Abstract
Bacteria have evolved regulatory traits to rapidly adapt to changing conditions. Two principal regulatory mechanisms to modulate gene expression consist of regulation via alternative sigma factors and phosphorylation-dependent response regulators. PhyR represents a recently discovered protein family combining parts of both systems: a sigma factor-like domain of the extracytoplasmic function (ECF) subfamily linked to a receiver domain of a response regulator. Here we investigated the mode of action of this key regulator of general stress response in Methylobacterium extorquens. Our results indicate that PhyR does not act as a genuine sigma factor but instead controls gene expression indirectly through protein-protein interactions. This is evident from the analysis of additional proteins involved in PhyR-dependent gene regulation. We demonstrated that the ECF sigma factor-like domain of PhyR interacts with a protein, designated NepR, upon phosphorylation of the PhyR receiver domain. Using transcriptome analysis and phenotypic assays, we showed that NepR is a negative regulator of PhyR response. Furthermore, we provide biochemical and genetic evidence that NepR exerts this inhibitory effect through sequestration of the ECF sigma factor σEcfG1. Our data support an unprecedented model according to which PhyR acts as a mimicry protein triggering a partner-switching mechanism. Such a regulation of general stress response clearly differs from the two known models operating via σS and σB. Given the absence of these master regulators and the concomitant conservation of PhyR in Alphaproteobacteria, the novel mechanism presented here is most likely central to the control of general stress response in this large subclass of Proteobacteria.
Keywords: Alphaproteobacterium, two-component system, ECF sigma factor, PhyR
Bacterial responses to environmental stresses are mainly regulated at the level of transcription initiation (1). This is primarily carried out by alternative sigma factors, which allow promoter recognition and initiation of transcription as a subunit of RNA polymerase (2, 3). Among them, the large and diverse group of extracytoplasmic function (ECF) sigma factors plays a key role in adaptation to environmental conditions (4, 5). The activity of these ECF sigma factors is often negatively regulated by direct interaction with cognate anti-sigma factors, which prevent their association with the core RNA polymerase or facilitate holoenzyme dissociation (6–9).
Alternatively, regulation of gene expression is achieved by transcriptional regulators that bind DNA and modulate RNA polymerase holoenzyme recruitment at specific promoters (1). One large class of transcriptional regulators consists of response regulators of His-Asp phosphorelays (10–12). In classic systems, response regulators work together with a cognate sensor histidine kinase, and both proteins communicate via phosphorylation. Perception of a stimulus causes sensor autophosphorylation on a histidine residue; the phosphoryl group is then transferred to an aspartate residue of the response regulator, which modulates gene expression.
Both systems often result in the transduction of an external signal into the cytoplasm to regulate gene expression. The mechanisms from stimulus to gene regulation, however, are completely different in both systems. Interestingly, we recently identified an original regulator, PhyR, which combines domains of both systems: an amino terminal ECF sigma factor-like domain and a carboxy terminal receiver domain of a response regulator. This raises the possibility that an ECF sigma factor could be directly regulated by phosphorylation (13). Our previous work indicated that PhyR is a key regulator of the general stress response in the methylotrophic plant colonizer Methylobacterium extorquens. This protein is responsible for the activation of many stress-related genes, and a phyR deletion mutant shows drastic phenotypes when faced with various stressful conditions, including temperature shifts, oxidative stress, and desiccation (13, 14), which are all of relevance to the natural habitat of the bacteria (i.e., plant surfaces). Interestingly, PhyR is conserved in Alphaproteobacteria (13, 15), which altogether lack rpoS homologues, the gene encoding the well-known regulator of the general stress response first described in Escherichia coli (16, 17). This repartition may suggest that the phylogenetic group of Alphaproteobacteria uses PhyR to regulate the general stress response. How such a regulator activates stress genes is not known. Data from recent studies, however, provide indirect evidence for the involvement of additional proteins in a PhyR-dependent transduction cascade. Microsynteny is observed at the phyR locus of various Alphaproteobacteria: a small gene, rmq12793 (named nepR; see Results) in M. extorquens, is transcribed divergently to phyR (14). Except in Methylobacterium spp., this gene precedes an ECF sigma factor-encoding gene. In Sinorhizobium meliloti and Caulobacter crescentus, both ECF sigma factors, RpoE2 and SigT, respectively, have been shown to be involved in the general stress response (18, 19). Genes regulated by RpoE2, SigT, and PhyR possess conserved ECF-type promoters (14, 18, 19). In addition, the product of the nepR homologue in S. meliloti, smc1505, was described as a negative regulator of the ECF sigma factor RpoE2 response (18). These findings suggest that these 3 proteins act in the same signal transduction cascade.
In the present work, we investigated how PhyR activates its target genes and the role of these additional proteins in the PhyR signal transduction cascade in M. extorquens using complementary genetic and biochemical approaches. Our findings suggest that upon activation, PhyR dissociates the sigma/anti-sigma interaction (i.e., the interaction between the anti-sigma factor NepR and the ECF sigma factor σEcfG1). This mechanism represents the first example of the use of sigma factor mimicry in a partner-switching mechanism.
Results
Characterization of PhyR as a Response Regulator.
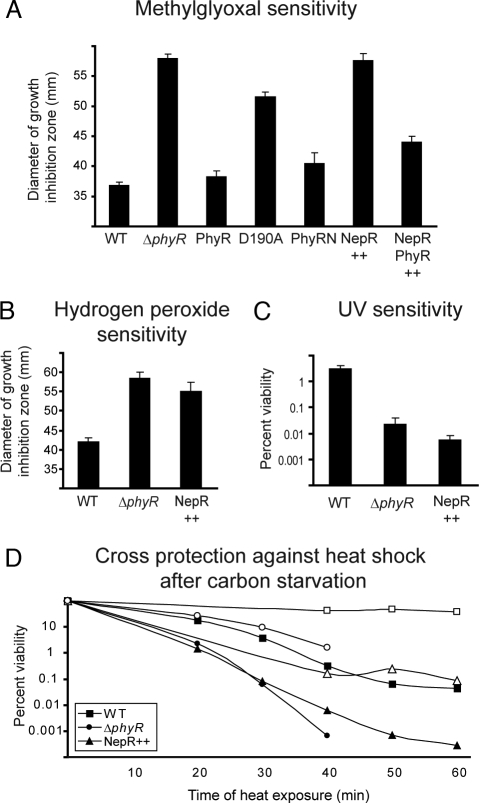
To characterize the mechanism of PhyR action, we first investigated whether PhyR was regulated by phosphorylation. Sequence analysis indicated that the PhyR receiver domain possessed the 5 highly conserved amino acids that constitute the active site of described response regulators (10, 20): Glu-144, Asp-145, Asp-190, Thr-218, and Lys-238 (corresponding to Asp-12, Asp-13, Asp-57, Thr-87, and Lys-109, respectively, in CheY of E. coli), with Asp-190 being the predicted site of phosphorylation. We changed aspartate residue 190 to an alanine and tested whether this allele could complement a phyR mutant. For this purpose, phyR and phyRD190A were cloned into pCM62 under the control of its native promoter and the resulting plasmids introduced into the ΔphyR mutant. Both strains were tested for their sensitivity to the electrophilic compound methylglyoxal in disk diffusion assays (14). As shown in Fig. 1A, expression of phyRD190A in the ΔphyR mutant did not restore the wild-type level of methylglyoxal resistance. This result suggests that phosphorylation of PhyR is required to activate the protein in vivo and that Asp-190 is the site of phosphorylation.
Fig. 1.
Phenotypic analysis of M. extorquens mutant and overexpressing strains. (A and B) Methylglyoxal and hydrogen peroxide resistance. Sensitivity was tested in disk diffusion assays. Data indicate the diameter of the growth inhibition zone and are the mean of 3 measurements from at least 3 independent experiments. Bars show the standard error. (C) UV tolerance. UV tolerance was tested by spotting a dilution series of exponential-phase cultures on plates and exposing the plates to UV light (254 nm). Data indicate the percentage viability after 40 s of UV exposure compared with nonexposed cells and are the mean of 3 independent experiments. Bars show the standard error. (D) Cross-protection against heat shock after carbon starvation. Cultures were subjected to heat shock (closed symbols) or exposed to overnight carbon starvation before heat shock (open symbols). Data of 1 representative experiment out of 3 independent experiments is shown. Strains are noted as follows: WT, wild type; ΔphyR; PhyR, ΔphyR/pCM62_phyR; D190A, ΔphyR/pCM62_phyRD190A; PhyRN, ΔphyR/pCM80_phyRNterm; NepR++, wild type/pCM80_nepR (rmq12793); NepR PhyR++, wild type/pCM80_nepR_phyR.
In some response regulators, the receiver domain inhibits the output domain, and this steric hindrance is suspended upon phosphorylation. Thus, such response regulators can be activated by removing their receiver domain (21–24). To test whether a similar mechanism was active for PhyR, we tested whether the N-terminal ECF sigma factor-like domain alone could complement a ΔphyR mutant. To this end, a truncated PhyR protein containing the first 132 aa was overproduced in a phyR mutant, which was then tested for its sensitivity to methylglyoxal in disk diffusion assays. As shown in Fig. 1A, the ECF sigma factor-like domain of PhyR restored the wild-type phenotype, indicating that this domain is active in vivo. This finding suggests that upon phosphorylation, PhyR undergoes conformational changes that release the inhibitory effect of the receiver domain on the output domain.
PhyR-Dependent Response Is Negatively Regulated by NepR.
The finding that PhyR is activated upon phosphorylation facilitates the study of the role of its output domain. We were unable to show a DNA-binding activity or an interaction with RNA polymerase of phosphorylated and unphosphorylated PhyR or of its N-terminal ECF sigma factor domain alone (data not shown). Therefore, we searched for other proteins that could be involved in the regulatory cascade directed by PhyR. As pointed out in the Introduction, nepR encodes a protein with unknown function and seemed to be a good candidate given that (i) it is conserved and found in synteny with phyR in Alphaproteobacteria (14), and (ii) the NepR homologue SMc01505 is a negative regulator of the ECF sigma factor RpoE2 in S. meliloti (18). Thus, this protein could represent a link between PhyR and another ECF sigma factor.
To analyze the role of this protein, a nepR-overexpressing strain was constructed and tested for its sensitivity to the broad spectrum of stresses that have been shown to affect a phyR mutant (14). As indicated in Fig. 1, this strain showed increased sensitivity to methylglyoxal (Fig. 1A), hydrogen peroxide (Fig. 1B), and UV exposure (Fig. 1C) compared with wild type. Furthermore, when exposed to drought stress for 3 days, the capacity of the nepR-overexpressing strain to form colonies was found to be 1,000-fold lower than wild type (data not shown). In addition, this strain was more sensitive to heat shock compared with wild type, and the protection against heat shock after carbon starvation seen in the wild type was partially lost when nepR was overexpressed (Fig. 1D). Thus, the phenotypes of the phyR mutant and the nepR-overexpressing strain were very similar, both qualitatively and quantitatively, involving the same stresses with comparable levels of sensitivity. These results suggest that NepR is a negative regulator of the PhyR-mediated response, and in light of this finding we named the protein NepR.
To confirm that PhyR and NepR regulate a common set of genes, we used microarray analyses to compare the transcriptional profile of the NepR-overexpressing strain with wild type and compared this regulon with the PhyR regulon, which we previously identified (14). Among 263 genes differentially regulated when NepR was overproduced, 114 were also part of the PhyR regulon (Table 1 and Table S1). These genes were found to be downregulated by NepR and upregulated by PhyR. This further supports the finding that NepR acts as a negative regulator of the PhyR response. Among the genes identified to be regulated by PhyR and NepR were several genes encoding typical stress proteins, including genes involved in oxidative stress (catalase), methylglyoxal resistance (lactoglutathione lyase), and heat shock response (RpoH) (Table 1). These findings are in agreement with the observed phenotypes.
Table 1.
Genes of PhyR, NepR, and σEcfG1 regulons*
| Function (gene homologue) | Gene | Fold change phyR OE/ΔphyR† | Fold change nepR OE/WT† | Fold change ΔecfG1/WT† |
|---|---|---|---|---|
| Detoxification/protection against stresses | ||||
| Catalase (katE) | RMQ09549 | 10.2 | −7.4 | −7.9 |
| Lactoylglutathione lyase (gloA) | RMQ01575 | 4.9 | −3.7 | −2.1 |
| Lactoylglutathione lyase (gloA) | RMQ02894 | 6.1 | −4.4 | −2.7 |
| Glutathione S-transferase (yqjG) | RMQ06706 | 4.7 | −2.2 | −3.2 |
| Glycogen debranching enzyme (glgX) | RMQ05904 | 7.0 | −4.2 | −4.4 |
| DNA protection during starvation protein (dps) | RMQ05258 | 12.1 | −7.7 | −5.7 |
| Regulation | ||||
| RNA polymerase σ 32 factor (rpoH) | RMQ12010 | 9.0 | −4.3 | −8.0 |
| Transcriptional regulator CRP/FNR family | RMQ08139 | 24.6 | −6.4 | −10.2 |
OE, overexpression; WT, wild type.
*Twenty-six additional genes are not presented in this table. For a complete list, see Table S1.
†Average changes of 3 experiments.
Both proteins also had specific regulons. The 133 genes specific to PhyR mainly encoded proteins of unknown function or putative enzymes with unassigned pathways (Table S1). Among 149 genes specific to the NepR regulon, we found genes encoding sulfur-oxidizing ability (25) and methanol oxidation to formaldehyde (26), as well as a region in the genome comprising approximately 50 genes, which seemed to correspond to a prophage (Table S1).
To further analyze whether PhyR and NepR belong to the same signaling cascade, we tested the possibility of suppressing the effects of nepR overexpression by coexpressing nepR and phyR. We constructed a pCM80_nepR_phyR plasmid, whereby both genes were under the control of the strong mxaF promoter, and introduced it in the wild-type strain. We then compared the methylglyoxal resistance of wild type, wild type/pCM80_nepR, and wild type/pCM80_nepR_phyR. As shown in Fig. 1A, the co-overexpression of phyR and nepR partially restored the wild-type phenotype relative to the overexpression of nepR alone. This finding clearly supports the hypothesis that both proteins act in the same signal transduction cascade.
Phosphorylated PhyR Interacts with NepR via Its ECF sigma Factor-Like Domain.
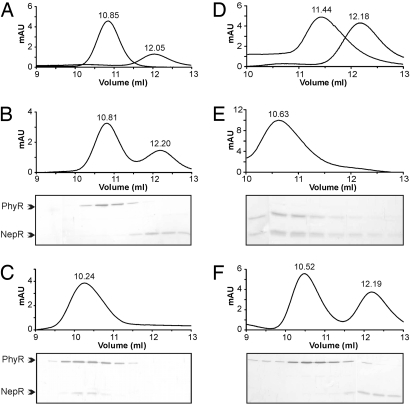
The functional relationship of PhyR and NepR could be explained by a physical interaction between the proteins. To test for the formation of such a complex in vitro, we performed analytic size-exclusion experiments. PhyR and NepR were overproduced in E. coli and purified by means of a histidine tag. When passed separately over a Superdex 75 10/300 column, the elution volume corresponded to an apparent molecular mass of PhyR of ≈32 kDa (predicted molecular mass, 29.5 kDa), suggesting that it was present as a monomer. An identical elution profile was obtained with phosphorylated PhyR using acetyl phosphate as phospho-donor (data not shown). Thus it can be concluded that PhyR remains in its monomeric form when phosphorylated. Under the same conditions, the elution profile of purified NepR (predicted molecular mass, 11.8 kDa) revealed an apparent molecular mass of ≈22 kDa, indicating homodimer formation (Fig. 2A). When nonphosphorylated PhyR was incubated with NepR, no complex formation between the 2 proteins was observed (Fig. 2B). PhyR phosphorylated by the addition of acetyl phosphate, however, directly interacted with NepR when both proteins were incubated in equimolar amounts before loading onto the column. The observed PhyR-P/NepR complex eluted with an apparent molecular mass of ≈42 kDa, indicating the formation of a heterodimer composed of phosphorylated PhyR and NepR (Fig. 2C). When we tested the PhyRD190A mutant, no interaction with NepR was observed, with or without the addition of acetyl phosphate (Fig. 2F). These results clearly show that binding of PhyR to NepR strongly depends on the phosphorylation status of PhyR in vitro and are in agreement with results obtained in vivo.
Fig. 2.
Phosphorylated PhyR directly interacts with NepR via its N-terminal ECF sigma factor-like domain. Protein complexes were resolved on a Superdex 75 10/300 column (flow rate, 0.5 mL/min). The elution volume is indicated above the absorption peak. Fractions were subsequently analyzed on a 15% SDS-polyacrylamide gel and silver stained. (A) The elution profile of purified PhyR and NepR (each 3 μM) passed separately over the column. (B) No interaction of PhyR and NepR was observed when incubated in equimolar amounts (each 3 μM) in the absence of acetyl phosphate. (C) Upon the addition of acetyl phosphate to the binding buffer (25 mM), the formation of a heterodimeric complex of phosphorylated PhyR and NepR was observed. (D) Separate gel filtration of the purified N-terminal ECF sigma factor-like domain of PhyR and NepR. (E) The ECF domain of PhyR and NepR showed a direct interaction, even in the absence of acetyl phosphate. (F) A PhyR D190A mutant could not form a complex with NepR in the presence of 25 mM acetyl phosphate.
To further characterize the interaction between PhyR and NepR, we tested whether the N-terminal ECF sigma factor domain of PhyR alone could bind to NepR. As showed in Fig. 2 D and E, when both proteins were incubated together, a heterodimeric complex of the N-terminal ECF sigma factor domain of PhyR (predicted mass, 18.9 kDa) and NepR was formed (apparent molecular mass of 35 kDa). This result indicates that the ECF sigma factor domain of PhyR is sufficient to interact with NepR. Altogether, these findings demonstrate that phosphorylation of PhyR enables interaction of its N-terminal ECF sigma factor-like domain with NepR.
NepR Is an Anti-Sigma Factor of σEcfG1.
The finding that PhyR acts through protein-protein interactions raises the question of how the incoming signal is transferred from PhyR and NepR to the level of transcriptional regulation. As mentioned in the Introduction, nepR homologues are located upstream of genes encoding the ECF sigma factors RpoE2 and SigT in S. meliloti and C. crescentus, respectively. In fact, many genes regulated by RpoE2, SigT, and PhyR possess the same ECF sigma factor-type promoter (14, 18, 19). Although no gene encoding a putative ECF sigma factor could be detected in the phyR genomic region in M. extorquens AM1, the complete genome sequence indicated the presence of several genes encoding putative ECF sigma factors. Thus, we pursued the possibility that an ECF sigma factor could be responsible for PhyR-dependent stress gene regulation in this Alphaproteobacterium and that NepR might exert its function as an anti-sigma factor. Among 14 putative ECF sigma factors, RMQ08147 (designated σEcfG1, G standing for general stress response) exhibited the highest sequence identity to RpoE2 of S. meliloti (18) and was therefore considered to be a prime candidate for a possible involvement in the general stress response in M. extorquens AM1.
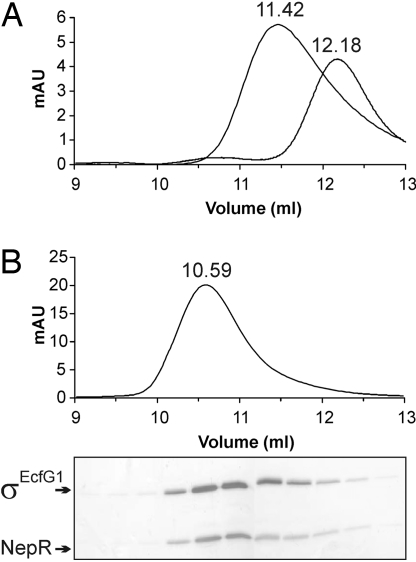
To address the question of whether NepR acts as an anti-sigma factor of σEcfG1, recombinant σEcfG1 containing an N-terminal histidine tag was purified from E. coli to study potential interactions with NepR by gel filtration. No interaction of NepR and full-length σEcfG1 was observed (data not shown). However, this could be due to solubility problems with purified σEcfG1, a problem that has been encountered for several sigma factors (27). In addition to the conserved regions 2 and 4 of sigma factors, σEcfG1 possesses an unusual ≈75-aa N-terminal extension. A truncated version of σEcfG1 missing the first 64 aa showed better solubility and was used for interaction studies. When incubated in equimolar amounts, NepR and truncated σEcfG1 formed a stable complex, which eluted from a Superdex 75 10/300 column corresponding to a molecular mass of 32 kDa, indicating heterodimer formation between NepR and σEcfG1 (predicted molecular mass: NepR, 11.8 kDa; truncated version of σEcfG1, 21.5 kDa) (Fig. 3). When σEcfG1, full-length or truncated version, was incubated with PhyR or phosphorylated PhyR no complex formation was observed (data not shown).
Fig. 3.
NepR forms a complex with σEcfG1. (A) Elution profile of purified σEcfG1 and NepR (4 μM) analyzed separately by gel filtration using a Superdex 75 10/300 column (flow rate, 0.5 mL/min). (B) Mixture of equimolar of each protein (8 μM each) led to the formation of a heterodimeric complex of σEcfG1 and NepR. Fractions were collected and analyzed on a 15% SDS-polyacrylamide gel and silver stained. The exact elution volume is indicated above the absorption peak.
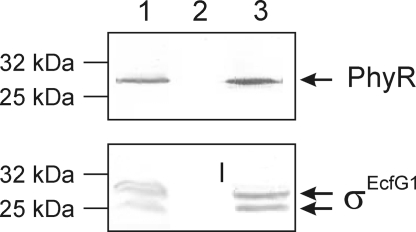
To test for an interaction of NepR with PhyR and σEcfG1 in vivo, NepR with a C-terminal histidine tag was overproduced in M. extorquens AM1/pCM80-nepRhis. When NepR and associated proteins were purified using Ni2+-activated agarose beads, both PhyR and σEcfG1 were detected in the elution fraction using specific antibodies against PhyR and σEcfG1, respectively (Fig. 4). Neither PhyR nor σEcfG1 were detected in the elution fraction for the negative control M. extorquens AM1/pCM80 (data not shown). In the case of σEcfG1, 2 specific bands were detected in Western blot analysis. N-terminal sequencing of the 2 σEcfG1 versions revealed that the upper band represented the full-length σEcfG1 with the annotated N terminus MRNDT, whereas the smaller protein was missing 14 aa at the N terminus (N terminus: TDGRP). This finding indicates that NepR is able to interact with full-length σEcfG1 in vivo.
Fig. 4.
NepR interacts in vivo with PhyR and σEcfG1. Recombinant NepR protein was overproduced in M. extorquens AM1/pCM80-nepRhis and purified by Ni-NTA via a C-terminal hexahistidine tag. Crude extract (lane 1), washing step (lane 2), and elution fraction (lane 3) were analyzed by Western blot using the polyclonal antibodies, anti-PhyR and anti-σEcfG1.
To test the involvement of σEcfG1 in the general stress response, we constructed an ecfG1 deletion mutant and tested its sensitivity to stresses. No increased sensitivity was observed upon exposure to methylglyoxal, hydrogen peroxide, or UV light (data not shown). To identify the genes regulated by σEcfG1, we used microarray analyses. We found 39 genes differentially regulated in the ecfG1 deletion mutant compared with wild type (Table S1); all but 1 were downregulated in the absence of ecfG1. A comparison of the PhyR, NepR, and σEcfG1 regulons indicated that essentially all σEcfG1-regulated genes are part of the PhyR and/or NepR regulons (Table 1 and Table S1). As shown in Table 1, genes found in all 3 regulons include genes involved in protection against stresses and genes encoding putative transcriptional regulators. Taken together, these findings suggest that NepR acts as an anti-sigma factor of σEcfG1, the latter being responsible for the regulation of a subset of PhyR-regulated genes.
Discussion
In the present study, we investigated the mechanism by which PhyR activates stress genes to trigger the general stress response in M. extorquens. Our results show that PhyR is a response regulator and is active in its phosphorylated form in vivo. Because no evidence of PhyR acting as a bona fide sigma factor was found, we investigated whether PhyR acts through protein-protein interaction and elucidated the role of 2 additional proteins in the regulatory pathway, NepR and σEcfG1. Our results indicate that the negative regulator of the general stress response NepR interacts with the ECF sigma factor σEcfG1 and the ECF sigma factor-like domain of phosphorylated PhyR. Thus our results are in agreement with a partner-switching mechanism in which NepR represents the anti-sigma factor of σEcfG1, and PhyR represents the antagonist or anti-anti-sigma factor (Fig. 5). According to this model, PhyR is inactive in unstressed cells, and the ECF sigma factor σEcfG1 is sequestered by its anti-sigma factor, NepR. In response to stress, PhyR is phosphorylated and interacts with NepR, thus releasing its interaction with σEcfG1, which can then associate with RNA polymerase to transcribe stress genes. Such a regulation involving a partner-switching mechanism to control the general stress response is reminiscent of the system found in Bacillus subtilis (28). That system consists of a sigma factor, SigB, an anti-sigma factor, RsbW, and an antagonist protein, RsbV, with protein communication occurring via serine-threonine phosphorylation. RsbW phosphorylates RsbV in growing cells to maintain RsbV in an inactive state. In response to stimuli, 2 phosphatases, RsbP and RsbU, dephosphorylate RsbV, which can dissociate the RsbW-SigB complex. Both the M. extorquens and the B. subtilis regulatory systems use phosphorylation/dephosphorylation mechanisms of the antagonist protein to achieve the release of the sigma/anti-sigma interaction. However, whereas in the B. subtilis system the anti-sigma factor phosphorylates the antagonist protein, there is no indication for a kinase activity of NepR, because NepR has no homology to RsbW and does not contain a conserved kinase domain. Thus we expect that the phosphoryl group is provided to PhyR by one or more not-yet-identified histidine kinases. Genes encoding histidine kinases are often found associated to the phyR locus in Alphaproteobacteria, which is, however, not the case in M. extorquens (14). Here, we propose that the anti-anti-sigma factor PhyR uses a sigma factor mimicry mechanism to compete with σEcfG1 for NepR binding. Although we cannot completely rule out that PhyR is a DNA-binding protein, sequence comparison of the PhyR sigma factor-like domain with ECF sigma factors suggests that this domain is a degenerate sigma factor, in which the region 2.4 involved in −10 binding is lost and the −35 binding region 4.2 is degenerate (Fig. S1; T. Mascher, unpublished data). Thus it may be assumed that only the determinants for the interaction with the anti-sigma factor are present, whereas those for DNA binding have been lost. Structure determination of NepR-PhyR and NepR-σEcfG1 complexes will be necessary to elucidate the binding sites of these interacting partners at the molecular level.
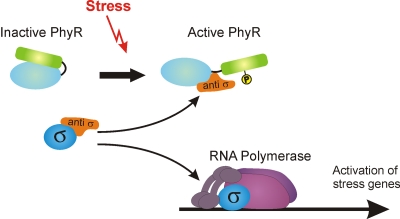
Fig. 5.
Model of partner-switching mechanism controlling general stress response in M. extorquens. In unstressed cells, PhyR is inactive, and the ECF sigma factor σEcfG1 is sequestered by its anti-sigma factor, NepR. In response to a stress, PhyR is phosphorylated and interacts with NepR, thus releasing σEcfG1 and allowing σEcfG1 to associate with RNA polymerase to transcribe stress genes. Note that additional σEcfG sigma factors are likely to be involved in PhyR dependent stress response (see Discussion for further details).
The next step in further characterizing this regulatory mechanism will be to demonstrate displacement of NepR from σEcfG1 by phosphorylated PhyR. In preliminary experiments, only the NepR-σEcfG1 complex was formed when NepR, σEcfG1, and PhyR were incubated together, even in the presence of acetyl phosphate (data not shown). As mentioned in Results, however, we could only show the interaction of σEcfG1 and NepR using the truncated form σEcfG1 Δ1–64. This truncated form apparently does not exist in vivo. The possibility that NepR interacts more strongly with σEcfG1Δ1–64 compared with the forms present in vivo could explain why PhyR cannot dissociate the complex. Surprisingly, this N-terminal extension shows similarity with NepR, leading to the hypothesis that some determinants for NepR binding in the σEcfG1 structure are hindered by the N-terminal domain interacting with the rest of the protein. On the basis of this sequence identity, even a direct interaction of PhyR with σEcfG1 is conceivable; however, this interaction was not observed in our in vitro studies. Again, structure data will be required to answer these questions as well as complementation experiments with σEcfG1 isoforms to understand the role of the N-terminal extension. In this context, it is interesting to note that although they are not typical, N-terminal extensions have been observed in other ECF sigma factors (29–31). In all of these studies, however, the role of the N-terminal extension has not yet been elucidated.
The observation that σEcfG1 regulates only a subset of PhyR-dependent genes suggests that additional sigma factors are part of the regulatory pathway. In support of this idea, M. extorquens AM1 possesses 5 other sigma factors belonging to the same subgroup of ECF sigma factors (T. Mascher, personal communication) that could fulfill this function. Further investigation of the regulon of each sigma factor will provide important insights with respect to the specificity and redundancy of these ECF sigma factors in M. extorquens. The possible involvement of several ECF sigma factors raises the question of whether NepR is the anti-sigma for all of them or whether several anti-sigma factors are involved. The observation that approximately 90 genes are PhyR and NepR dependent but not regulated by σEcfG1 suggests that NepR could control the activity of several sigma factors. In addition, the finding that specific regulons exist for PhyR and NepR supports the notion of a more complex regulatory network. The elucidation of this system together with the identification of proteins involved in the perception of activating stimuli will require more detailed analyses. Indeed, signal perception and transmission in the general stress response in the well-studied organisms E. coli and B. subtilis have proven to be highly complex (28, 32, 33).
In conclusion, central to the regulation of the general stress response in M. extorquens is the response regulator PhyR, which carries as its output domain a paralogue of an ECF sigma factor. This domain interacts with NepR, the anti-sigma factor of a bona fide sigma factor in a proposed model of partner switching. Such a core mechanism can be expected to exist in other Alphaproteobacteria as well. Thus, besides the well-described σS- and σB-dependent pathways, the PhyR-dependent cascade would represent a novel way to control general stress response in this large and diverse group of bacteria.
Experimental Procedures
Bacterial Strains and Growth Conditions.
Strains and plasmids used in this study are listed in Table S2. M. extorquens AM1 strains were grown at 28 °C on minimal medium (MM) (34) containing 18.5 mM sodium succinate and 120 mM methanol. E. coli DH5α was used for all cloning purposes, and E. coli BL21 (DE3) was used for overproduction of recombinant proteins. E. coli strains were cultivated aerobically in LB medium at 37 °C (DH5α) or 30 °C [BL21(DE3)]. When appropriate, the media contained kanamycin (50 μg/mL), ampicillin (100 μg/mL), or tetracycline (10 μg/mL).
Recombinant DNA Work.
The oligonucleotides used in this study are listed in Table S3 and were obtained from Microsynth. All DNA manipulations were performed according to standard protocols (35).
Strain Constructions.
For complementation and overexpression studies in M. extorquens AM1, the different genes were introduced in plasmid pCM62 under the control of their own promoter or in its derivative pCM80, which contains the strong pmxaF promoter (36).
To construct ecfG1 deletion mutant, the broad-host-range sacB-based vector for unmarked allelic exchange pCM433 was used (37). Plasmids were introduced into M. extorquens AM1 by electroporation (38) or were transferred between E. coli S17 (39) and M. extorquens AM1 by conjugative transfer (40). Details of strain constructions can be found in SI Experimental Procedures.
Phenotypic Assays.
For all assays, bacteria were grown at 28 °C in MM supplemented with methanol and succinate to an OD600 of 1. All phenotypic assays were performed as described previously (14).
RNA Sample Preparation and Transcriptome Analysis.
Overnight cultures of wild type/pCM80, wild type/pCM80_nepR, wild type, and ΔecfG1 were diluted 1:10 in fresh MM supplemented with methanol and succinate and grown to mid-exponential phase (OD600 = 1). RNA was prepared as described previously (14).
Sixty-mer oligonucleotide microarrays of M. extorquens AM1 were described previously (41). Hybridization probe preparation, microarray hybridization, scanning, and basic data analyses were performed by MOgene. A significance analysis of microarrays [1 class, default settings, false discovery rate (FDR) 0.5% and FDR 2% for NepR and σEcfG1 experiments, respectively] was performed on normalized data of 3 independent replicates. From this list, genes with a fold change average of at least 2.5 for NepR or 2 for σEcfG1 and an initial P value <0.005 for each replicate were considered to be significantly differentially expressed.
Overproduction and Purification of PhyR, NepR, and σEcfG1.
The PhyR, NepR, and σEcfG1 constructs were produced as His-tagged versions in E. coli BL21 (DE3) and purified using Ni-NTA and gel filtration. Detailed protein purification protocols can be found in SI Experimental Procedures. Purified PhyR, NepR, and σEcfG1 were used to raise polyclonal antibodies from rabbits (Neoclone and Biogenes).
Analytic Gel Filtration.
For complex formation, equimolar amounts of proteins (3 μM) were mixed in binding buffer [50 mM Tris/HCl (pH 8.0), 40 mM KCl, 5 mM MgCl2, 5% glycerol, and 1 mM DTT] and incubated at room temperature for 10 min. For phosphorylation of PhyR, 25 mM acetyl phosphate was added to the incubation buffer. Subsequently, samples (total volume, 500 μL) were loaded onto a Superdex 75 10/300 column (Amersham Pharmacia), equilibrated with buffer A [50 mM Tris/HCl (pH 8.0) and 150 mM NaCl] at a flow rate of 0.5 mL/min, using an Äkta purifier (Amersham Pharmacia). Apparent molecular masses were estimated by comparison to a low molecular mass standard (Amersham Pharmacia). After gel filtration, fractions were analyzed on a 15% SDS-polyacrylamide gel, and proteins were visualized by silver staining (42).
In Vivo Interaction Studies.
For in vivo interaction studies, recombinant NepR containing a C-terminal hexahistidine tag was overproduced in M. extorquens AM1 using the pCM80-nepRhis vector (Table S2). M. extorquens AM1 transformed with an empty pCM80 vector was used as a negative control. Both strains were grown in 1 L of minimal medium containing succinate and methanol and harvested at an OD600 of 2.0. For cell extract preparation, cells were washed once and resuspended in 8 mL of TNI5 buffer [20 mM Tris/HCl (pH 8.0), 150 mM NaCl, and 5 mM imidazol] supplemented with a protease inhibitor mixture (Complete, EDTA-free; Roche). Subsequently, the suspension was passed 3 times through a French pressure cell. After centrifugation (1 h, 4 °C, 100,000 × g), the supernatant was incubated with 0.5 mL of Ni-NTA agarose beads (Qiagen). The resin was then washed with 5 mL of TNI5 buffer before eluting bound proteins with 3 mL of TNI200 buffer (TNI5 with 200 mM imidazol). The fractions were separated on a 15% SDS-polyacrylamide gel, electrotransferred to a nitrocellulose membrane, and immunoblotted with anti-PhyR and anti-σEcfG1. Bound antibody was visualized with an alkaline phosphatase-conjugated goat antirabbit secondary immunoglobin (Bio-Rad).
Supplementary Material
Acknowledgments.
We thank Thorsten Mascher, University of Karlsruhe, for sharing unpublished data and for helpful discussion; and Nathalie Schmitt for technical assistance. This work was supported by the Swiss National Foundation, the European Molecular Biology Organization (fellowship to J.F.), and Eidgenössiche Technische Hochschule Zurich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810291106/DCSupplemental.
References
- 1.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 2.Murakami KS, Darst SA. Bacterial RNA polymerases: The wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 3.Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 5.Butcher BG, Mascher T, Helmann JD. In: Bacterial Physiology: A Molecular Approach. El Sharoud W, editor. Berlin: Springer Verlag; 2008. pp. 233–261. [Google Scholar]
- 6.Helmann JD. Anti-sigma factors. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 7.Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 8.Brooks BE, Buchanan SK. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim Biophys Acta. 2008;1778:1930–1945. doi: 10.1016/j.bbamem.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: A structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 11.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 12.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 13.Gourion B, Rossignol M, Vorholt JA. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci USA. 2006;103:13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourion B, Francez-Charlot A, Vorholt JA. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J Bacteriol. 2008;190:1027–1035. doi: 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galperin MY. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 17.McCann MP, Kidwell JP, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauviac L, Philippe H, Phok K, Bruand C. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J Bacteriol. 2007;189:4204–4216. doi: 10.1128/JB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Martinez CE, Lourenco RF, Baldini RL, Laub MT, Gomes SL. The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol Microbiol. 2007;66:1240–1255. doi: 10.1111/j.1365-2958.2007.06005.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee SY, et al. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- 21.Grimsley JK, et al. Subunit composition and domain structure of the Spo0A sporulation transcription factor of Bacillus subtilis. J Biol Chem. 1994;269:16977–16982. [PubMed] [Google Scholar]
- 22.Huala E, Stigter J, Ausubel FM. The central domain of Rhizobium leguminosarum DctD functions independently to activate transcription. J Bacteriol. 1992;174:1428–1431. doi: 10.1128/jb.174.4.1428-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn D, Ditta G. Modular structure of FixJ: Homology of the transcriptional activator domain with the -35 binding domain of sigma factors. Mol Microbiol. 1991;5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 24.Simms SA, Keane MG, Stock J. Multiple forms of the CheB methylesterase in bacterial chemosensing. J Biol Chem. 1985;260:10161–10168. [PubMed] [Google Scholar]
- 25.Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol. 2003;185:2980–2987. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Decatur A, Sorokin A, Helmann JD. The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 29.Bibb MJ, Buttner MJ. The Streptomyces coelicolor developmental transcription factor sigmaBldN is synthesized as a proprotein. J Bacteriol. 2003;185:2338–2345. doi: 10.1128/JB.185.7.2338-2345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakur KG, Joshi AM, Gopal B. Structural and biophysical studies on two promoter recognition domains of the extra-cytoplasmic function sigma factor sigma(C) from Mycobacterium tuberculosis. J Biol Chem. 2007;282:4711–4718. doi: 10.1074/jbc.M606283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dona V, et al. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor sigmaE in Mycobacterium tuberculosis. J Bacteriol. 2008;190:5963–5971. doi: 10.1128/JB.00622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 33.Klauck E, Typas A, Hengge R. The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci Prog. 2007;90:103–127. doi: 10.3184/003685007X215922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harder W, Attwood MM, Quayle JR. Methanol assimilation by Hyphomicrobium-sp. J Gen Microbiol. 1973;78:155–163. [Google Scholar]
- 35.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 36.Marx CJ, Lidstrom ME. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology. 2001;147:2065–2075. doi: 10.1099/00221287-147-8-2065. [DOI] [PubMed] [Google Scholar]
- 37.Marx CJ. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyama H, Anthony C, Lidstrom ME. Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol Lett. 1998;166:1–7. doi: 10.1111/j.1574-6968.1998.tb13175.x. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic-engineering—transposon mutagenesis in Gram-negative bacteria. Bio-Technology. 1983;1:784–791. [Google Scholar]
- 40.Fulton GL, Nunn DN, Lidstrom ME. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp strain AM1, a facultative methylotroph. J Bacteriol. 1984;160:718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okubo Y, Skovran E, Guo X, Sivam D, Lidstrom ME. Implementation of microarrays for Methylobacterium extorquens AM1. Omics. 2007;11:325–340. doi: 10.1089/omi.2007.0027. [DOI] [PubMed] [Google Scholar]
- 42.Nesterenko MV, Tilley M, Upton SJ. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J Biochem Biophys Methods. 1994;28:239–242. doi: 10.1016/0165-022x(94)90020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.