 18%–22% [20, 35] of OC cases.
In contrast to most previous studies, this study included examination of DNA methylation of a large number of normal ovarian tissues from women without OC (N=68) as well as tissues from OCs (N=100), permitting a direct assessment of the risk of OC associated with gene specific DNA hypermethylation. This is important both for understanding the pathogenesis of OC as well as for assessment of the potential of aberrantly methylated genes as biomarkers for OC. Of the over 90 studies that have previously reported on aberrant methylation in OC, most have not included tissue from women without OC [10, 36, 38–41]. In the largest study (n=215) of methylation in OC reported, no normal controls were included [40]. Further, in some studies, “normal” tissue controls consisted of adjacent non-cancerous tissue obtained from women with OC [5, 15, 39, 42–44]. Only three studies have included normal tissues from women without OC [20, 35, 45].
In our study, methylation of MINT31, HIC1, RASSF1A, APC, or BRCA1 each identified a large percentage (>20%) of OC cases and also had high specificity (>85%) for OC, while CDH1, TERC, CDH13, or CABIN1 were less sensitive, each identifying 14–18% of OC cases, while remaining specific (>90%). Rathi et al [20] similarly reported hypermethylation of RASSF1A, HIC1, CDH1 (E-cadherin), APC, and CDH13 (H-cadherin) in 18–41% of 49 OC tissue and 15% of 39 nonmalignant tissues. Makarla et al [35] detected hypermethylation of CDKN2A (p16), CDH1 (E-cadherin), CDH13 (H-cadherin), RASSF1A, and APC in 22–30% of 23 OC cases but in less than 15% of 23 benign cystadenomas and 16 normal tissues. Most recently, Tam et al. [45] detected high rates of hypermethylation of HIC1 (52%), MINT31 (51%), APC (47%) in 89 OC tissues, while hypermethylation of these three genes was less frequent in 19 benign tumors (21%, 16%, and 26%, respectively) and 16 normal ovarian tissues (13%, 0%, and 25%, respectively).
This study is unique as it is the first to examine the relationship between genetic alterations and DNA methylation in OC. Interestingly, within tumor specimens, hypermethylation of three different genes (MINT31, RASSF1 and CDH13) was associated with overexpression of Her-2/neu. Recently, several studies of other cancers have noted associations between methylation of specific loci and genetic mutations (38, 39, 21), with methylation of CDKN2A associated with K-ras mutations in non-small-cell lung cancer [12], methylation of 5 genes (CDKN2A, MINT1, MINT2, MINT31 and MLH1) associated with mutations of BRAF in colon cancer [14], and methylation of CDH13, PGR and HSD17B4 associated with overexpression of Her-2/neu in breast cancer [13]. The mechanistic relationship between such mutations and epigenetic changes is unclear. Whether the existence of such associations is associated with specific OC histologic subtypes or a specific clinical course is unknown, although such associations have been proposed for NSCLC [12], colon [14] and breast cancers [13].
In the present study we found that (after adjusting for stage of disease) methylation of CABIN1 was strongly associated with responsiveness to chemotherapy. A number of previous studies have examined associations between hypermethylation of specific genes or global methylation patterns and response to chemotherapy. Methylation of hMLH1 [46], MCJ [38], IGFBP3 [10], p16 [47] and BRCA1 [48] have been associated with poor response to platinum-based chemotherapy and poor survival. Wei et al examined the overall pattern of methylation and found that, as compared to drug-sensitive cell lines, drug-resistant OC cell lines had increased number of methylated loci, and identified a group of OCs associated with poor survival [25]. Clearly, whether methylation of any of these genes can predict therapy responsiveness and survival needs further investigation.
Our study has a number of limitations. We used a qualitative MSP assay to assess aberrant methylation, which does not allow for quantitative of methylation and may be associated with a lack of specificity [49]. However, if conventional MSP lacks specificity, the potential non-specificity of the MSP assay would most likely have resulted in non-differential misclassification, and thus may have attenuated our study risk estimates. Therefore, due to misclassification, the relationship between aberrant methylation and ovarian cancer may be stronger than what we observed. Due to the qualitative nature of our assay, we could not assess the possibility that aberrant methylation of the genes studied is also at lower levels in normal as compared to OC tissues, as methylation quantity cannot be determined in the present study. This limitation is common to most previous studies as well. However, while quantitative MSP may be more sensitive under many conditions, conventional MSP may be more sensitive to detect methylation changes in samples with limited amount of DNA without losing specificity [50]. Another potential weakness of this study is that the tissues analyzed included both ovarian surface epithelial cells as well as underlying ovarian stromal cells. However, while previously it was widely assumed that the pathogenesis of OCs was limited to changes in the ovarian surface epithelial cells, several studies have demonstrated that similar changes also occurred in adjacent stromal cells during the pathogenesis of epithelial OC. For example, SPARC has been reported to be up-regulated in stroma adjacent to epithelial OC [51]. COX2 and iNOS expressing macrophages were not only present in OC associated stroma, but also in stroma associated with benign tumors [52]. Further, similar genetic alterations were detected in both epithelial and stromal cells of OC [53, 54]. Thus far there has not been a study examining epigenetic changes in stromal cells in ovaries containing benign or malignant ovarian neoplasia. In the present study, we were not able to determine whether the aberrant methylation had occurred in epithelial or stromal cells. However, identifying the cell of origin of DNA hypermethylation will enhance our understanding the tumorigenesis of OC. Further, if changes in stromal cells are shown to be important in the early pathogenesis of OC, the use of histologically normal tissue adjacent to OC would make identification of such early epigenetic changes impossible.
Recent research shows that DNA methylation studies not only identify potential biomarkers for cancer diagnosis and prognosis, but also provide insight for tumorigenesis. Our current study confirms that this is also the case for ovarian cancer: DNA methylation is frequent in ovarian cancer, and certain methylation is associated with chemosensitivity. We also provide evidence for the complex interplay between genetic and epigenetic changes during tumorigenesis. Future studies are warranted to determine whether this interaction implies causal effect, or merely tumor specific association.
Acknowledgment
We thank Hiep Lu and Steve Cherne for technical support on DNA methylation analysis, Janice Morihara for immunohistochemistry analysis, Troy Wood for DNA sequencing analysis, Kathy O’briant and Donna Kenney for their assistance on the identification of clinical specimens, Alicia Young, Akhila Balasubramanian, Martin Mcintosh for advice on statistical analysis. We also thank Ingegerd Hellstrom, Karl Eric Hellstrom and Pavel Sova for helpful discussion and critical review on the manuscript.
Informed consent was obtained according to procedures approved by the Human Subjects Committee of the University of Washington and Fred Hutch Cancer Research Institute Supported by grants from the National Institute of Health/National Cancer Institute (CA85050)
The authors have no commercial or other associations that might pose a conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::aid-ssu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Boyd J, Rubin SC. Hereditary ovarian cancer: molecular genetics and clinical implications. Gynecol Oncol. 1997;64:196–206. doi: 10.1006/gyno.1996.4572. [DOI] [PubMed] [Google Scholar]
- 4.Fraser M, Leung B, Jahani-Asl A, Yan X, Thompson WE, Tsang BK. Chemoresistance in human ovarian cancer: the role of apoptotic regulators. Reprod Biol Endocrinol. 2003;1:66. doi: 10.1186/1477-7827-1-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94:212–217. doi: 10.1002/ijc.1466. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Campbell EJ, Shepherd TG, Nachtigal MW. Epigenetic Regulation of Proprotein Convertase PACE4 Gene Expression in Human Ovarian Cancer Cells. Mol Cancer Res. 2003;1:569–576. [PubMed] [Google Scholar]
- 7.Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D, Brown I, Williams AR, Bates PA, Smyth JF, Gabra H. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34:337–343. doi: 10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 8.Terasawa K, Sagae S, Toyota M, Tsukada K, Ogi K, Satoh A, Mita H, Imai K, Tokino T, Kudo R. Epigenetic inactivation of TMS1/ASC in ovarian cancer. Clin Cancer Res. 2004;10:2000–2006. doi: 10.1158/1078-0432.ccr-0932-03. [DOI] [PubMed] [Google Scholar]
- 9.Takada T, Yagi Y, Maekita T, Imura M, Nakagawa S, Tsao SW, Miyamoto K, Yoshino O, Yasugi T, Taketani Y, Ushijima T. Methylation-associated silencing of the Wnt antagonist SFRP1 gene in human ovarian cancers. Cancer Sci. 2004;95:741–744. doi: 10.1111/j.1349-7006.2004.tb03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley A, Katsaros D, Fracchioli S, Yu H. Methylation of the insulin-like growth factor binding protein-3 gene and prognosis of epithelial ovarian cancer. Int J Gynecol Cancer. 2006;16:210–218. doi: 10.1111/j.1525-1438.2006.00299.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Li M, Lu S, Zhang Y, Wang H. Promoter Hypermethylation of FANCF Plays an Important Role in the Occurrence of Ovarian Cancer through Disrupting Fanconi Anemia-BRCA Pathway. Cancer Biol Ther. 2006;5:256–260. doi: 10.4161/cbt.5.3.2380. [DOI] [PubMed] [Google Scholar]
- 12.Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, Tomii K, Ichihara S, Suzuki M, Aoe M, Date H, Gazdar AF, Shimizu N. Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66:1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- 13.Fiegl H, Millinger S, Goebel G, Muller-Holzner E, Marth C, Laird PW, Widschwendter M. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res. 2006;66:29–33. doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- 14.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 15.Strathdee G, Appleton K, Illand M, Millan DW, Sargent J, Paul J, Brown R. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhillon VS, Young AR, Husain SA, Aslam M. Promoter hypermethylation of MGMT, CDH1, RAR-beta and SYK tumour suppressor genes in granulosa cell tumours (GCTs) of ovarian origin. Br J Cancer. 2004;90:874–881. doi: 10.1038/sj.bjc.6601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi T, Liang SB, Matsuyoshi N, Zhou S, Miyachi Y, Sonobe H, Ohtsuki Y. Loss of T-cadherin (CDH13, H-cadherin) expression in cutaneous squamous cell carcinoma. Lab Invest. 2002;82:1023–1029. doi: 10.1097/01.lab.0000025391.35798.f1. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61:5558–5561. [PubMed] [Google Scholar]
- 20.Rathi A, Virmani AK, Schorge JO, Elias KJ, Maruyama R, Minna JD, Mok SC, Girard L, Fishman DA, Gazdar AF. Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin Cancer Res. 2002;8:3324–3331. [PubMed] [Google Scholar]
- 21.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 22.Hattori M, Sakamoto H, Satoh K, Yamamoto T. DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c-erbB-2 and survivin genes. Cancer Lett. 2001;169:155–164. doi: 10.1016/s0304-3835(01)00499-2. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–4518. [PubMed] [Google Scholar]
- 24.Du Y, Carling T, Fang W, Piao Z, Sheu JC, Huang S. Hypermethylation in Human Cancers of the RIZ1 Tumor Suppressor Gene, a Member of a Histone/Protein Methyltransferase Superfamily. Cancer Res. 2001;61:8094–8099. [PubMed] [Google Scholar]
- 25.Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8:2246–2252. [PubMed] [Google Scholar]
- 26.Tobias ES, Hurlstone AF, MacKenzie E, McFarlane R, Black DM. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene. 2001;20:2844–2853. doi: 10.1038/sj.onc.1204433. [DOI] [PubMed] [Google Scholar]
- 27.O'Doherty AM, Church SW, Russell SE, Nelson J, Hickey I. Methylation status of oestrogen receptor-alpha gene promoter sequences in human ovarian epithelial cell lines. Br J Cancer. 2002;86:282–284. doi: 10.1038/sj.bjc.6600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 29.Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, King MC. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193:662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 30.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2002;64:479–498. [Google Scholar]
- 31.Ludbrook J. Multiple inferences using confidence intervals. Clinical and Experimental Pharmacology and Physiology. 2000;27:212–215. doi: 10.1046/j.1440-1681.2000.03223.x. [DOI] [PubMed] [Google Scholar]
- 32.McCullach P, Nelder J. Generalized linear models. London, United Kingdom: Chapman & Hall; 1989. [Google Scholar]
- 33.Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, Hosoe S, Lerman MI, Minna JD, Maher ER, Latif F. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 34.Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, Edelson MI, Bergman C, Ehya H, Eisenberg BL, Cairns P. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64:6476–6481. doi: 10.1158/0008-5472.CAN-04-1529. [DOI] [PubMed] [Google Scholar]
- 35.Makarla PB, Saboorian MH, Ashfaq R, Toyooka KO, Toyooka S, Minna JD, Gazdar AF, Schorge JO. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res. 2005;11:5365–5369. doi: 10.1158/1078-0432.CCR-04-2455. [DOI] [PubMed] [Google Scholar]
- 36.Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G, Brown R. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 37.Yuecheng Y, Hongmei L, Xiaoyan X. Clinical Evaluation of E-cadherin Expression and its Regulation Mechanism in Epithelial Ovarian Cancer. Clin Exp Metastasis. 2006;23:65–74. doi: 10.1007/s10585-006-9020-3. [DOI] [PubMed] [Google Scholar]
- 38.Strathdee G, Vass JK, Oien KA, Siddiqui N, Curto-Garcia J, Brown R. Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol Oncol. 2005;97:898–903. doi: 10.1016/j.ygyno.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer. 2006;6:212. doi: 10.1186/1471-2407-6-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiley A, Katsaros D, Chen H, Rigault de la Longrais IA, Beeghly A, Puopolo M, Singal R, Zhang Y, Amoako A, Zelterman D, Yu H. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107:299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 41.Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, Li L, Wang Z, Wan JC, Davuluri RV, Karlan BY, Gifford G, Brown R, Kim S, Huang TH, Nephew KP. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 42.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–7733. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- 44.Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, Nagata Y, Nakao M, Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005 doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 45.Tam KF, Liu VW, Liu SS, Tsang PC, Cheung AN, Yip AM, Ngan HY. Methylation profile in benign, borderline and malignant ovarian tumors. J Cancer Res Clin Oncol. 2006 doi: 10.1007/s00432-006-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 47.Katsaros D, Cho W, Singal R, Fracchioli S, Rigault De La Longrais IA, Arisio R, Massobrio M, Smith M, Zheng W, Glass J, Yu H. Methylation of tumor suppressor gene p16 and prognosis of epithelial ovarian cancer. Gynecol Oncol. 2004;94:685–692. doi: 10.1016/j.ygyno.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 48.Chiang JW, Karlan BY, Cass L, Baldwin RL. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol Oncol. 2006;101:403–410. doi: 10.1016/j.ygyno.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 49.Shames DS, Minna JD, Gazdar AF. Methods for detecting DNA methylation in tumors: From bench to bedside. Cancer Lett. 2006 doi: 10.1016/j.canlet.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Jeronimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, Sidransky D. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 51.Brown TJ, Shaw PA, Karp X, Huynh MH, Begley H, Ringuette MJ. Activation of SPARC expression in reactive stroma associated with human epithelial ovarian cancer. Gynecol Oncol. 1999;75:25–33. doi: 10.1006/gyno.1999.5552. [DOI] [PubMed] [Google Scholar]
- 52.Klimp AH, Hollema H, Kempinga C, van der Zee AG, de Vries EG, Daemen T. Expression of cyclooxygenase-2 and inducible nitric oxide synthase in human ovarian tumors and tumor-associated macrophages. Cancer Res. 2001;61:7305–7309. [PubMed] [Google Scholar]
- 53.Tuhkanen H, Anttila M, Kosma VM, Heinonen S, Juhola M, Helisalmi S, Kataja V, Mannermaa A. Frequent gene dosage alterations in stromal cells of epithelial ovarian carcinomas. Int J Cancer. 2006;119:1345–1353. doi: 10.1002/ijc.21785. [DOI] [PubMed] [Google Scholar]
- 54.Tuhkanen H, Anttila M, Kosma VM, Yla-Herttuala S, Heinonen S, Kuronen A, Juhola M, Tammi R, Tammi M, Mannermaa A. Genetic alterations in the peritumoral stromal cells of malignant and borderline epithelial ovarian tumors as indicated by allelic imbalance on chromosome 3p. Int J Cancer. 2004;109:247–252. doi: 10.1002/ijc.11733. [DOI] [PubMed] [Google Scholar]