Abstract
The Maillard reaction, starting from the glycation of protein and progressing to the formation of advanced glycation end-products (AGEs), is implicated in the development of complications of diabetes mellitus, as well as in the pathogenesis of cardiovascular, renal, and neurodegenerative diseases. In this perspective review, we provide an overview on the relevance of the Maillard reaction in the pathogenesis of chronic disease and discuss traditional approaches and recent developments in the analysis of glycated proteins by mass spectrometry. We propose that proteomics approaches, particularly bottom-up proteomics, will play a significant role in analyses of clinical samples leading to the identification of new markers of disease development and progression.
INTRODUCTION
In 1912, the French scientist Louis-Camille Maillard (1878–1936) published a paper describing the reaction between amino acids and reducing sugars during heating that resulted in discoloration (browning) of the reaction mixture.1 This network of reactions between amino acids and reducing sugars came to be known as the Maillard reaction. For the next 60+ years, work on the Maillard reaction focused on foods and food-like model systems, and the Maillard reaction became recognized as an important member of the group of browning reactions that take place in foods and beverages. This complex reaction not only occurs in virtually all heat processed and stored foods but also takes place in the paper,2 textile,2 and biopharmaceutical industries.3–5 In addition, it plays a role in the formation of humic substances in the soil.2 However, it is only since the 1970s that substantial attention has been given to the Maillard reaction in vivo. In the body, glycation – the reaction of glucose or its autoxidation products with amines, amino acids, peptides, and proteins – is generally considered the first step in this complex reaction. Later stages in the reaction lead to the formation of sugar-derived protein adducts and crosslinks known as advanced glycation end-products (AGEs). In this perspective review, we provide on overview on the relevance of the Maillard reaction in the pathogenesis of chronic disease and discuss traditional approaches and recent developments in the analysis of glycated and AGE-modified proteins by mass spectrometry.
History and Overview of the Maillard Reaction
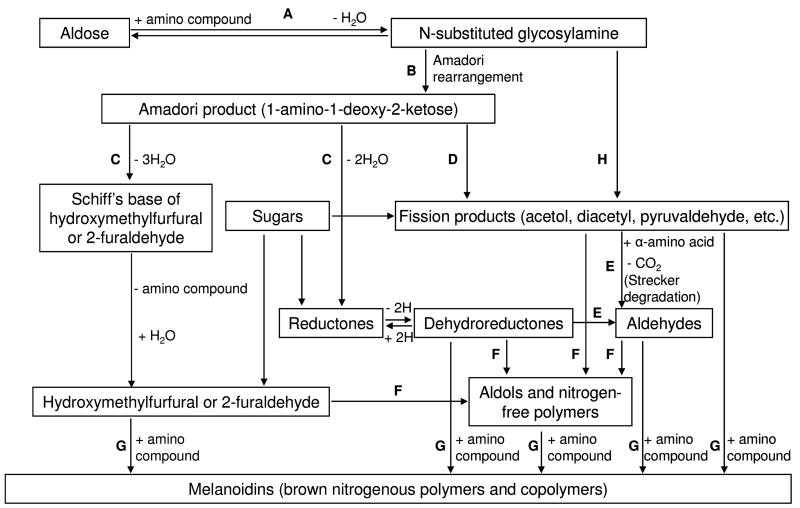
The Maillard reaction encompasses a complex network of reactions; it is not a typical organic “named reaction” and it is difficult to summarize succinctly. Nevertheless, the American scientist, John Hodge (1914–1996), in his seminal review of the Maillard reaction,6 constructed a diagram (Figure 1) that attempts to simplify the reaction network; this diagram still holds good today and is widely quoted. The original Hodge scheme divides the Maillard reaction into 7 steps (A–G). In 1986, Nursten recognized the work of the Japanese scientists, Namiki and co-workers,7, 8 concerning the free radical degradation of Maillard intermediates, by adding Step H to the original scheme (Figure 1).9 Step H is also known as the Namiki pathway and involves free radical-mediated formation of carbonyl fission products from the reducing sugar.7, 8
Figure 1. The Hodge Diagram.
A) The initial reaction between a reducing sugar and amino group forms an unstable Schiff base; B) The Schiff base slowly rearranges to form the Amadori product; C) Degradation of the Amadori product; D) Formation of reactive carbonyl and dicarbonyl compounds; E) Formation of Strecker aldehydes of amino acids and aminoketones; F) Aldol condensation of furfurals, reductones, and aldehydes produced in Steps C, D, and E without intervention of amino compounds; G) Reaction of furfurals, reductones, and aldehydes produced in Steps C, D, and E with amino compounds to form melanoidins; H) Free radical-mediated formation of carbonyl fission products from the reducing sugar (Namiki pathway).7, 8 Reproduced from Trends in Food Science and Technology, 1, Ames JM, Control of the Maillard reaction in food systems, pp. 150–154, 1990, with permission from Elsevier.
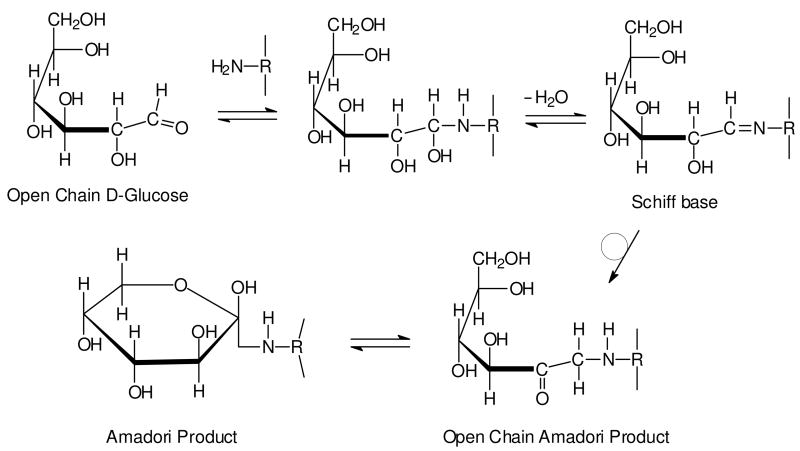
The initial stage of the Maillard reaction (Figure 1, Step A) involves the condensation of a carbonyl group, for example from a reducing sugar such as glucose, with a free amino group, typically the epsilon amino group of lysine residues within proteins. This glycation reaction results in the formation of an unstable Schiff base (aldimine) that spontaneously rearranges (Figure 1, Step B) to form the more stable 1-amino-1-deoxy-2-ketose (ketoamine), which is also known as the Amadori product (Figure 2) after the Italian scientist Mario Amadori.10 When the initial sugar is glucose, the Amadori product is commonly known as fructoselysine (FL). The equilibrium constant for the formation of the Amadori product lies strongly in the forwards direction at physiological pH,2 but reversibility increases with increasing pH and also in the presence of phosphate.2
Figure 2. Reaction between glucose and amino group of protein to form the Amadori product.
Nucleophilic attack by a free amino group of protein on the aldehyde of glucose initially forms a carbinolamine, which subsequently dehydrates to a Schiff base. The Schiff base then undergoes a slow rearrangement to form the Amadori product. While only a single cyclic isoform of the Amadori product is shown, it is important to note that it exists as a mixture of several isoforms.
Amadori products are degraded via various pathways (Figure 1, Steps C and D),11, 12 leading to the formation of furfurals, reductones and fragmentation products (carbonyl and hydroxycarbonyl compounds). All of these intermediates can also form directly from the sugar in uncatalyzed reactions, i.e. without the intervention of an amino compound (Figure 1). In Step C, furfural formation is favoured under acidic conditions, while alkaline media favor the production of reductones. These conjugated enediol intermediates possess moderate reducing power; they may catalyze redox reactions dependent on recycling of transition metals (e.g. Fe, Cu), but, like ascorbate, they may also contribute to antioxidant activity.
Sugar fragmentation (Figure 1, Step D) occurs mainly by retroaldolisation. α-Dicarbonyl compounds, e.g., butanedione, glyoxal, methylglyoxal, formed in Step D, are able to react with amino acids via the Strecker degradation (Figure 1, Step E) (named after the German chemist Adolph Strecker) to give Strecker aldehydes of the amino acids and aminoketones; the latter subsequently condense to form pyrazines. Strecker aldehydes and pyrazines contribute to aroma in heated foods.
Steps F and G in Figure 1 summarize the final stage of the Maillard reaction, and it is here that the majority of the compounds contributing color are formed. These may be relatively small molecules or much larger polymeric materials.13, 14 Step F involves aldol condensation of the furfurals, reductones and aldehydes produced in steps C, D and E without the intervention of amino compounds. Step G represents reactions between the same intermediates with amino compounds and that lead to the ultimate reaction products, known in food science as melanoidins. These poorly defined compounds chelate redox-active, transition metal ions and thereby possess antioxidant activity. Hodge defined melanoidins as ‘brown, nitrogenous polymers and copolymers’.6 Polymers are generally considered to contain repeating units but, since the structures of melanoidins are unknown and the presence of true repeating units uncertain, melanoidins are sometimes described more generally as macromolecular materials.
Role of the Maillard Reaction in Food Quality
The vast majority of research on and characterization of the Maillard reaction up to the 1980s came from studies by food scientists on the flavour, color, texture, digestibility, and nutritional value of foods – these properties are profoundly affected by the Maillard reaction and may be controlled by modifying food composition, processing, or storage conditions. Since the 1980s, work from biomedical laboratories (below) has made a tremendous impact on knowledge in the field, while contributions from food chemists have continued unabated.
The Maillard reaction modifies the nutritional value of food, especially as a consequence of reactions involving the e-amino group of lysine residues in protein (lysine is nutritionally essential). In addition, following oxidation to dehydroascorbic acid, vitamin C (ascorbic acid) can react with free amino groups in place of the reducing sugar, leading to losses of this vitamin. Furthermore, nutritionally important metals such as copper, zinc, and iron may form complexes with Maillard reaction products, including Amadori products and melanoidins.15 Finally, the Maillard reaction may lead to impaired nutritional value of food through protein crosslinking mediated by recognized AGEs or other peptide-bound adducts, such as those contributing to color.
The Maillard reaction is also responsible for an increase in antioxidant activity in food systems, including coffee16, 17 and malt.18, 19 Both reductones and melanoidins may contribute to antioxidant activity, and their formation may prolong food shelf-life by inhibiting oxidative reactions that result in rancidity and off-flavour. Furthermore, the Maillard reaction can lead to acrylamide formation in food, which has received much attention in the last six years.20
Role of the Maillard Reaction in Biopharmacy
Currently, all U.S. Food and Drug Administration approved monoclonal antibodies are produced from mammalian cell culture.5 During the 10–14 day cell culturing process required for antibody production, proteins are incubated with a large molar excess of reducing sugar per mole protein product. The high concentration of reducing sugar provides energy to the cell, as well as a means for modulating enzymatic glycosylation. Thus, modification of the protein product by the reducing sugar during cell culture is very likely and may compromise the pharmaceutical activity of therapeutic antibodies and proteins.4, 5
Alternatively, protein modification may occur during subsequent formulation, packaging, long term storage, or clinical administration steps, where sugars are commonly used as excipients in liquid or lyophilized formulations of therapeutic proteins.3–5, 21–26 For example, sucrose-containing formulations have been described to undergo browning with time – likely considered to be a result of Maillard-like reactions.27 Although sucrose is a non-reducing disaccharide, it can undergo hydrolysis – e.g. at low pH – to glucose and fructose.
Thus, modification of therapeutic proteins by reducing sugars may change their charge, target binding capacities, and degradation rates.4 These in turn could result in reduced efficacy and stability, and increased toxicity. Protein stability, in particular, is a major challenge in the development of bio-therapeutics, as these products must be maintained through multiple processing steps to market.
THE MAILLARD REACTION IN VIVO
While fructose and lactose are important reducing sugars in food products, glucose is the major free sugar in the body. Thus, glycation of proteins by glucose is considered the first step in the Maillard reaction in vivo. Glycation is the most general IUPAC term for adduction of a sugar to another biomolecule. For proteins, glycation is frequently used in apposition to glycosylation, with glycation being a nonenzymatic process and glycosylation an enzymatic process forming a glycosidic bond. Sometimes the terms nonenzymatic glycation and enzymatic glycosylation are used for greater precision, but, in general, the term glycation is not used in a precise manner: it may refer to the formation of a Schiff base and/or Amadori product to an amino group in protein; to the adduction of dicarbonyl compounds, such as methylglyoxal, to arginine; or to the formation of AGEs, including both adducts and crosslinks. In the following discussion, we define glycation as the nonenzymatic reaction of glucose with amino groups in protein to form the Amadori product.
Glycation of Hemoglobin
Interest in glycation in biological systems began with the discovery and characterization of hemoglobin A1c (HbA1c, 4–6% total hemoglobin) in the late 1970s. The discovery was somewhat serendipitous because formation of the Amadori product on the amino terminal valine residue in the β-chain (βV1) of hemoglobin caused an acidic shift in the pKa of the protein; this led to an anionic shift in mobility of the protein in electrophoresis buffers then used for screening for genetic variants of hemoglobin. Chromatographic fractionation of hemoglobin revealed a number of other anionic forms of hemoglobin that were also glycated at βV1, including HbA1a1 (fructose-1,6-P2, <1% total hemoglobin), HbA1a2 (glucose-6-P, <1% total hemoglobin), and HbA1b (pyruvate ketimine, <1% total hemoglobin).28–30 It also became clear that there was significant glycation of lysine residues in the main, hemoglobin A1 fraction of Hb; in fact, there are ~2.5 glycated lysines (FLs) per glycated valine residue in total Hb.31 Glycation of the lysine residues does not affect the chromatographic or electrophoretic mobility of hemoglobin at pH 6–9. Today the term HbA1c is used in reference to the specific species containing glucose at the βV1 residue. The terms glycated hemoglobin or glycohemoglobin (GHb, GlcHb) refer to the full spectrum of glycated hemoglobins, including those containing glycated valine and/or lysine residues.
Kinetics and Extent of Protein Glycation
Most studies on glycation in vivo focus on measurement of FL in hemoglobin, plasma proteins, crystallins, plasma proteins, or extracellular matrix proteins (primarily collagen). These proteins are accessible, turnover relatively slowly, and Amadori products accumulate gradually on them. Glycation is a slow reaction (about 0.2% of Hb/day or 0.01% of amino groups/day) at normal (5 mM) blood glucose concentration. GlcHb accounts for ~24% of total hemoglobin at the end of the erythrocyte lifespan (~120 days), yielding an average of 12% GlcHb, including 4% HbA1c in a normoglycemic person.31 There is some evidence for differences in rates of glycation of proteins – the overall rate of glycation of albumin, for example, is estimated to be about 9-times faster than that of hemoglobin (Garlick and Mazer, 1983). The measurement of both HbA1c and glycated albumin can be used to determine an individuals long- and medium-term glycemia, respectively. HbA1c reflects blood glucose concentrations over the previous 6–8 weeks, whereas glycated albumin reflects the same over the previous 2–3 weeks.32 Alternatively, measurement of glycation of ApoB/LDL, a shorter-lived plasma protein, can be used to estimate mean blood glucose concentrations over the previous week.32
The extent of glycation of proteins varies from trace amounts of FL in short-lived proteins, to 20 – 30% glycated albumin,33 hemoglobin (tetramer), and collagen (monomer),34, 35 representing about 3 – 6 mmol FL/mol Lys in these proteins. There is a slight gradient in the extent of glycation of proteins vs. age; for both skin and articular collagen, for example, glycation increases from about 5 mmol FL/mol Lys at age 20 to 7 mmol FL/mol Lys at age 70.34, 35 A similar increase has been reported for mean HbA1c, from 3.8 to 4.4% between ages 20 and 70.36 These slight increases in glycation with age are commonly attributed to a decline in glucose tolerance (insulin sensitivity) and an increase in mean glycemia with age. Thus, glycation is not a function of age in a healthy population.
Advanced Glycation End-Products (AGEs)
The extent of glycation of a protein reaches a steady state, depending on the rate of formation of the Amadori product and its rate of reversal or conversion to other products; there are also deglycating enzymes that limit glycation of intracellular proteins.37 The nonenzymatic reverse hydrolysis reaction is straightforward, releasing glucose and its C-2 epimer mannose, without much consequence. The enzymatic reverse reaction involves phosphorylation of the Amadori product, followed by release of 3-deoxyglucosone. In contrast, the forward reactions are more complex, generating a wide range of reactive carbonyl and dicarbonyl compounds, both free and protein-bound, in non-oxidative and oxidative (glycoxidative) reactions analogous to those occurring in food systems during cooking. These intermediate glycation products (IGPs) may then react with proteins to form stable, irreversible adducts and crosslinks, known as AGEs. It is important to note here that, in addition to the traditional definition of glycation as a process leading to the formation of the Amadori product, it is common today to refer to glycation of proteins by dicarbonyl compounds. However, in this review, we consider glycation to be the reaction between glucose and amino groups on protein, and we will focus on the Amadori product as a gateway to formation of IGPs and AGEs.
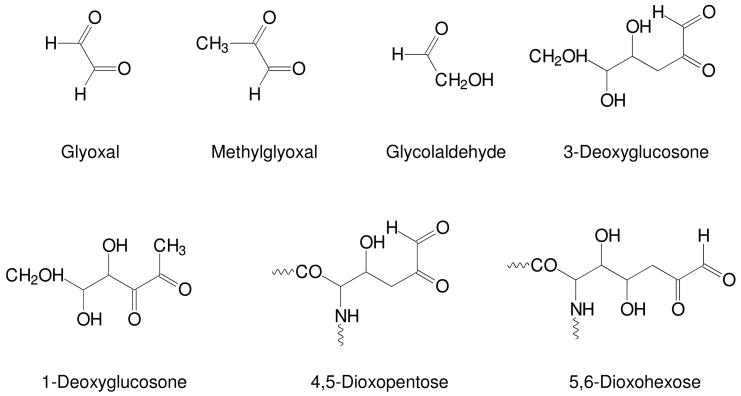
IGPs (Figure 3) include free glycolaldehyde and dicarbonyl compounds, such as 1- and 3-deoxyglucosones (DG), methylglyoxal (MGO) and glyoxal (GO). Protein-bound IGPs include 4,5-dioxopentose and 5,6-dioxohexose derivatives, indicating substantial carbonyl mobility along the carbohydrate chain.38 These dicarbonyl compounds serve as precursors for more complex AGE crosslinks, such as pentosidine and glucosepane.
Figure 3. Representative intermediate glycation products.
Oxidative decomposition of the Amadori product leads to the formation of a wide range of reactive carbonyl and dicarbonyl compounds. These intermediate glycation products (IGPs) include glyoxal, methylglyoxal, glycolaldehyde, 3-deoxyglucosone, 1-deoxyglucosone, 4,5-dioxopentose, and 5,6-dioxohexose. These IGPs can also be produced by autoxidation of glucose or, in the case of glyoxal and methylglyoxal, by peroxidation of lipids.
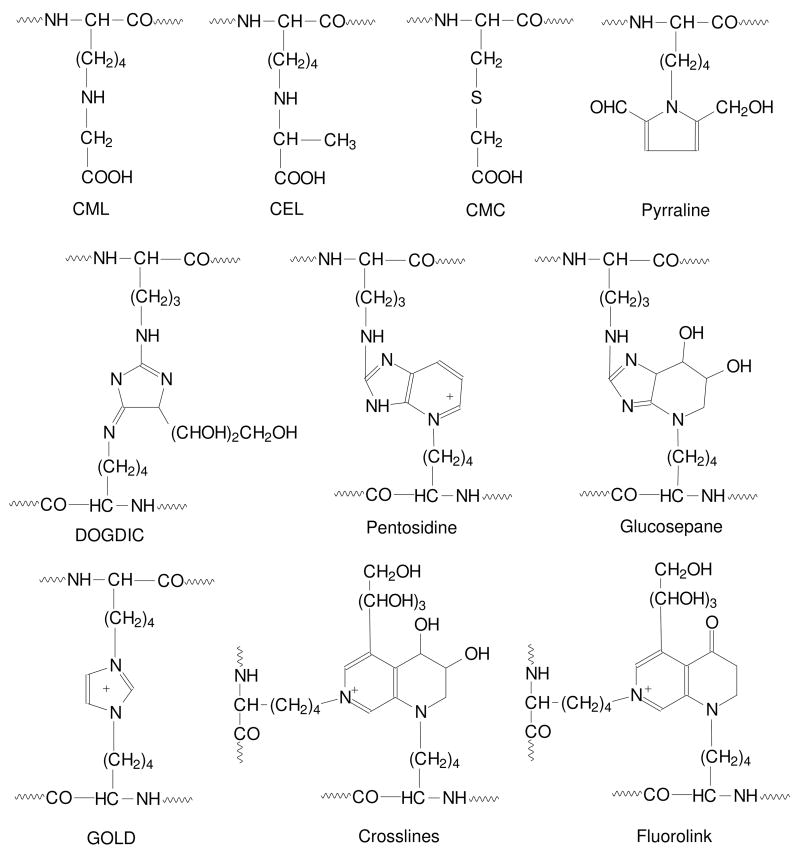
AGEs (Figure 4) include adducts to protein, such as Nε-(carboxymethyl)lysine (CML), Nε-(carboxyethyl)lysine (CEL) and pyrraline, and intra- and inter-molecular crosslinks, such as pentosidine, glucosepane and imidazolium compounds;39 among these, glucosepane is the major Maillard crosslink formed in proteins.40 All of these modifications involve lysine and/or arginine residues, plus the α-amino group in proteins. S-(carboxymethyl)cysteine (CMC) has also been identified in proteins; it is present at approximately the same level as CML in muscle protein,41 but no other S-AGEs have been identified. Because of their acid stability, CML and pentosidine are the most frequently measured AGEs. Despite the fact that these two compounds differ by over 100-fold in their concentrations in proteins, their concentrations are strongly correlated with one another and with concentrations of other AGEs in tissue proteins.
Figure 4. Representative advanced glycation end-products.
Oxidative decomposition of the Amadori product or reaction of tissue proteins with reactive carbonyl and dicarbonyl compounds can lead to the formation of advanced glycation end-products (AGEs). AGEs include Nε-(carboxymethyl)lysine (CML), Nε-(carboxylethyllysine) (CEL), S-(carboxymethyl)cysteine (CMC), pyrraline, 3-deoxyglucosone-derived imidazolium crosslink (DOGDIC), pentosidine, glucosepane, glyoxal lysine dimer (GOLD), crosslines, and fluorolink.
More than 20 AGEs have now been identified in tissue protein – most, if not all, of them are also present in the crust of bread and pretzels.42 Some of these compounds, such as pyrraline, pentosidine and glucosepane are clearly AGEs - they have 5 or 6 carbons derived exclusively from sugars. However, others such as CML and CEL are produced by multiple pathways. CML, for example, may be formed by oxidative cleavage of the Amadori product, or by reaction of glycolaldehyde or GO with lysine residues in protein. CML (and CEL) are also formed during lipid peroxidation reactions and during autoxidation of ascorbate. They are not unique biomarkers of glycation reactions and are sometimes described as AGE/ALEs, i.e. advanced glycation or lipoxidation end products.43 Similar reservations apply to imidazole derivatives formed from GO and MGO; the dicarbonyls may be derived from oxidation of Amadori products or lipids, or may be formed directly by decomposition of triose phosphates or by enzymatic degradation of serine or threonine. For these compounds, their origins may not be as important as their consequences, since AGEs and ALEs, and other protein oxidation and nitration products, tend to accumulate together in tissues often with detrimental effects in response to oxidative stress, diseases, and aging.44, 45
Glycation and AGEs in Aging and Disease
The most common symptom of aging at the molecular level is the accumulation of altered proteins, both intra- and extra-cellular.46 However, as noted above, glycation is relatively constant with age in a healthy population. In diabetes, it increases to a new steady-state level, roughly proportional to the increase in blood glucose concentration.47 In contrast, AGEs tend to accumulate in protein with age; this is true for AGEs formed by both non-oxidative (e.g. glucosepane) and oxidative (e.g. pentosidine) reactions. The highest concentrations of CML and pentosidine are found in long-lived proteins, such as lens crystallins and collagens. Crystallins, which are the longest-lived of body proteins, have the highest levels of CML, much of which is derived from ascorbate.48 Articular collagen has a higher concentration of AGEs, compared to skin collagen, consistent with the longer half-life of articular collagen.49
The accumulation of colored, fluorescent, and crosslinking AGEs contributes to the insolubilization of crystallins and the yellow color and decreased transparency of the lens with age. Through modification of collagen and elastin, AGEs affect the elasticity of the extracellular matrix (ECM),50–52 the binding of cells,53–55 and the turnover of ECM proteins.55 AGEs also interact with various cell receptors to induce oxidative stress and to activate inflammatory cascades. For example, the receptor for AGE (RAGE), a 35 KDa protein56 and multi-ligand member of the immunoglobulin superfamily of cell surface molecules,56,57 interacts with a diverse class of ligands, including AGEs, S100/calgranulins, amphoterin and amyloid-β peptide.58 This receptor is expressed on the surface of various cells including endothelial,59 kidney,60 and Caco-2 (human colon adenocarcinoma).61 Ligation of AGEs by RAGE induces the generation of reactive oxygen species (ROS) by an NAD(P)H oxidase. The free radicals in turn activate a Ras-MAP-kinase pathway which eventually leads to the activation and translocation of NF-κB.62 Induction of NF-κB leads to the transcriptional activation of many genes including a variety with roles in inflammation such as tumour necrosis factors (e.g. TNF-α) and interleukins 1, 6 and 8. This inflammatory cascade ultimately contributes to the increase in oxidative stress, which is implicated in the pathogenesis of a range of chronic diseases. While this sequence of RAGE-mediated reactions has been elaborated in numerous studies, there are still questions regarding the identity of the actual AGEs that initiate the inflammatory response, as well as evidence that the detailed interpretation of some studies is compromised by endotoxin contamination of AGE protein preparations.63, 64
The accumulation of AGEs is implicated in the development of vascular, renal, retinal and neural complications of diabetes.65–67 In particular, the results of the Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) study, a 10-year prospective study of protein glycation and diabetic complications, indicated that glycated and CML-modified skin collagen predicted the progression of retinopathy (p < 0.0001) and nephropathy (p = 0.0001) after adjustment for mean HbA1c.65 Since diabetes is characterized by dyslipidemia and hyperlipidemia, as well as hyperglycemia, lipoxidation reactions, or the interplay between glycoxidation and lipoxidation, may also contribute to the accumulation of AGE/ALEs, such as CML and CEL, in tissue proteins, both in diabetes and atherosclerosis.43 The chronic accumulation of these compounds in tissue proteins may provide the basis for the glycemic memory observed in the DCCT/EDIC trial, i.e. early periods of poor metabolic control increased the long-term risk for development of diabetic complications, despite institution of improved control later in life. In addition to their physical and biological effects, AGEs may also affect the chemistry of tissues. For example, MGO adducts to protein may catalyze the production of reactive oxygen species,68 and accumulation of MGO adducts in tissue proteins is implicated in the development of oxidative stress and damage.69–71
In addition to diabetes, there are other diseases characterized by accumulation of AGEs and oxidative stress in tissues. Uremia accelerates the accumulation of AGEs, in part because of compromised clearance of IGPs and AGE-peptides from circulation – a condition described as carbonyl stress.72 The accumulation of AGEs is also increased at sites of inflammation, e.g. the vascular wall in atherosclerosis and in plaque in neurodegenerative diseases.44, 45 There is also evidence that dietary AGEs may contribute to the accumulation of AGEs in tissues. Diets rich in AGEs affect vascular tone and renal function, exacerbating the development of pathologies in diabetes and uremia.73
Absorption, Metabolic Fate, and Bioactivity of Dietary Amadori Products and AGEs
Both Amadori products and AGEs have been investigated and reviewed74 for their absorption, metabolic rate, and bioactivity. Components may be (1) released from protein by digestive enzymes or gut bacteria and absorbed; (2) metabolized by bacteria in the gut; or (3) excreted in the feces. Absorbed components may be excreted in urine or accumulate in tissues without being metabolized. Alternatively, the products of their metabolism may accumulate in plasma or tissues or be excreted. It appears that the human body handles these compounds quite differently, no doubt due to the diversity of chemical structures involved.
Several studies suggest that dietary Amadori products are largely metabolized by the colonic microflora,75–77 but most work on the bioavailability and metabolic fate of dietary AGEs has involved CML and pyrraline. A study with rats showed that large amounts of dietary protein-bound CML were recovered in urine and feces; only low levels were recovered from organs. About half of the CML consumed was unaccounted for78 and may have been degraded by the colonic microflora or metabolized post-absorption. In humans, almost all (~80%) dietary pyrraline is absorbed and then rapidly excreted via the kidneys within 48 h without being metabolized.79 In contrast to CML and pyrraline, the bioavailability of pentosidine depends upon whether it is present in the diet in the free or protein-bound form. Absorption and excretion rates are much higher for the free form.80, 81
Unfortunately, there is a lack of well-designed studies concerning the bioactivity of dietary AGEs. One group of papers resulting from acute and chronic feeding studies with well-defined diets82–84 in both animals and human subjects report (1) positive correlations between both dietary and plasma AGEs and insulin resistance; (2) positive correlations between postprandial serum AGE levels and markers of inflammation; and (3) increased retention of dietary AGEs in plasma and tissues of renal patients compared to healthy controls. However, the minor components in these diets and antioxidant capacity of the high and low AGE diets were not determined but may be expected to differ. Also, there is uncertainty about the immunological methods used in these studies and doubts about the linearity of response of the assay over a wide AGE concentration range, the methodology for measuring AGEs in lipid-rich foods (e.g. butter), and the effects of other matrix components on response.
Some very recent studies on the possible effects of dietary AGEs or Maillard reaction products (MRPs) are beginning to shed light on bioactivity. A mouse study involving a low AGE mouse chow with and without supplementation with MGO-modified BSA led to increased serum levels of AGEs and increased plasma levels of isoprostanes.85 In contrast, a dietary intervention randomized crossover study involving healthy subjects fed either a low MRPs or a high MRPs diet for two weeks demonstrated that there was no effect on various plasma markers of oxidative damage or antioxidant defense.86 In fact, the high MRP diets possessed higher in vitro antioxidant activity, and post-intervention erythrocyte samples subjected to oxidation revealed higher antioxidant capacity following the high MRP diet. Thus, more well-designed studies are required to fully understand the in vivo effects of dietary AGEs and MRPs, particularly their role in stimulating the glycation and advanced glycation of endogenous proteins.
TRADITIONAL METHODS FOR MEASURING FL AND AGES ON TISSUE PROTEINS
In foods, a primary characteristic of the Maillard reaction is color formation due to the final reaction products. Thus, the simplest means of assessing the degree of reaction is to measure absorbance, transmission, or reflectance of light in the visible region. In vivo, color formation is not a primary symptom in most cases. Instead, an alternative is to measure fluorescence, which develops prior to color and is more sensitive. Various crosslinking AGEs, such as pentosidine, crosslines, and vesperlysines, are fluorescent (Figure 4).2
While absorption and fluorescence spectrophotometry may be used to follow the course of the Maillard reaction, they give no indication about the nature of specific reaction products. In contrast, liquid or gas chromatography (LC, GC) coupled with relatively non-specific detectors (e.g., fluorescence in the case of LC and flame ionization in the case of GC) allows the accurate quantification of FL and AGEs when authentic standards are employed. If LC or GC is coupled with mass spectrometry (MS) and stable isotope standards are utilized, then these compounds may be quantified with a high degree of specificity and sensitivity.47, 87–89
However, tissue proteins must be hydrolyzed to release free FL and AGEs before applying these chromatographic methods. Hydrolysis is most easily achieved by heating in strong acid (e.g., 6M HCl at 110°C for 24 h). A disadvantage to total protein hydrolysis is that FL and some AGEs, such as the hydroimidazolones, are not stable to these conditions. A modified approach is to reduce protein-bound Amadori products to their corresponding alcohols (e.g., with sodium borohydride) and to quantify hexitollysine after acid hydrolysis. Alternatively, hydrolysis procedures based on enzymatic digestion of proteins have been developed, enabling comprehensive measurement of both acid stable and labile molecules.47, 90
PROTEOMIC METHODS FOR MEASURING AMADORI PRODUCTS AND AGES ON TISSUE PROTEINS
While the overall levels of the Amadori product and AGEs in tissues can be measured by LC- or GC-MS, any information regarding the identity of the modified proteins and their sites of modification is lost. Knowledge of which proteins are modified and their extent of modification would provide new insight on the broad impact of protein modification, which may be more important than fasting blood glucose or HbA1c alone as a means of predicting risk for myocardial infarction.32 Analysis of specific proteins could lead to new and more sensitive markers of risk for or progression of diabetic complications.
In the following sections, we describe how MS-based proteomics approaches may be utilized in the analysis of the extent of protein glycation and AGE formation, using methodology specific to Amadori-modified protein as an example.
Enrichment of Glycated Proteins by Boronate Affinity Chromatography
As discussed above, the extent of glycation of proteins varies from trace amounts in short-lived proteins to 20 – 30% in long-lived proteins.33–35 However, by their very nature, MS-based proteomics methods tend to provide measurements of the most abundant proteins in a sample due to the finite dynamic range of the instrumentation and, to a lesser extent, the ionization suppression of low abundance proteins or peptides present in complex mixtures such as plasma, particularly when matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI) are employed.91–95 Thus, in order to obtain as comprehensive coverage of the glycated proteome as possible, Amadori-modified proteins should be separated from non-modified peptides prior to MS-based proteomics analyses in order to reduce the overall sample complexity.
Single and two dimensional gel electrophoreses (1- and 2-DGE) are widely used in proteomics research to separate proteins into bands or spots. Coupled with Western blotting with specific antibodies raised against the Amadori product on lysine or its reduced equivalent (hexitollysine), these size or charge-based protein separations can give an overview of the number of glycated proteins in a sample. Although not widely used to separate glycated from unmodified proteins, 1- and 2-DGE have been used to study the level of protein glycation in cerebrospinal fluid from individuals with Alzheimer’s disease,96 as well as in the analysis of glycated plasma proteins in type 2 diabetic subjects.97 Bands or spots corresponding to glycated proteins can be removed from the gels and subsequently analyzed using a MS-based proteomics approach. Although widely used, gel-based approaches for protein separation suffer from low throughput and moderate sensitivity and will not be discussed further.
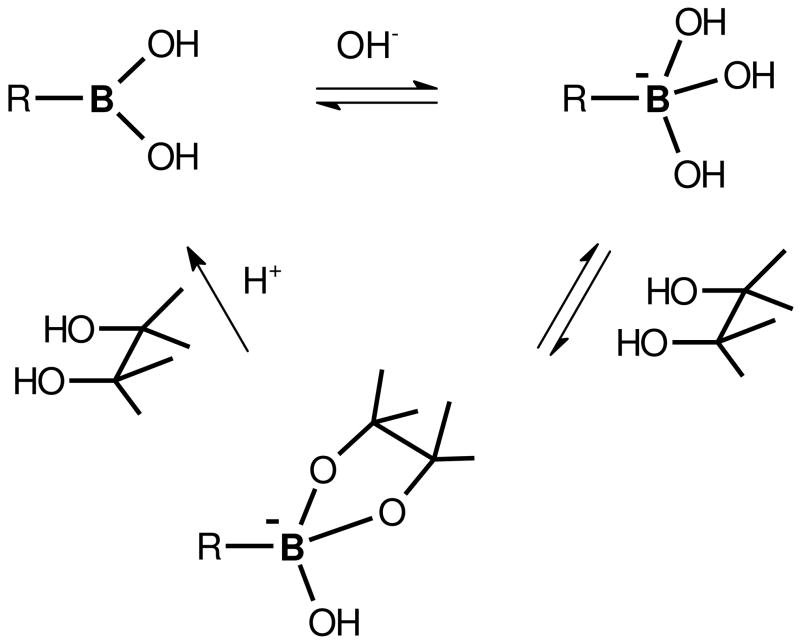
Boronate affinity chromatography is an alternative to gel-based methods for separating glycated from unmodified proteins and can be easily automated using commercial or in-house packed HPLC columns. Thus, boronate affinity chromatography represents a high throughput method for selective enrichment of glycated proteins from complex protein mixtures prior to MS-based proteomics analyses. This method relies on the highly specific and reversible interaction between the tetrahedral anion formed from boronic acid at alkaline pH and the 1,2-cis-diols typically found in sugars. Non-glycated species can be washed from the polymer-bound boronate resin, while retained glycated proteins can be eluted by lowering the buffer pH or by adding a source of competing hydroxyl groups, such as a sorbitol buffer (Figure 5).
Figure 5. The equilibria between boronic acid and CIS-diol-containing compounds.
Affinity attachment, elution, and regeneration of boronic acid are shown. Reproduced from the Journal of Proteome Research, 7, Zhang Q et al., Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry, pp. 2323–2330, 2007, with permission from the American Chemical Society.
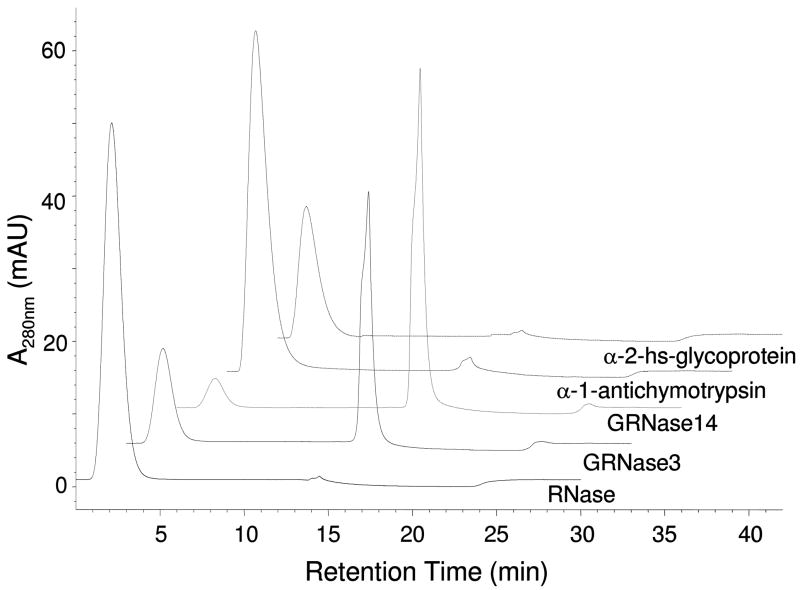
The utility of boronate affinity chromatography for enrichment of glycated proteins lies in its specificity. We recently evaluated the binding affinity of boronate resin for nonenzymatically glycated RNase and the enzymatically glycosylated proteins α-1-antichymotrypsin and α-2-hs-glycoprotein, which contain 6 N-linked and 2 N-linked and 3 O-linked glycans, respectively.98 The results showed that glycated RNase bound to the boronate resin, while the glycosylated proteins did not (Figure 6). Failure of the glycosylated proteins to bind to the boronate resin was attributed to electrostatic repulsion between the boronate anion and negatively charged sialic acid residues in the these proteins, as well as possible steric effects due to the complex structures of the larger glycans. In addition, the coplanar cis-diols present in the Amadori product likely bind with higher affinity to the boronate resin compared to the equatorial or non-coplanar cis-diols in the glycans.98
Figure 6. Boronate affinity chromatography with different standard proteins.
The traces are labeled with the respective sample name. GRNase 3, RNase lycated in vitro for 3 days; GRNase 14, RNase glycated in vitro for 14 days. Peaks at approximately 2 min are non-glycated proteins, and glycated proteins elute near 15 min. Reproduced from the Journal of Proteome Research, 7, Zhang Q et al., Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry, pp. 2323–2330, 2007, with permission from the American Chemical Society.
In practice, the use of boronate affinity chromatography is not without problems – steric factors may limit the binding of some glycated proteins and, depending on the ligand density, cationic and serine/threonine-rich proteins may bind non-specifically to the boronate resin. Nevertheless, boronate affinity chromatography was successfully used in a recent study to enrich glycated proteins from human serum. In this study, we evaluated a two-step enrichment scheme to selectively isolate first glycated proteins and then glycated, tryptic peptides from human serum glycated in vitro.98 This work indicated that the two-step enrichment procedure resulted in slightly better specificity compared to enrichment of only glycated, tryptic peptides. However, the two-step enrichment procedure also incurs more sample loss compared to enrichment on only the peptide level. Thus, when the initial sample amount is limited, enrichment of glycated, tryptic peptides is recommended over the two-step procedure.
In addition to boronate affinity chromatography, a boronate affinity chip was recently developed as a high throughput method for analysis of glycated proteins in conjunction with MALDI-time of flight (TOF) MS. Gontarev et al. used agarose hydrogel to create a high-capacity affinity layer on a modified aluminum surface to which 3-aminophenylboronic acid was immobilized as a ligand for affinity sorption of glycated proteins.99 The immobilized proteins were then analyzed directly using MALDI-TOF MS. The spot capacity of the affinity chip was quite high due to the three-dimensional gel structure, which allowed the researchers to obtain spectra with high abundance peaks and to analyze minor components of complex biological liquids.99
It is worth noting that the boronate affinity method is not specific to the Amadori product alone, but may also bind other forms of modified proteins, e.g. those containing deoxyglucosones and phosphorylated sugars. There is some evidence, based on chromatographic techniques, that glycation of hemoglobin increases in erythrocytes in response to oxidative stress.100–103 The fact that glycation is not an oxidative process suggests that other carbohydrates formed in oxidatively stressed erythrocytes may be reacting with protein. Isolation of these products by boronate affinity chromatography and analysis by MS-methods may provide a unique insight into the chemical consequence of oxidative stress.
Non-Sequencing MS Approaches for Measuring Protein Glycation
The characteristic mass shift incurred to protein by the Amadori product (Δm 162 Da) can be exploited for the identification of glycated proteins.104–106 Lapolla et al. used MALDI-TOF MS to quantify the degree of glycation of human serum albumin (HSA) in individuals with type 1 diabetes. They found a Δm of 439 to 2403 Da in HSA from 20 patients with poorly controlled diabetes, which roughly corresponds to the addition of 3 to 15 Amadori products.104 In contrast, HSA from 10 individuals with well controlled diabetes contained only 2 to 3 Amadori products, while HSA from healthy control subjects was essentially free of the Amadori modification.104 The same authors reported a similar trend for glycation of IgG in healthy and diabetic subjects, which raised doubts on the normal immunological function of IgG in the patients.106
The primary advantage of this approach, particularly when using MALDI-TOF MS, is high sample throughput and minimal sample pretreatment. However, the use of this method for the analysis of glycated proteins has several disadvantages. Many post-translational modifications, both enzymatic and nonenzymatic, may be present on the same protein; thus, the total mass shift of a modified protein will reflect the sum of all modifications. In addition, the mass shift characteristic of the Amadori product, Δm 162 Da, can also result from incomplete or variable incorporation of galactose or mannose residues into the oligosaccharides of glycosylated proteins. These disadvantages seriously complicate the analysis of glycated protein data. In addition, this approach does not result in any information regarding the actual modification site within the amino acid sequence of the protein.
Bottom-up Proteomics Approaches for Measuring Protein Glycation
Modern MS-based proteomics can be divided into two approaches: top-down and bottom-up. Top-down proteomics involves the ionization of intact proteins using either ESI or MALDI, followed by their gas-phase fragmentation by tandem MS (MS/MS) to yield the masses of both the protein precursor and its fragments.107 High mass resolution instrumentation, such as TOF or Fourier transform ion cyclotron resonance (FTICR) MS, is required for this approach in order to separate multiple isoforms of the same protein and, for proteins ionized by ESI, to resolve individual isotopes of highly charged proteins. To date, no studies involving top-down proteomics analysis of glycated proteins have been reported. Therefore, our discussion of MS-based proteomics will focus on bottom-up approaches.
In contrast to top-down proteomics, bottom-up proteomics involves the enzymatic or chemical cleavage of complex protein mixtures; the resulting peptide mixtures are then analyzed by MS.107 In order to minimize the effects of sample dynamic range, ionization suppression, and undersampling during data-dependent MS/MS analyses, the peptide mixtures are normally separated by LC prior to introduction to the mass spectrometer. Several approaches and technologies have been developed or applied for the identification of glycated peptides and their corresponding proteins using bottom-up proteomics. One method, peptide mass fingerprinting, relies only on MS analysis of glycated peptides; the remaining methods utilize MS/MS analysis of glycated peptides. All of these methods require enzymatic digestion of glycated proteins into their constituent peptides.
Peptide mass fingerprinting
Peptide mass fingerprinting methods are based on the mass shift (Δm 162 Da) imparted to peptides by the Amadori product. Both LC-MS108, 109 and MALDI-TOF MS110, 111 have been used in this approach, and glycated peptides are identified based on comparison of the observed peptide masses to the masses of peptides produced from in silico digestion. Using LC-MS-based peptide mass fingerprinting, Brock et al. determined that the major sites of glycation in RNase glycated in vitro were K41, K7, K1, and K37.108 While peptide mass fingerprinting has been used to identify certain specific glycation sites in incubations of model proteins with glucose, the identifications of glycation sites were ambiguous in many cases – based only on Amadori product-induced mass increases. This ambiguity presents a problem when enzymatic digestion results in peptides containing more than one lysine residue, each capable of carrying the Amadori modification. Therefore, it is difficult to extend this approach to more complex, proteome-level glycation analyses. However, because it has been traditionally difficult to obtain the sequence of glycated peptides using MS/MS approaches, peptide mass fingerprinting methods are still relied on to identify and quantify glycated peptides.
Collision-induced dissociation MS/MS
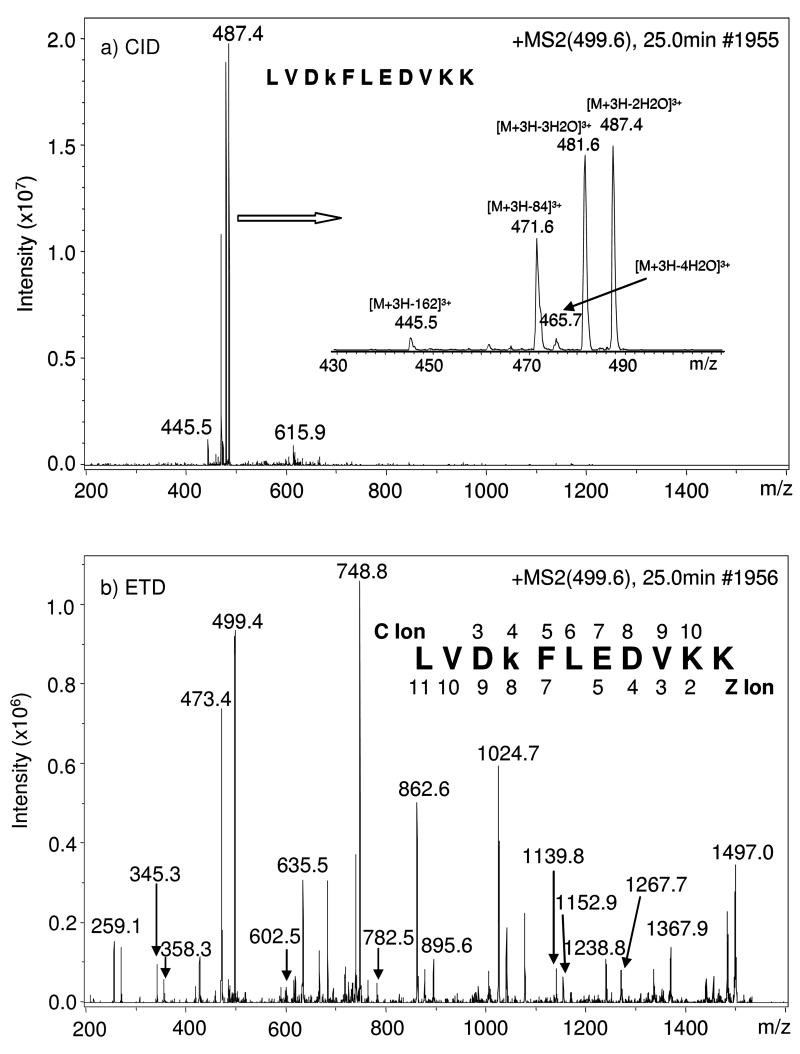
Recently, collision-induced dissociation (CID)-MS/MS was used to sequence glycated peptides. Unfortunately, high abundance ions corresponding to various neutral-losses (i.e. H2O, 2 H2O, 3 H2O, 4 H2O, 3 H2O + HCHO, and C6H10O5) dominated the MS/MS spectra and very limited and weak peptide backbone fragmentation was observed (Figure 7).112–114 During CID-MS/MS, intramolecular vibrational energy redistribution occurs prior to bond cleavage. Thus, the weakest bonds on the peptide side-chain modifications tend to dissociate preferentially, resulting in neutral-losses for modifications with labile functional groups (e.g. hydroxyl groups on the Amadori product). While certain neutral-losses (e.g. various neutral-losses of water in conjunction with formation of the furylium ion from neutral-loss of 3 H2O + HCHO) can be used to indicate the presence of a glycated peptide, information leading to a confident identification of the glycated peptide sequence will be unavailable.112–114
Figure 7. Comparison of spectra obtained under CID- and ETD-MS/MS.
MS/MS spectra obtained under CID (a) and ETD (b) fragmentation modes, respectively, of m/z 499.6, the [M + 3H]3+ of peptide LVDkFLEDVKK from α-1-antitrypsin precursor. “k” represents Amadori modification of lysine glucose. Inset in (a) is the zoom in view of the ions between m/z 445.5 and m/z 487.4; the identified c and z ions were labeled above and below the sequence in (b). Reproduced from the Journal of Proteome Research, 7, Zhang Q et al., Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry, pp. 2323–2330, 2007, with permission from the American Chemical Society.
To overcome the generation of neutral-losses and obtain sequence information during CID-MS/MS of glycated peptides, Brady et al. recently developed a new method involving the reduction of the Amadori product with sodium borohydride prior to enzymatic digestion and analysis of peptides.115 The borohydride reduced the ketone of the Amadori product, which stabilized the linkage between the peptide and sugar residue. As a result, MS/MS spectral quality was dramatically improved and the authors were able to identify a number of glycation sites that were distributed across both light and heavy chain subunits in force-degraded and antibody bulk-drug substances.115 A disadvantage of this method is that, in complex protein samples, various functional groups in other molecules could also be reduced, complicating the peptide identification process through the introduction of artifacts.
Precursor-ion scanning CID-MS/MS
FL generates an immonium ion at m/z 192.1, which is characteristic of the Amadori modification. This ion was recently exploited to map glycation sites on bovine serum albumin (BSA) using quadrupole-TOF (Q-TOF) MS and a precursor-ion scanning method; complete sequence coverage was obtained for Amadori-modified BSA peptides, based on singly and doubly charged furylium y-ions, as well as doubly charged pyrylium y-ions.113 However, due to the low mass cut-off limitation of all commercial cylindrical or linear ion trap instruments, this precursor-ion scanning method is limited for applications in bottom-up proteomics.
Alternatively, a neutral-loss of 162 Da, corresponding to the entire Amadori moiety, has been observed in addition to the consecutive neutral-losses of water during CID-MS/MS analysis of glycated peptides by Q-TOF MS at moderate collision energies.24 Because the neutral-loss of 162 Da is unique to glycated peptides, a prescreening and sequencing method comprised of two segments was developed based on a neutral-loss scan to identify sites of glycation in proteins. Peptides carrying the Amadori product were initially identified by neutral-loss of 162 Da during the first segment. In the second segment, identified glycated peptides were fragmented at higher collision energies in order to obtain peptide sequence and glycation site information. The two-segment neutral-loss method, in addition to retention time shift and accurate mass information, was then used to identify glycation sites in HSA; 31 of 59 total lysine residues were identified as partially glycated in vitro.24 Due to its selectivity, this method is capable of detecting and sequencing low-abundance glycated peptides that may coelute with nonglycated species.
While promising, both of the above precursor-ion scanning methods relied on Q-TOF instruments employing features unavailable on ion trap instruments; the latter are more commonly used for LC-MS-based bottom-up proteomics experiments. An additional limitation is that both were evaluated only for a single model protein; it is unclear how these approaches would perform with a complex peptide mixture produced from enzymatic digestion of biofluids or cell extracts.
Neutral-loss triggered MS3 and multi-stage activation CID-MS/MS
Alternative advanced CID-MS/MS methods have been developed to more fully utilize the MSn capability of ion trap instruments for sequencing peptides containing labile modifications. For example, data-dependent neutral-loss triggered MS3 (NLMS3) and multi-stage activation (MSA) methods were developed for the analysis of phosphopeptides, due to the intense neutral-loss of phosphoric acid produced by this modification during CID-MS/MS.116, 117 Data-dependent NLMS3 functions by first isolating a product-ion produced during the MS2 scan and corresponding to a specified neutral-loss, which is then activated and dissociated in the following MS3 stage. Similarly, MSA, or pseudo-MS3, was developed to avoid the loss of sequence informative ions from the backbone of peptides containing labile post-translational modifications in the MS2 stage. In this approach, a precursor-ion is first fragmented in the MS2 stage, and product-ions corresponding to specified neutral-losses are further activated and fragmented in the MS3 stage. The product-ions (MS2) from the initial precursor-ion and the activated neutral-loss product-ions (MS3) are then simultaneously stored, and a composite spectrum is generated containing ions from all of the activated precursors.
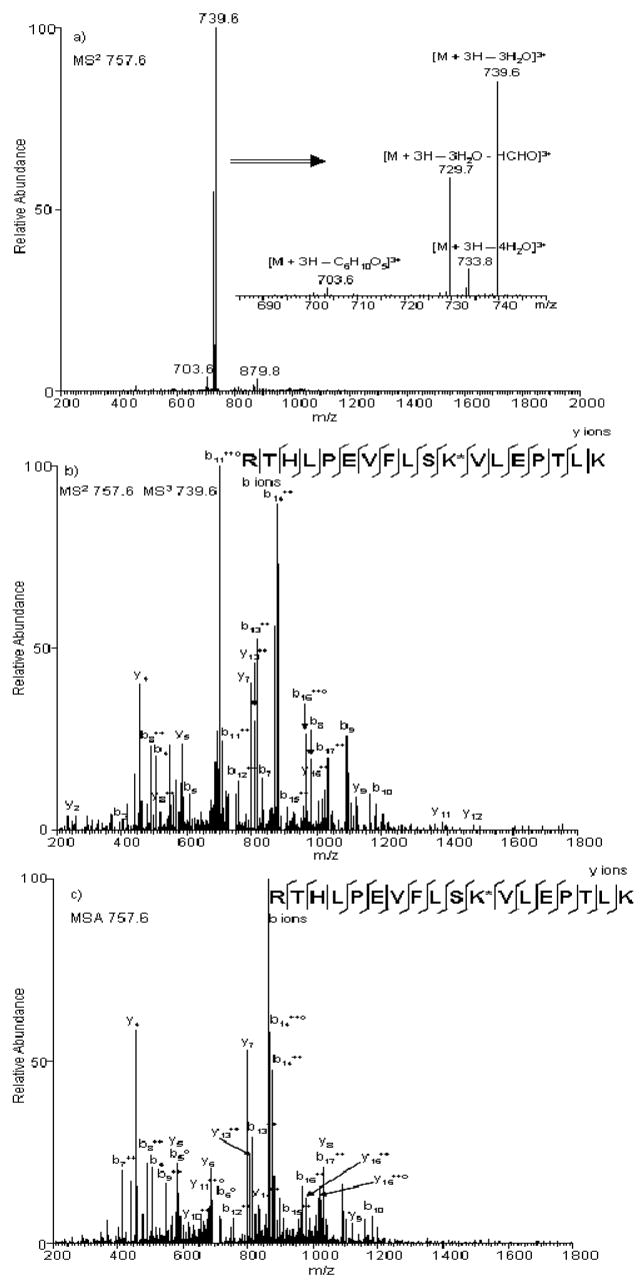
We recently evaluated different NLMS3 and MSA approaches during LC-MSn analyses of Amadori-modified peptides enriched from human serum proteins glycated in vitro.118 During neutral-loss triggered MS3 experiments, MS3 scans triggered by neutral-losses of 3 H2O or 3 H2O + HCHO produced similar results in terms of glycated peptide identifications (Figure 8). However, neutral-losses of 3 H2O resulted in significantly more glycated peptide identifications during MSA experiments. Overall, the MSA approach produced more glycated peptide identifications, while the NLMS3 approach resulted in much higher specificity. Almost all of the peptides identified by NLMS3 were Amadori-modified, which provided an additional level of confidence in the results. In addition, approximately 90% less time was required for database searching of NLMS3 datasets compared with MSA datasets, because only the MS3 scans are searched when using these approaches. In general, a significant amount of overlap in identifications using these two advanced CID-MS/MS strategies did not clearly indicate which method should be used when sequencing Amadori-modified peptides.
Figure 8. Comparison of spectra obtained under standard and advanced CID-MS/MS.
(a) Product-ion spectrum produced from CID-MS/MS fragmentation of the [M + 3H]3+ ion of peptide RTHLPEVFLSK*VLEPTLK, where * represents the Amadori modification site. Different neutral-losses are shown in the zoomed inset. Product-ion spectra produced from the [M + 3H]3+ ion of peptide RTHLPEVFLSK*VLEPTLK under b) NLMS3 triggered by neutral-loss of 3 H2O or c) MSA triggered by neutral-loss of 3 H2O, where * represents the Amadori adduct modification site.
Electron transfer dissociation MS/MS
The implementation of electron capture dissociation (ECD) on FTICR mass spectrometers represents a significant advance in tandem mass spectrometry.119 Being non-ergodic in nature, (i.e. bond dissociation occurs before energy redistribution) ECD provides more extensive sequence fragmentation and allows labile modifications to remain intact. It has been applied successfully for sequencing of peptides containing labile post-translational modifications, including large peptides and even intact proteins. Recently, electron transfer dissociation (ETD) fragmentation was developed by Hunt and colleagues using a modified linear ion trap mass spectrometer.120 ETD is similar to ECD but relies on aromatic anions as an electron vehicle. It has produced abundant peptide backbone c- and z-ions during MS/MS analyses of phosphopeptides, resulting in almost complete sequence coverage in the ETD fragmentation spectra. Like ECD, ETD is well-suited for the characterization of peptides containing labile post-translational modifications.
We reported the first application of ETD-MS/MS in the analysis of glycated, synthetic peptides.114 Contrary to what is typically observed during CID-MS/MS analysis of glycated species, ions corresponding to neutral-losses of water or furylium ion production were not observed in the ETD-MS/MS spectra. Instead, an abundant and almost complete series of c- and z-ions was observed (Figure 7). This study strongly suggests that ETD-MS/MS be especially useful for sequencing of glycated peptides, compared to the traditional method of CID-MS/MS; the labile Amadori compound remains intact during ETD-MS/MS, facilitating the identification of the peptide sequence and the Amadori modification site.
We further evaluated the performance of ETD- and CID-MS/MS during bottom-up proteomics analysis of a complex sample. Modified peptides produced from tryptic digestion of human serum glycated in vitro were enriched first on the protein level and then on the peptide level with boronate affinity chromatography, followed by LC-MS/MS analysis. ETD- and CID-MS/MS scans of the same precursor-ions were performed alternately, in order to provide a fair comparison between the two fragmentation techniques.98 ETD-MS/MS yielded significantly more glycated peptide identifications (156 identified spectra) compared with CID-MS/MS (15 identified spectra), based on automated MS/MS spectral identification. An additional advantage of ETD-MS/MS analysis of glycated peptides is higher confidence in the identifications; the scores from automated spectral identification were much higher for glycated peptides identified with ETD-MS/MS than with CID-MS/MS. This translated to a lower glycated peptide false discovery rate (FDR) for the species identified by ETD-MS/MS (mean FDR 3.1%) than by CID-MS/MS (mean FDR 6.7%).98 The difference in FDRs for the two techniques is due to the significantly better sequence coverage of glycated peptides obtained with ETD-MS/MS compared with CID-MS/MS. These results further demonstrate the usefulness of ETD-MS/MS for analysis of glycated peptides in bottom-up proteomics studies.
We recently reported the first high throughput, bottom-up proteomics study of glycated proteins in human plasma and erythrocyte membranes from individuals with normal glucose tolerance (NGT), impaired glucose tolerance (IGT), and type 2 diabetes mellitus (T2DM).121 Boronate affinity chromatography was used to enrich glycated proteins and glycated, tryptic peptides from both human plasma and erythrocyte membranes, followed by LC-ETD-MS/MS analysis in duplicate. This approach resulted in the identification of 260 and 75 unique glycated peptides (glycated peptide FDR <1%) from human plasma and erythrocyte membranes, respectively, corresponding to 76 and 31 unique glycated proteins – the largest number of unique glycated proteins yet identified in plasma proteins. Although most of the glycated proteins could be identified in samples from individuals with NGT, slightly higher numbers of glycated proteins and more glycation sites were identified in samples from individuals with IGT and T2DM.
While ETD-MS/MS is the most suitable technique for sequencing glycated peptides during bottom-up proteomics experiments, it is not without its own limitations. For example, it is less effective for doubly charged ions and for ions with m/z over 850.122, 123 In addition, it is not yet widely available on commercial mass spectrometers; in the absence of ETD-MS/MS, the NLMS3 and MSA methods discussed above would be viable alternatives during high throughput, bottom-up proteomics studies of glycated proteins.
FUTURE STUDIES ON THE RELEVANCE OF THE MAILLARD REACTION IN CHRONIC DISEASE
As discussed above, the accumulation of AGEs is implicated in the development of vascular, renal, retinal, and neural complications of diabetes65–67, as well as in the development of inflammation in atherosclerosis and neurodegenerative diseases.44, 45 However, the true impact of AGEs in chronic disease may not yet be fully understood, since traditional methods for their measurement result in the loss of any information regarding which proteins are modified, where those modifications occur, and to what extent. For example, the observation of an increase in CML in plasma proteins may not be useful if the majority of modification occurs on albumin. However, if several important signaling proteins are found to be modified at sites vital to their function, then their modification may have a greater impact on the development and progression of disease. We propose that future study of the Maillard reaction in chronic disease lies in advanced proteomics methods – particularly in bottom-up approaches. As demonstrated in the previous sections for Amadori-modified proteins, bottom-up proteomics methods can be used to definitively identify the sequence of the modified peptide, the site of the modification, and the corresponding protein which contains the modified peptide. The potential of bottom-up proteomics methods for analysis of the site specificity of protein modification will be discussed in the following section.
Site Specificity of Glycation and AGE Formation
Glycation is not a random reaction - amino groups on protein vary widely in their rate and extent of glycation. For hemoglobin, with 22 lysine residues per α/β dimer, the major sites of glycation in vivo are, in order: βV1, βK66, αK61, βK17, and αV1. Lysines 7, 16, 120, and possibly 144 are also glycated during incubation of carboxyhemoglobin with glucose in vitro.124 Thus, in addition to clear evidence of specificity of glycation, glycation in vitro is not necessarily the same as glycation in vivo. The specificity of glycation is determined by both the structure of the protein and endogenous ligands, and both acidic and basic neighboring groups affect the specificity of glycation of protein,125 either through effects on the pKa of the amino group, which enhances its nucleophilicity and the kinetics of formation of the Schiff base, or through catalysis of the Amadori rearrangement, which is the rate limiting step in glycation. Anionic ligands are also potent catalysts of glycation at specific sites on protein; the preferential, 10-fold greater glycation of βV1 vs. αV1 in Hb, despite their similar pKa, is attributed to catalysis of the Amadori rearrangement by 2,3-bisphosphoglycerate bound in the allosteric site near βV1; phosphate and bicarbonate exert similar effects on the specificity of glycation of hemoglobin in vitro.126 The anionic buffers phosphate, arsenate, and bicarbonate also catalyze the preferential glycation of K41 in the cationic, RNA-binding site of ribonuclease. Overall, glycation is less specific than glycosylation, which may be determined by an amino acid sequence in the protein, but glycation is not random. It often occurs at specialized sites on protein, e.g. the allosteric site of hemoglobin, the substrate binding site of RNase, and fatty acid and drug binding sites on albumin. While these modifications have a demonstrable effect on protein or enzyme function in vitro, there is little evidence that glycation itself is pathologically significant.
In contrast, accumulation of AGEs is strongly implicated in the pathogenesis of chronic disease. However, much less is known about the distribution of protein targets and site specificity of AGE formation. Methylglyoxal (MGO), a dicarbonyl intermediate and AGE precursor formed during the Maillard reaction, reacts primarily with arginine residues in protein, forming an imidazole derivative. Arg-410 was identified as a major site of modification of albumin by MGO in vitro and in vivo,127 while Arg-39 is the primary site of modification of RNase by MGO in vitro.128 Modification of Arg-410 inhibited the esterase activity of albumin, and Arg-39 is in the substrate binding/active site of RNase, suggesting that MGO may target functionally important residues in proteins.129 Indeed, two recent studies have reported MGO modification of arginine residues in the Arg-Gly-Asp and Gly-Phe-Hyp-Gly-Glu-Arg integrin-binding sites of type IV collagen, causing both endothelial cell detachment54 and inhibition of endothelial cell adhesion.53
While it is clear that glycation and AGEs increase in aging and disease, there are few studies on the specificity of AGE adduct and crosslink formation in the broader array of tissue proteins, yet this is where the action, i.e. pathology, takes place. As discussed above, most of what we know about glycation and AGE formation is based on studies with hemoglobin, albumin and RNase. Recent studies with RNase suggest that oxidation of the Amadori adduct is the primary source of the AGE CML in glycated proteins, i.e. that the specificity of carboxymethylation is the same as that of glycation.108 Similarly, both intra- and inter-molecular crosslinking of RNase by the AGE glucosepane occurs primarily at the lysine and arginine residues which are most reactive with glucose and MGO.128, 130 Although the specificity of AGE formation appears to track that of glycation, glycoxidation reactions depend on metal-catalyzed oxidation of glycated proteins. However, little is known in detail about the relationship between metal-binding sites, e.g. at histidine residues and anionic centers in protein, and the kinetics or specificity of formation of AGEs.
The application of bottom-up proteomics in a comprehensive study of protein glycation could yield additional information regarding the site specificity of modification of proteins by Amadori products and AGEs. The large dataset produced from such a study could be used in efforts to model hot spots for modification of proteins during Maillard reactions. Johansen et al. recently used artificial neural networks to investigate glycation of lysine residues in protein. From 89 glycated and 126 nonglycated lysine residues in 20 experimentally examined proteins, the authors trained a sequence-based glycation prediction algorithm.125 Application of the algorithm in an in silico evaluation of the human proteome revealed many important biological processes and molecular functions that could be affected by glycation. A comprehensive bottom-up proteomics study of protein modification by AGEs could lead to a similar predictive algorithm and also define the relationship between glycation and AGE formation.
Quantitation of Protein Glycation and AGE Formation in Clinical Samples Using Bottom-Up Proteomics
Bottom-up proteomics methods are increasingly gaining attention due to the power of these approaches for analyzing the complex protein mixtures present in clinical samples. However, the application of these methods for the analysis of AGE-modified protein in clinical samples will not be trivial.
Similar to protein glycation, AGEs will be present in low abundance on proteins in vivo. Therefore, some form of enrichment will be required in order to increase the concentration of modified proteins prior to analysis by LC-MS/MS or LC-MS methods. Due to their chemical diversity, it is unlikely that a single method will result in the enrichment of all AGEs. However, work by Vlassara and colleagues showed that lysozyme and lactoferrin bind proteins bearing AGEs.131 In later work, they demonstrated that AGE-modified proteins could be removed from diabetic uremic serum by affinity chromatography on an immobilized lysozyem column.132 Thus, lysozyme could be linked to a solid support and placed in an HPLC column for high throughput global enrichment of AGE-modified protein from clinical samples. Unfortunately, neither CML- nor pentosidine-modified BSA bound to the lysozyme column;132 free CML and free pentosidine also did not bind, indicating that these AGEs were not the major AGEs recognized by lysozyme. A more likely approach to the enrichment of AGE-modified protein will be by immunoaffinity methods, using either immobilized antibodies to specific AGEs or immobilized receptors, such as RAGE. Important considerations here will be the overall quality and specificity of the antibody, as well as its immobilization to a solid support for enrichment purposes.
In addition to the enrichment of AGE-modified proteins, the MS/MS analysis of AGE-peptides produced by enzymatic digestion of proteins may require special methods. For glycated peptides, we reported that either advanced CID-MS/MS (NLMS3 or MSA)118 methods or ETD-MS/MS98, 114 were amenable to sequencing glycated peptides due to the labile hydroxyl groups present on the Amadori product. It is conceivable that some AGEs may require similar approaches. For example, AGEs derived from either 1- or 3-deoxyglucosone may also carry labile hydroxyl groups, which fragment preferentially during traditional CID-MS/MS experiments. Other AGEs, such as CML, CEL, and pentosidine, are expected to be stable during CID-MS/MS. Crosslinks, such as the crosslines, fluorolink, and DOGDIC (Figure 4), will also require special considerations during automated identification of peptide MS/MS spectra by commercial software (e.g. SEQUEST, SpectrumMill, Mascot, X!Tandem).
Once a suitable method is established for the enrichment and MS/MS analysis of AGE-modified proteins, the next step is application of the method to the analysis of clinical samples. For plasma, the extreme dynamic range of protein concentration becomes a limiting factor for bottom-up proteomics approaches. Albumin, the most abundant plasma protein, accounts for ~60% of the plasma protein mass. This number increases to 96% when the 12 most abundant plasma proteins are considered and to ~99% when the 22 most abundant proteins are considered. Therefore, a measurement dynamic range of >107 is required for the detection of many clinically relevant proteins present at ng/mL levels or lower. The application of immunoaffinity multi-protein depletion (e.g. Agilent Mixed Affinity Removal System or Beckman-Coulter IgY-12 technology) is a common approach for increasing the dynamic range of bottom-up proteomics methods.133 We utilized a Beckman-Coulter IgY-12 immunodepletion column to deplete the 12 most abundant proteins in human plasma prior to the enrichment of glycated proteins. As a result, we were able to identify 76 unique glycated proteins, many previously undetected in the plasma glycated proteome. However, even after immunodepletion, we were only able to identify 76 glycated plasma proteins.121
To further increase the dynamic range of MS/MS-based proteomics experiments, multidimensional separations have been employed. These include 2-D LC, free-flow electrophoresis, solution-based isoelectric focusing, and gel-based approaches for fractionation at the intact protein level and at the peptide level.134–136 For example, Washburn et al. utilized multidimensional protein identification technology (MudPIT) for large scale proteomic analysis of Saccharomyces cerevisiae.137 This approach includes strong cation exchange (SCX) chromatography coupled with reversed-phase chromatography and greatly improves the peptide measurement dynamic range. Similarly, Liu et al. used 2D-LC-MS/MS in conjunction with immunodepletion to identify 300 plasma proteins.138 While such fractionation approaches have greatly enhanced the sensitivity of MS/MS-based proteomics studies, they exhibit significant limitations in both quantitation and throughput.
Although routinely used for peptide identification, data-dependent MS/MS has several inherent limitations, including the inability to select all chromatographic co-eluting species for fragmentation (“under sampling”) and the overall low duty-cycle of analysis (i.e. one MS scan followed by serial MS/MS scans). To overcome these limitations, Smith and colleagues developed an accurate mass and time (AMT) tag strategy that exploits capillary LC and high mass measurement accuracy MS (e.g. FTICR) to achieve high throughput proteomic analyses; this method includes high-resolution LC separations, large dynamic range of detection, and increased sensitivity.139 In the AMT tag approach, peptides from a tryptic digest of a complex protein mixture are first fractionated by SCX followed by reversed-phase capillary LC-MS/MS analysis of each SCX fraction. The calculated mass (based on the peptide sequence) and observed normalized elution time of each identified peptide is incorporated into a database of AMT tags; subsequent lower throughput LC-MS/MS analyses are no longer required and the AMT tag database essentially serves as a “look-up table” for subsequent LC-FTICR MS analyses of the same or similar samples. A peptide is identified from an LC-FTICR MS analysis when its measured mass normalized elution time match that of an AMT tag. This strategy can be extended to any high resolution mass spectrometer, such as TOF MS and the new generation of hydrid instruments. The AMT tag strategy provides a number of advantages over conventional MS/MS-based proteomics approaches, including (1) higher throughput by avoiding routine 2D-LC-MS/MS measurements, (2) increased quantification accuracy due to the higher resolution and signal-to-noise ratio of the FTICR measurements, and (3) enhanced sensitivity and dynamic range for greater coverage of the proteome. This strategy is amenable for high throughput quantitative studies of protein glycation and AGE formation in bottom-up proteomics, provided an AMT tag database of glycated peptides or AGE-modified peptides is developed.
CONCLUSIONS
We predict a shift in the methodology employed to measure protein glycation in clinical samples over the next few years, and the approach with highest risk and highest payoff is MS-based proteomics. This approach is high risk due to the investment of time and money for development of enrichment and MS/MS analysis methods; it is high payoff due to the potential identification of novel markers of impending or progressing disease. Indeed, bottom-up proteomics has proven to be especially useful for identifying proteins in the last decade, due to the advances made in reversed-phase LC separations of tryptic peptides and in MS instrumentation. Importantly, the bottom-up approach has been applied with success in the last two years in proteomics studies of glycated proteins.98, 114, 118, 121 However, the relatively small peptides generated during enzymatic digestion create a challenge: how to correlate the peptide level information to the protein level, particularly when dealing with post-translational modifications and protein alternative splice isoforms.
This challenge may be best met by top-down proteomics, an approach that has also seen great progress in recent years. If a sufficient number of informative protein fragment ions are observed, this method can potentially provide a complete description of the primary structure of the protein and reveal all of its modifications. To this end, McLafferty and coworkers have demonstrated highly informative fragmentation for proteins with molecular masses up to 200 kDa, using prefolding dissociation, heated vaporization, and separation of noncovalent and covalent bond dissociation.140 In addition, Kelleher et al. have successfully used the top-down approach to decipher the complex modification patterns of histones.141 The limitation of this method is its relative incompatibility with LC and the requirement of high resolution mass spectrometers, such as FTICR or Orbitrap MS.
Alternatively, middle-down proteomics may be a more practical method for analyzing proteins with post-translation modifications, including glycation, due to the recent improvement of ion fragmentation methods (e.g. ECD and ETD) and the introduction of user-friendly hybrid mass spectrometers (e.g. LTQ-Orbitrap and LTQ-FT).142, 143 The middle-down approach involves very limited digestion or chemical cleavage of proteins to produce relatively large peptides.
Although both top-down and middle-down proteomics are in their infancy relative to bottom-up proteomics, they will likely become important tools in the future for accurate determination and quantification of proteins with post-translational modifications and their isoforms. The bottom-up approach will continue to be used in glycated protein analyses during the next few years, but its application may fade.
No matter the proteomics method employed for the analysis of glycated and AGE-modified proteins, some form of enrichment of the target modification will be required in order to increase the sensitivity of the measurement. In addition, novel MS/MS approaches, such as ETD, may be necessary for labile AGEs.
Acknowledgments
This work was supported by NIH grants DK071283 to R.D.S. (PI) and T.O.M. (co-PI) and DK19971 to J.W.B. Portions of the work were performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility located at Pacific Northwest National Laboratory (PNNL) and supported by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
References
- 1.Maillard LC. Action of amino acids on sugars. Formation of melanoidins in a methodical way. Compt Rend. 1912;154:66–68. [Google Scholar]
- 2.Nursten HE. The Maillard Reaction. Chemistry, Biochemistry, and Implications. Royal Society of Chemistry; Cambridge: 2005. [Google Scholar]
- 3.Li S, Patapoff TW, Overcashier D, Hsu C, Nguyen TH, Borchardt RT. Effects of reducing sugars on the chemical stability of human relaxin in the lyophilized state. J Pharm Sci. 1996;85(8):873–877. doi: 10.1021/js950456s. [DOI] [PubMed] [Google Scholar]
- 4.Quan C, Alcala E, Petkovska I, Matthews D, Canova-Davis E, Taticek R, Ma S. A study in glycation of a therapeutic recombinant humanized monoclonal antibody: where it is, how it got there, and how it affects charge-based behavior. Anal Biochem. 2008;373(2):179–191. doi: 10.1016/j.ab.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Yang Y, Yuk I, Pai R, McKay P, Eigenbrot C, Dennis M, Katta V, Francissen KC. Unveiling a glycation hot spot in a recombinant humanized monoclonal antibody. Anal Chem. 2008;80(7):2379–2390. doi: 10.1021/ac701810q. [DOI] [PubMed] [Google Scholar]
- 6.Hodge JE. Chemistry of browning reactions in model systems. J Agric Food Chem. 1953;1(15):928–943. [Google Scholar]
- 7.Namiki M, Hayashi T, Kawakishi S. Free radicals developed in the amino-carbonyl reaction of sugars with amino acids. Agric Biol Chem. 1973;37:2935–2936. [Google Scholar]
- 8.Namiki M, Hayashi T. Development of novel free radicals during amino-carbonyl reaction of sugars with amino acids. J Agric Food Chem. 1975;23:487–491. [Google Scholar]
- 9.Nursten HE. Maillard browning reaction in dried food. In: MacCarthy D, editor. Concentration and Drying of Foods. Elsevier Applied Science; London: 1986. pp. 53–68. [Google Scholar]
- 10.Amadori M. Products of condensation between glucose and p-phenetidine. Atti Accad Naz Lincei. 1925;2:337–342. [Google Scholar]
- 11.Ames JM. The Maillard Reaction. In: Hudson BJF, editor. Progress in Food Proteins - Biochemistry. Elsevier Applied Science; London: 1992. pp. 99–153. [Google Scholar]
- 12.Ames JM. Nonenzymatic browning. In: Caballero B, Trugo L, Finglas P, editors. Encyclopedia of Food Sciences and Nutrition. Academi Press; London: 2003. pp. 665–672. [Google Scholar]
- 13.Ledl F, Schleicher E. New Aspects of the Maillard Reaction in Foods and in the Human-Body. Angew Chem Int Ed Engl. 1990;29(6):565–594. [Google Scholar]
- 14.Hofmann T. Characterization of chemical structure of novel coloured Maillard reaction products from furan-2-carboxaldehyde and amino acids. J Agric Food Chem. 1998;46:932–940. [Google Scholar]
- 15.Borrelli RC, Fogliano V, Monti SM, Ames JM. Characterization of melanoidins from a glucose-glycine model system. Eur Food Res Technol. 2002;215:210–215. [Google Scholar]
- 16.Borrelli RC, Visconti A, Mennella C, Anese M, Fogliano V. Chemical characterization and antioxidant properties of coffee melanoidins. J Agric Food Chem. 2002;50(22):6527–6533. doi: 10.1021/jf025686o. [DOI] [PubMed] [Google Scholar]
- 17.del Castillo MD, Ames JM, Gordon MH. Effect of roasting on the antioxidant activity of coffee brews. J Agric Food Chem. 2002;50(13):3698–3703. doi: 10.1021/jf011702q. [DOI] [PubMed] [Google Scholar]
- 18.Woffenden HM, Ames JM, Chandra S. Relationships between antioxidant activity, color, and flavor compounds of crystal malt extracts. J Agric Food Chem. 2001;49(11):5524–5530. doi: 10.1021/jf010583b. [DOI] [PubMed] [Google Scholar]
- 19.Woffenden HM, Ames JM, Chandra S, Anese M, Nicoli MC. Effect of kilning on the antioxidant and pro-oxidant activities of pale malts. J Agric Food Chem. 2002;50(17):4925–4933. doi: 10.1021/jf020312g. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y. Formation and reduction of acrylamide in Maillard reaction: a review based on the current state of knowledge. Crit Rev Food Sci Nutr. 2007;47(5):521–542. doi: 10.1080/10408390600920070. [DOI] [PubMed] [Google Scholar]
- 21.Andya JD, Maa YF, Costantino HR, Nguyen PA, Dasovich N, Sweeney TD, Hsu CC, Shire SJ. The effect of formulation excipients on protein stability and aerosol performance of spray-dried powders of a recombinant humanized anti-IgE monoclonal antibody. Pharm Res. 1999;16(3):350–358. doi: 10.1023/a:1018805232453. [DOI] [PubMed] [Google Scholar]
- 22.Quan CP, Wu S, Dasovich N, Hsu C, Patapoff T, Canova-Davis E. Susceptibility of rhDNase I to glycation in the dry-powder state. Anal Chem. 1999;71(20):4445–4454. doi: 10.1021/ac9900580. [DOI] [PubMed] [Google Scholar]
- 23.Zheng X, Wu SL, Hancock WS. Glycation of interferon-beta-1b and human serum albumin in a lyophilized glucose formulation. Part III: application of proteomic analysis to the manufacture of biological drugs. Int J Pharm. 2006;322(1–2):136–145. doi: 10.1016/j.ijpharm.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 24.Gadgil HS, Bondarenko PV, Treuheit MJ, Ren D. Screening and sequencing of glycated proteins by neutral loss scan LC/MS/MS method. Anal Chem. 2007;79(15):5991–9. doi: 10.1021/ac070619k. [DOI] [PubMed] [Google Scholar]
- 25.Fischer S, Hoernschemeyer J, Mahler HC. Glycation during storage and administration of monoclonal antibody formulations. Eur J Pharm Biopharm. 2008;70(1):42–50. doi: 10.1016/j.ejpb.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Kozlowski S, Swann P. Current and future issues in the manufacturing and development of monoclonal antibodies. Adv Drug Deliv Rev. 2006;58(5–6):707–722. doi: 10.1016/j.addr.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Townsend MW, DeLuca PP. Use of lyoprotectants in the freeze-drying of a model protein, ribonuclease A. J Parenter Sci Technol. 1988;42(6):190–199. [PubMed] [Google Scholar]
- 28.McDonald MJ, Shapiro R, Bleichman M, Solway J, Bunn HF. Glycosylated minor components of human adult hemoglobin. Purification, identification, and partial structural analysis. J Biol Chem. 1978;253(7):2327–2332. [PubMed] [Google Scholar]
- 29.Prome D, Blouquit Y, Ponthus C, Prome JC, Rosa J. Structure of the human adult hemoglobin minor fraction A1b by electrospray and secondary ion mass spectrometry. Pyruvic acid as amino-terminal blocking group. J Biol Chem. 1991;266(20):13050–13054. [PubMed] [Google Scholar]
- 30.Schwartz JG. The role of glycohemoglobin and other proteins in diabetes management. Diabetes Rev. 1995;3(2):269–287. [Google Scholar]
- 31.Bunn HF, Shapiro R, McManus M, Garrick L, McDonald MJ, Gallop PM, Gabbay KH. Structural heterogeneity of human hemoglobin A due to nonenzymatic glycosylation. J Biol Chem. 1979;254(10):3892–3898. [PubMed] [Google Scholar]
- 32.Misciagna G, De Michele G, Trevisan M. Non enzymatic glycated proteins in the blood and cardiovascular disease. Curr Pharm Des. 2007;13(36):3688–3695. doi: 10.2174/138161207783018545. [DOI] [PubMed] [Google Scholar]
- 33.Garlick RL, Mazer JS. The principal site of nonenzymatic glycosylation of human serum albumin in vivo. J Biol Chem. 1983;258(10):6142–6146. [PubMed] [Google Scholar]
- 34.Verzijl N, DeGroot J, Oldehinkel E, Bank RA, Thorpe SR, Baynes JW, Bayliss MT, Bijlsma JW, Lafeber FP, Tekoppele JM. Age-related accumulation of Maillard reaction products in human articular cartilage collagen. Biochem J. 2000;350(Pt 2):381–387. [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, Thorpe SR, Jenkins AJ, Shaw JN, Sochaski MA, McGee D, Aston CE, Orchard TJ, Silvers N, Peng YG, McKnight JA, Baynes JW, Lyons TJ. Advanced glycation end-products and methionine sulphoxide in skin collagen of patients with type 1 diabetes. Diabetologia. 2006;49(10):2488–2498. doi: 10.1007/s00125-006-0355-8. [DOI] [PubMed] [Google Scholar]
- 36.Kilpatrick ES, Dominiczak MH, Small M. The effects of ageing on glycation and the interpretation of glycaemic control in Type 2 diabetes. QJM. 1996;89(4):307–312. doi: 10.1093/qjmed/89.4.307. [DOI] [PubMed] [Google Scholar]
- 37.Van Schaftingen E, Delpierre G, Collard F, Fortpied J, Gemayel R, Wiame E, Veiga-da-Cunha M. Fructosamine 3-kinase and other enzymes involved in protein deglycation. Adv Enzyme Regul. 2007;47:261–269. doi: 10.1016/j.advenzreg.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Biemel KM, Conrad J, Lederer MO. Unexpected carbonyl mobility in aminoketoses: the key to major Maillard crosslinks. Angew Chem Int Ed Engl. 2002;41(5):801–804. doi: 10.1002/1521-3773(20020301)41:5<801::aid-anie801>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 39.Thorpe SR, Baynes JW. Maillard reaction products in tissue proteins: new products and new perspectives. Amino Acids. 2003;25(3–4):275–281. doi: 10.1007/s00726-003-0017-9. [DOI] [PubMed] [Google Scholar]
- 40.Biemel KM, Friedl DA, Lederer MO. Identification and quantification of major maillard cross-links in human serum albumin and lens protein. Evidence for glucosepane as the dominant compound. J Biol Chem. 2002;277(28):24907–24915. doi: 10.1074/jbc.M202681200. [DOI] [PubMed] [Google Scholar]
- 41.Alt N, Carson JA, Alderson NL, Wang Y, Nagai R, Henle T, Thorpe SR, Baynes JW. Chemical modification of muscle protein in diabetes. Arch Biochem Biophys. 2004;425(2):200–206. doi: 10.1016/j.abb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Henle T. Protein-bound advanced glycation endproducts (AGEs) as bioactive amino acid derivatives in foods. Amino Acids. 2005;29(4):313–322. doi: 10.1007/s00726-005-0200-2. [DOI] [PubMed] [Google Scholar]
- 43.Baynes JW, Thorpe SR. Glycoxidation and lipoxidation in atherogenesis. Free Radic Biol Med. 2000;28(12):1708–1716. doi: 10.1016/s0891-5849(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 44.Pamplona R, Portero-Otin M, Requena J, Gredilla R, Barja G. Oxidative, glycoxidative and lipoxidative damage to rat heart mitochondrial proteins is lower after 4 months of caloric restriction than in age-matched controls. Mech Ageing Dev. 2002;123(11):1437–1446. doi: 10.1016/s0047-6374(02)00076-3. [DOI] [PubMed] [Google Scholar]
- 45.Pamplona R, Naudi A, Gavin R, Pastrana MA, Sajnani G, Ilieva EV, Del Rio JA, Portero-Otin M, Ferrer I, Requena JR. Increased oxidation, glycoxidation, and lipoxidation of brain proteins in prion disease. Free Radic Biol Med. 2008 doi: 10.1016/j.freeradbiomed.2008.07.009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 46.Hipkiss AR. Accumulation of altered proteins and ageing: causes and effects. Exp Gerontol. 2006;41(5):464–73. doi: 10.1016/j.exger.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem J. 2003;375(Pt 3):581–592. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan X, Reneker LW, Obrenovich ME, Strauch C, Cheng R, Jarvis SM, Ortwerth BJ, Monnier VM. Vitamin C mediates chemical aging of lens crystallins by the Maillard reaction in a humanized mouse model. Proc Natl Acad Sci U S A. 2006;103(45):16912–16917. doi: 10.1073/pnas.0605101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verzijl N, DeGroot J, Thorpe SR, Bank RA, Shaw JN, Lyons TJ, Bijlsma JW, Lafeber FP, Baynes JW, TeKoppele JM. Effect of collagen turnover on the accumulation of advanced glycation end products. J Biol Chem. 2000;275(50):39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 50.Bruel A, Oxlund H. Changes in biomechanical properties, composition of collagen and elastin, and advanced glycation endproducts of the rat aorta in relation to age. Atherosclerosis. 1996;127(2):155–65. doi: 10.1016/s0021-9150(96)05947-3. [DOI] [PubMed] [Google Scholar]
- 51.Corman B, Duriez M, Poitevin P, Heudes D, Bruneval P, Tedgui A, Levy BI. Aminoguanidine prevents age-related arterial stiffening and cardiac hypertrophy. Proc Natl Acad Sci U S A. 1998;95(3):1301–6. doi: 10.1073/pnas.95.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konova E, Baydanoff S, Atanasova M, Velkova A. Age-related changes in the glycation of human aortic elastin. Exp Gerontol. 2004;39(2):249–54. doi: 10.1016/j.exger.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Pedchenko VK, Chetyrkin SV, Chuang P, Ham AJ, Saleem MA, Mathieson PW, Hudson BG, Voziyan PA. Mechanism of perturbation of integrin-mediated cell-matrix interactions by reactive carbonyl compounds and its implication for pathogenesis of diabetic nephropathy. Diabetes. 2005;54(10):2952–60. doi: 10.2337/diabetes.54.10.2952. [DOI] [PubMed] [Google Scholar]
- 54.Dobler D, Ahmed N, Song L, Eboigbodin KE, Thornalley PJ. Increased dicarbonyl metabolism in endothelial cells in hyperglycemia induces anoikis and impairs angiogenesis by RGD and GFOGER motif modification. Diabetes. 2006;55(7):1961–9. doi: 10.2337/db05-1634. [DOI] [PubMed] [Google Scholar]
- 55.Chong SA, Lee W, Arora PD, Laschinger C, Young EW, Simmons CA, Manolson M, Sodek J, McCulloch CA. Methylglyoxal inhibits the binding step of collagen phagocytosis. J Biol Chem. 2007;282(11):8510–20. doi: 10.1074/jbc.M609859200. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt AM, Vianna M, Gerlach M, Brett J, Ryan J, Kao J, Esposito C, Hegarty H, Hurley W, Clauss M, et al. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J Biol Chem. 1992;267(21):14987–14997. [PubMed] [Google Scholar]
- 57.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267(21):14998–15004. [PubMed] [Google Scholar]
- 58.Yan SF, Ramasamy R, Naka Y, Schmidt AM. Glycation, inflammation, and RAGE: a scaffold for the macrovascular complications of diabetes and beyond. Circ Res. 2003;93(12):1159–1169. doi: 10.1161/01.RES.0000103862.26506.3D. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt AM, Yan SD, Yan SF, Stern DM. The biology of the receptor for advanced glycation end products and its ligands. Biochim Biophys Acta. 2000;1498(2–3):99–111. doi: 10.1016/s0167-4889(00)00087-2. [DOI] [PubMed] [Google Scholar]
- 60.Skolnik EY, Yang Z, Makita Z, Radoff S, Kirstein M, Vlassara H. Human and rat mesangial cell receptors for glucose-modified proteins: potential role in kidney tissue remodelling and diabetic nephropathy. J Exp Med. 1991;174(4):931–939. doi: 10.1084/jem.174.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zill H, Gunther R, Erbersdobler HF, Folsch UR, Faist V. RAGE expression and AGE-induced MAP kinase activation in Caco-2 cells. Biochem Biophys Res Commun. 2001;288(5):1108–1111. doi: 10.1006/bbrc.2001.5901. [DOI] [PubMed] [Google Scholar]
- 62.Basta G, Schmidt AM, De Caterina R. Advanced glycation end products and vascular inflammation: implications for accelerated atherosclerosis in diabetes. Cardiovasc Res. 2004;63(4):582–592. doi: 10.1016/j.cardiores.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Valencia JV, Mone M, Koehne C, Rediske J, Hughes TE. Binding of receptor for advanced glycation end products (RAGE) ligands is not sufficient to induce inflammatory signals: lack of activity of endotoxin-free albumin-derived advanced glycation end products. Diabetologia. 2004;47(5):844–52. doi: 10.1007/s00125-004-1392-9. [DOI] [PubMed] [Google Scholar]
- 64.Buetler TM, Leclerc E, Baumeyer A, Latado H, Newell J, Adolfsson O, Parisod V, Richoz J, Maurer S, Foata F, Piguet D, Junod S, Heizmann CW, Delatour T. N(epsilon)-carboxymethyllysine-modified proteins are unable to bind to RAGE and activate an inflammatory response. Mol Nutr Food Res. 2008;52(3):370–8. doi: 10.1002/mnfr.200700101. [DOI] [PubMed] [Google Scholar]
- 65.Genuth S, Sun W, Cleary P, Sell DR, Dahms W, Malone J, Sivitz W, Monnier VM. Glycation and carboxymethyllysine levels in skin collagen predict the risk of future 10-year progression of diabetic retinopathy and nephropathy in the diabetes control and complications trial and epidemiology of diabetes interventions and complications participants with type 1 diabetes. Diabetes. 2005;54(11):3103–3111. doi: 10.2337/diabetes.54.11.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmed N, Thornalley PJ. Advanced glycation endproducts: what is their relevance to diabetic complications? Diabetes Obes Metab. 2007;9(3):233–245. doi: 10.1111/j.1463-1326.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- 67.Goh SY, Cooper ME. Clinical review: The role of advanced glycation end products in progression and complications of diabetes. J Clin Endocrinol Metab. 2008;93(4):1143–1152. doi: 10.1210/jc.2007-1817. [DOI] [PubMed] [Google Scholar]
- 68.Lee C, Yim MB, Chock PB, Yim HS, Kang SO. Oxidation-reduction properties of methylglyoxal-modified protein in relation to free radical generation. J Biol Chem. 1998;273(39):25272–8. doi: 10.1074/jbc.273.39.25272. [DOI] [PubMed] [Google Scholar]
- 69.Rosca MG, Mustata TG, Kinter MT, Ozdemir AM, Kern TS, Szweda LI, Brownlee M, Monnier VM, Weiss MF. Glycation of mitochondrial proteins from diabetic rat kidney is associated with excess superoxide formation. Am J Physiol Renal Physiol. 2005;289(2):F420–F430. doi: 10.1152/ajprenal.00415.2004. [DOI] [PubMed] [Google Scholar]
- 70.Rabbani N, Thornalley PJ. Dicarbonyls linked to damage in the powerhouse: glycation of mitochondrial proteins and oxidative stress. Biochem Soc Trans. 2008;36(Pt 5):1045–1050. doi: 10.1042/BST0361045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morcos M, Du X, Pfisterer F, Hutter H, Sayed AA, Thornalley P, Ahmed N, Baynes J, Thorpe S, Kukudov G, Schlotterer A, Bozorgmehr F, El Baki RA, Stern D, Moehrlen F, Ibrahim Y, Oikonomou D, Hamann A, Becker C, Zeier M, Schwenger V, Miftari N, Humpert P, Hammes HP, Buechler M, Bierhaus A, Brownlee M, Nawroth PP. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell. 2008;7(2):260–269. doi: 10.1111/j.1474-9726.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 72.Miyata T, van Ypersele de Strihou C, Kurokawa K, Baynes JW. Alterations in nonenzymatic biochemistry in uremia: origin and significance of “carbonyl stress” in long-term uremic complications. Kidney Int. 1999;55(2):389–399. doi: 10.1046/j.1523-1755.1999.00302.x. [DOI] [PubMed] [Google Scholar]
- 73.Peppa M, Uribarri J, Vlassara H. Aging and glycoxidant stress. Hormones (Athens) 2008;7(2):123–132. doi: 10.1007/BF03401503. [DOI] [PubMed] [Google Scholar]
- 74.Mori B, Nakatsuji H. Studies on Nutrition of Browned Protein1. Utilization in Rats of L-Lysine-C-14-Labeled Casein Browned by Amino-Carbonyl Reaction. Agric Biol Chem. 1977;41(2):345–350. [Google Scholar]
- 75.Finot PA, Magnenat E. Metabolic transit of early and advanced Maillard products. Prog Food Nutr Sci. 1981;5(1–6):193–207. [PubMed] [Google Scholar]
- 76.Faist V, Erbersdobler HF. Metabolic transit and in vivo effects of melanoidins and precursor compounds deriving from the Maillard reaction. Ann Nutr Metab. 2001;45(1):1–12. doi: 10.1159/000046699. [DOI] [PubMed] [Google Scholar]
- 77.Wiame E, Delpierre G, Collard F, Van Schaftingen E. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J Biol Chem. 2002;277(45):42523–42529. doi: 10.1074/jbc.M200863200. [DOI] [PubMed] [Google Scholar]
- 78.Faist V, Wenzel E, Erbersdobler HF. In vitro and in vivo studies on the metabolic transit of Nε-carboxymethyllysine. Czech J Food Sci. 2000;18:116–119. [Google Scholar]
- 79.Forster A, Henle T. Glycation in food and metabolic transit of dietary AGEs (advanced glycation endproducts): Studies on the urinary excretion of pyrraline. Biochem Soc Trans. 2003;31:1383–1385. doi: 10.1042/bst0311383. [DOI] [PubMed] [Google Scholar]
- 80.Forster A, Kuhne Y, Henle T. Studies on absorption and elimination of dietary maillard reaction products. Ann N Y Acad Sci. 2005;1043:474–481. doi: 10.1196/annals.1333.054. [DOI] [PubMed] [Google Scholar]
- 81.Miyata T, Ueda Y, Horie K, Nangaku M, Tanaka S, van Ypersele de Strihou C, Kurokawa K. Renal catabolism of advanced glycation end products: the fate of pentosidine. Kidney Int. 1998;53(2):416–422. doi: 10.1046/j.1523-1755.1998.00756.x. [DOI] [PubMed] [Google Scholar]
- 82.Koschinsky T, He CJ, Mitsuhashi T, Bucala R, Liu C, Buenting C, Heitmann K, Vlassara H. Orally absorbed reactive glycation products (glycotoxins): an environmental risk factor in diabetic nephropathy. Proc Natl Acad Sci U S A. 1997;94(12):6474–6479. doi: 10.1073/pnas.94.12.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vlassara H, Cai W, Crandall J, Goldberg T, Oberstein R, Dardaine V, Peppa M, Rayfield EJ. Inflammatory mediators are induced by dietary glycotoxins, a major risk factor for diabetic angiopathy. Proc Natl Acad Sci U S A. 2002;99(24):15596–15601. doi: 10.1073/pnas.242407999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hofmann SM, Dong HJ, Li Z, Cai W, Altomonte J, Thung SN, Zeng F, Fisher EA, Vlassara H. Improved insulin sensitivity is associated with restricted intake of dietary glycoxidation products in the db/db mouse. Diabetes. 2002;51(7):2082–2089. doi: 10.2337/diabetes.51.7.2082. [DOI] [PubMed] [Google Scholar]
- 85.Cai W, He JC, Zhu L, Chen X, Zheng F, Striker GE, Vlassara H. Oral glycotoxins determine the effects of calorie restriction on oxidant stress, age-related diseases, and lifespan. Am J Pathol. 2008;173(2):327–336. doi: 10.2353/ajpath.2008.080152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seiquer I, Ruiz-Roca B, Mesias M, Munoz-Hoyos A, Galdo G, Ochoa JJ, Navarro MP. The antioxidant effect of a diet rich in Maillard reaction products is attenuated after consumption by healthy male adolescents. In vitro and in vivo comparative study. J Sci Food Agric. 2008;88(7):1245–1252. [Google Scholar]
- 87.Alderson NL, Chachich ME, Youssef NN, Beattie RJ, Nachtigal M, Thorpe SR, Baynes JW. The AGE inhibitor pyridoxamine inhibits lipemia and development of renal and vascular disease in Zucker obese rats. Kidney Int. 2003;63(6):2123–2133. doi: 10.1046/j.1523-1755.2003.00027.x. [DOI] [PubMed] [Google Scholar]
- 88.Anderson MM, Heinecke JW. Production of N(epsilon)-(carboxymethyl)lysine is impaired in mice deficient in NADPH oxidase: a role for phagocyte-derived oxidants in the formation of advanced glycation end products during inflammation. Diabetes. 2003;52(8):2137–43. doi: 10.2337/diabetes.52.8.2137. [DOI] [PubMed] [Google Scholar]
- 89.Synold T, Xi B, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Advanced Glycation End Products of DNA: Quantification of N(2)-(1-Carboxyethyl)-2′-deoxyguanosine in Biological Samples by Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry. Chem Res Toxicol. 2008 doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delatour T, Fenaille F, Parisod V, Richoz J, Vuichoud J, Mottier P, Buetler T. A comparative study of proteolysis methods for the measurement of 3-nitrotyrosine residues: enzymatic digestion versus hydrochloric acid-mediated hydrolysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851(1–2):268–276. doi: 10.1016/j.jchromb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 91.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21(6):1104–15. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 92.Beaudry F, Vachon P. Electrospray ionization suppression, a physical or a chemical phenomenon? Biomed Chromatogr. 2006;20(2):200–205. doi: 10.1002/bmc.553. [DOI] [PubMed] [Google Scholar]
- 93.Tang KQ, Smith RD. Physical/chemical separations in the break-up of highly charged droplets from electrosprays. J Am Soc Mass Spectrom. 2001;12(3):343–347. doi: 10.1016/S1044-0305(01)00222-7. [DOI] [PubMed] [Google Scholar]
- 94.Tang KQ, Page JS, Smith RD. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2004;15(10):1416–1423. doi: 10.1016/j.jasms.2004.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cech NB, Enke CG. Relating electrospray ionization response to nonpolar character of small peptides. Anal Chem. 2000;72(13):2717–2723. doi: 10.1021/ac9914869. [DOI] [PubMed] [Google Scholar]
- 96.Shuvaev VV, Laffont I, Serot JM, Fujii J, Taniguchi N, Siest G. Increased protein glycation in cerebrospinal fluid of Alzheimer’s disease. Neurobiol Aging. 2001;22(3):397–402. doi: 10.1016/s0197-4580(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 97.Jaleel A, Halvatsiotis P, Williamson B, Juhasz P, Martin S, Nair KS. Identification of Amadori-modified plasma proteins in type 2 diabetes and the effect of short-term intensive insulin treatment. Diabetes Care. 2005;28(3):645–652. doi: 10.2337/diacare.28.3.645. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J Proteome Res. 2007;6(6):2323–30. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gontarev S, Shmanai V, Frey SK, Kvach M, Schweigert FJ. Application of phenylboronic acid modified hydrogel affinity chips for high-throughput mass spectrometric analysis of glycated proteins. Rapid Commun Mass Spectrom. 2007;21(1):1–6. doi: 10.1002/rcm.2793. [DOI] [PubMed] [Google Scholar]
- 100.Donde UM, Baxi AJ, el Tawil H, Ghwarsha K. Glycosylated haemoglobin in glucose-6-phosphate dehydrogenase deficiency. Lancet. 1985;(2):8445–47. doi: 10.1016/s0140-6736(85)90102-3. [DOI] [PubMed] [Google Scholar]
- 101.Jain SK, Palmer M. The effect of oxygen radicals metabolites and vitamin E on glycosylation of proteins. Free Radic Biol Med. 1997;22(4):593–596. doi: 10.1016/s0891-5849(96)00377-2. [DOI] [PubMed] [Google Scholar]
- 102.Selvaraj N, Bobby Z, Koner BC, Das AK. Reassessing the increased glycation of hemoglobin in nondiabetic chronic renal failure patients: a hypothesis on the role of lipid peroxides. Clin Chim Acta. 2005;360(1–2):108–113. doi: 10.1016/j.cccn.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 103.Sathiyapriya V, Bobby Z, Vinod Kumar S, Selvaraj N, Parthibane V, Gupta S. Evidence for the role of lipid peroxides on glycation of hemoglobin and plasma proteins in non-diabetic asthma patients. Clin Chim Acta. 2006;366(1–2):299–303. doi: 10.1016/j.cca.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 104.Lapolla A, Fedele D, Seraglia R, Catinella S, Baldo L, Aronica R, Traldi P. A new effective method for the evaluation of glycated intact plasma proteins in diabetic subjects. Diabetologia. 1995;38(9):1076–1081. doi: 10.1007/BF00402178. [DOI] [PubMed] [Google Scholar]
- 105.Lapolla A, Fedele D, Plebani M, Garbeglio M, Seraglia R, D’Alpaos M, Arico CN, Traldi P. Direct evaluation of glycated and glyco-oxidized globins by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 1999;13(1):8–14. doi: 10.1002/(SICI)1097-0231(19990115)13:1<8::AID-RCM438>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 106.Lapolla A, Fedele D, Garbeglio M, Martano L, Tonani R, Seraglia R, Favretto D, Fedrigo MA, Traldi P. Matrix-assisted laser desorption/ionization mass spectrometry, enzymatic digestion, and molecular modeling in the study of nonenzymatic glycation of IgG. J Am Soc Mass Spectrom. 2000;11(2):153–159. doi: 10.1016/S1044-0305(99)00134-8. [DOI] [PubMed] [Google Scholar]
- 107.Chen CH. Review of a current role of mass spectrometry for proteome research. Anal Chim Acta. 2008;624(1):16–36. doi: 10.1016/j.aca.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 108.Brock JW, Hinton DJ, Cotham WE, Metz TO, Thorpe SR, Baynes JW, Ames JM. Proteomic analysis of the site specificity of glycation and carboxymethylation of ribonuclease. J Proteome Res. 2003;2(5):506–513. doi: 10.1021/pr0340173. [DOI] [PubMed] [Google Scholar]
- 109.Ames JM. Application of semiquantitative proteomics techniques to the maillard reaction. Ann N Y Acad Sci. 2005;1043:225–235. doi: 10.1196/annals.1333.028. [DOI] [PubMed] [Google Scholar]
- 110.Farah MA, Bose S, Lee JH, Jung HC, Kim Y. Analysis of glycated insulin by MALDI-TOF mass spectrometry. Biochim Biophys Acta. 2005;1725(3):269–82. doi: 10.1016/j.bbagen.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 111.Wa C, Cerny RL, Clarke WA, Hage DS. Characterization of glycation adducts on human serum albumin by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2007;385(1–2):48–60. doi: 10.1016/j.cca.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J Am Soc Mass Spectrom. 2004;15(4):496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 113.Frolov A, Hoffmann P, Hoffmann R. Fragmentation behavior of glycated peptides derived from D-glucose, D-fructose and D-ribose in tandem mass spectrometry. J Mass Spectrom. 2006;41(11):1459–69. doi: 10.1002/jms.1117. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Q, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides. Rapid Commun Mass Spectrom. 2007;21(5):661–6. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brady LJ, Martinez T, Balland A. Characterization of nonenzymatic glycation on a monoclonal antibody. Anal Chem. 2007;79(24):9403–9413. doi: 10.1021/ac7017469. [DOI] [PubMed] [Google Scholar]
- 116.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76(13):3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 117.Yu LR, Zhu Z, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6(11):4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Q, Petyuk VA, Schepmoes AA, Orton DJ, Monroe ME, Yang F, Smith RD, Metz TO. Analysis of non-enzymatically glycated peptides: neutral-loss-triggered MS(3) versus multi-stage activation tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(19):3027–3034. doi: 10.1002/rcm.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 120.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Q, Tang N, Schepmoes AA, Phillips LS, Smith RD, Metz TO. Proteomic profiling of nonenzymatically glycated proteins in human plasma and erythrocyte membranes. J Proteome Res. 2008;7(5):2025–2032. doi: 10.1021/pr700763r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem. 2007;79(2):477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6(11):1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 124.Shapiro R, McManus MJ, Zalut C, Bunn HF. Sites of nonenzymatic glycosylation of human hemoglobin A. J Biol Chem. 1980;255(7):3120–3127. [PubMed] [Google Scholar]
- 125.Johansen MB, Kiemer L, Brunak S. Analysis and prediction of mammalian protein glycation. Glycobiology. 2006;16(9):844–853. doi: 10.1093/glycob/cwl009. [DOI] [PubMed] [Google Scholar]
- 126.Watkins NG, Neglia-Fisher CI, Dyer DG, Thorpe SR, Baynes JW. Effect of phosphate on the kinetics and specificity of glycation of protein. J Biol Chem. 1987;262(15):7207–7212. [PubMed] [Google Scholar]
- 127.Ahmed N, Dobler D, Dean M, Thornalley PJ. Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J Biol Chem. 2005;280(7):5724–5732. doi: 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- 128.Brock JWC, Cotham WE, Thorpe SR, Baynes JW, Ames JM. Detection and identification of arginine modifications on methylglyoxal-modified ribonuclease by mass spectrometric analysis. J Mass Spectrom. 2007;42(1):89–100. doi: 10.1002/jms.1144. [DOI] [PubMed] [Google Scholar]
- 129.Rabbani N, Thornalley PJ. The dicarbonyl proteome: proteins susceptible to dicarbonyl glycation at functional sites in health, aging, and disease. Ann N Y Acad Sci. 2008;1126:124–127. doi: 10.1196/annals.1433.043. [DOI] [PubMed] [Google Scholar]
- 130.Dai Z, Wang B, Sun G, Fan X, Anderson VE, Monnier VM. Identification of glucose-derived cross-linking sites in ribonuclease A. J Proteome Res. 2008;7(7):2756–2768. doi: 10.1021/pr700874a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li YM, Tan AX, Vlassara H. Antibacterial activity of lysozyme and lactoferrin is inhibited by binding of advanced glycation-modified proteins to a conserved motif. Nat Med. 1995;1(10):1057–1061. doi: 10.1038/nm1095-1057. [DOI] [PubMed] [Google Scholar]
- 132.Mitsuhashi T, Li YM, Fishbane S, Vlassara H. Depletion of reactive advanced glycation endproducts from diabetic uremic sera using a lysozyme-linked matrix. J Clin Invest. 1997;100(4):847–854. doi: 10.1172/JCI119600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5(10):1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3(7):1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 135.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3(4):311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 136.Lee HJ, Lee EY, Kwon MS, Paik YK. Biomarker discovery from the plasma proteome using multidimensional fractionation proteomics. Curr Opin Chem Biol. 2006;10(1):42–49. doi: 10.1016/j.cbpa.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 137.Washburn MP, Wolters D, Yates JR. 3rd, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 138.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5(11):2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25(3):450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314(5796):109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 141.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4(6):487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 142.Han X, Aslanian A, Yates JR. 3rd, Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.07.024. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone h3 variants. J Proteome Res. 2008;7(10):4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]