Abstract
The mechanisms responsible for the virulence of the highly pathogenic avian influenza (HPAI) and of the 1918 pandemic influenza virus in humans remain poorly understood. To identify crucial components of the early host response during these infections by using both conventional and functional genomics tools, we studied 34 cynomolgus macaques (Macaca fascicularis) to compare a 2004 human H5N1 Vietnam isolate with 2 reassortant viruses possessing the 1918 hemagglutinin (HA) and neuraminidase (NA) surface proteins, known conveyors of virulence. One of the reassortants also contained the 1918 nonstructural (NS1) protein, an inhibitor of the host interferon response. Among these viruses, HPAI H5N1 was the most virulent. Within 24 h, the H5N1 virus produced severe bronchiolar and alveolar lesions. Notably, the H5N1 virus targeted type II pneumocytes throughout the 7-day infection, and induced the most dramatic and sustained expression of type I interferons and inflammatory and innate immune genes, as measured by genomic and protein assays. The H5N1 infection also resulted in prolonged margination of circulating T lymphocytes and notable apoptosis of activated dendritic cells in the lungs and draining lymph nodes early during infection. While both 1918 reassortant viruses also were highly pathogenic, the H5N1 virus was exceptional for the extent of tissue damage, cytokinemia, and interference with immune regulatory mechanisms, which may help explain the extreme virulence of HPAI viruses in humans.
Keywords: 1918 pandemic, functional genomics, H5N1
Since 2003, the mortality for highly pathogenic avian influenza (HPAI) of the H5N1 virus subtype has been 63% of reported cases (www.who.int/csr/disease/avian_influenza/). Regardless of age or prior health, infected individuals have died within 10 days after onset of symptoms from a fast-progressing pneumonia, variably complicated by intestinal and CNS symptoms, often leading to respiratory distress syndrome and multiorgan failure (1–3). Mild infections or asymptomatic seroconversion in high-risk groups, such as health-care or poultry workers, has been rare (4). The extreme virulence and rapidly fatal clinical outcomes of human H5N1 virus infections are reminiscent of the 1918 pandemic virus, which reportedly caused up to 100 million fatalities (5).
Clinical and pathological features in H5N1- and 1918-infected humans and animal models (6–12) suggest that high levels of viral replication with early robust host responses play a key role in pneumonia severity and outcome. Recent nonhuman primate experiments comparing HPAI H5N1 and a reconstructed 1918 virus suggest many similarities in early host responses to these viruses. The goal of the present study, conducted in our well-characterized macaque and systems biology model of influenza (12–15), was to refine our understanding by using a 2004 human H5N1 Vietnam isolate and 2 1918 reassortant viruses possessing the 1918 hemagglutinin (HA) and neuraminidase (NA) surface proteins, based on the role surface glycoproteins play in the high virulence of the 1918 virus in the mouse model (6). One of the reassortants also contained the 1918 nonstructural (NS1) protein, an inhibitor of the host interferon (IFN) response (13, 16–22). This study revealed important similarities but also critical differences between the H5N1 and 1918-reassortant viruses, highlighting aspects of the host–pathogen interface caused by highly virulent influenza viruses.
Results
Differential Replication, Lung Tissue Tropism, and Inflammatory and Innate Immune Responses in Macaques Infected with H5N1 and 1918 Reassortant Influenza Viruses.
To perform a systematic comparison of several highly pathogenic influenza viruses, we inoculated cynomolgus macaques (8 animals per group) with either influenza A/Vietnam/1203/2004 (referred to as H5N1), 1918HA/NA:A/Texas/36/91 (1918HANA), 1918HA/NA/NS:A/Texas/36/91 (1918HANANS), or the parental H1N1 A/Texas/36/91 (Texas) virus. Inoculation was via the tracheal, nasal, conjunctival, and tonsillar routes as described (13–15). On days 1, 2, 4, and 7 postinfection (PI), 2 animals per group were euthanized. One H5N1 animal died between days 6 and 7, because of extensive pulmonary damage. Despite having similar pathology, the remaining animal lived until its endpoint on day 7. Finally, 2 mock-infected animals were euthanized on day 7. Viral load was quantified by median tissue culture infectious dose (TCID50) on 4 lung lobes from each animal (Fig. 1) and tissue distribution of virus antigen indicative of actively replicating virus (Fig. S1A). All showed that the H5N1 virus replicated to a much higher extent and with a wider distribution within the lungs compared with the other viruses, particularly at the early time points (days 1 and 2 PI). Interestingly, there was a rebound in viral replication after day 4 in the H5N1 animal euthanized on day 7. Although definitive conclusions cannot be based on a single animal, this may suggest failure of the host response to control infection, despite the relative lack of target cells. Moreover, intense viral antigen staining was observed in type II pneumocytes in the H5N1 virus-infected animals throughout the course of infection (Fig. 2). A semiquantitative assessment of infected cell types was performed, based on morphological and distribution criteria: (i) type II pneumocytes were identified as large, rounded to cuboidal cells with large, round nuclei and a vacuolated cytoplasm; and (ii) type I pneumocytes were identified as squamous cells with small round to oval nuclei and mostly imperceptible cytoplasm. This analysis revealed a ratio of type I to type II in the reassortant virus-infected animals of 11: 1 or higher; in the H5N1-infected animals the ratio was 1:3 or higher. In addition, more macrophage-like cells and endothelial cells appeared to be productively infected in the H5N1-challenged animals compared to the reassortant virus-challenged animals. In the Texas-infected animals, where only few virus positive cells were detected on days 1–4 PI, the majority of virus-antigen positive cells were macrophages. While not approaching the levels of the H5N1 virus, the 1918HANA and 1918HANANS viruses replicated to higher titers than the Texas virus, confirming the role of the 1918 viral glycoproteins in enhancing viral replication in the nonhuman primate model (Fig. 1). As in prior studies (13–15), the Texas infection was only detectable by real-time PCR and viral antigen staining in lung tissue.
Fig. 1.
Log (TCID50/mL) averaged from all lung lobes (4 out of 7, including left cranial, middle, caudal and accessory lobes) harvested. Detection limit of the plaque assay was 100 TCID50/mL. Note: Tissues from day 6 death were not suitable for viral isolation.
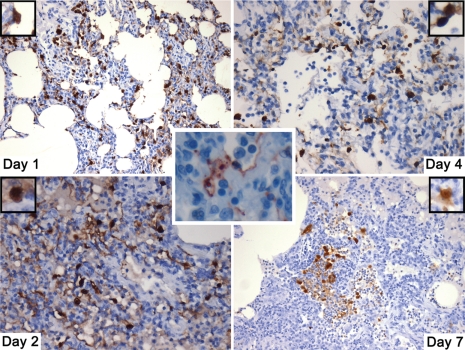
Fig. 2.
Influenza antigen staining in lung tissues shows prolonged targeting of type II pneumocytes during the H5N1 infection (50×). In contrast, the middle insert illustrates targeting of type I pneumocytes in 1 of the 1918 recombinant groups (150×).
The H5N1 virus replicated to moderate levels in the nasopharynx through day 2 PI (2.24 × 102 to 2.8 × 104 TCID50) and in trachea through day 7 PI (1.31 × 102 to 2.8 × 104 TCID50), demonstrating that the H5N1 virus can be detected in the upper respiratory tract of macaques. By immunolabeling, widespread H5N1 replication was also detected in tonsils on days 1–4 PI. Viral replication, as determined by the TCID50 assay, remained below the detection level (100 TCID50) in the upper respiratory tract of the other groups, with the exception of 1 animal in the Texas group on day 1. At several time points, the H5N1 virus was also found by a combination of viral culture and immunolabeling in tracheobronchial lymph nodes, but only to a small extent in macrophage- and dendritic cell-like cells in the retropharyngeal lymph nodes and spleen, and in 1 animal, killed on day 4, in cells of the leptomeninges. The presence of virus or viral antigens, however limited, in spleen and meninges suggests that viremia did occur, supported by detection of low levels of viral mRNA in blood samples of animals in the H5N1 group on days 1 and 2. Extrapulmonary spread of the virus through alternate routes, such as the gastrointestinal or olfactory tracts, did not appear to be a feature in these animals. This finding is consistent with the patterns observed in a number of human patients (1, 2, 23–27). In the reassortant 1918 and Texas virus groups, virus antigen was only found in lymph nodes and tonsils on day 1.
The H5N1 infection resulted in very high transcriptional induction in affected lung tissue (Fig. 3) of type I IFNs, including several IFNα variants and IFNβ; IFN-induced genes, such as IFN-induced proteins with tetratricopeptide repeats (IFITs); IFN-inducible guanylate binding protein (GBP); and IFN-stimulated exonuclease (ISG20). Additionally, the H5N1 group exhibited dramatic induction of genes important during the acute phase response, such as interleukin (IL)-1 and 6, tumor necrosis factor (TNF)-α, and several members of the complement pathway. Finally, genes coding for chemokines were also strongly and, in many cases, continuously induced in the H5N1 group. These genes included CXCL10, recently singled out as being highly expressed in H5N1-infected ferrets (28). While most of these genes were highly induced in the 1918 reassortant groups as well, particularly on days 1 and 2, expression in the H5N1-infected animals was often above the limit of detection of the array analysis software. Furthermore, in this group, a slight relative decline in induction on day 4 of some, but not all, of these genes, was followed by a rebound on day 7 to levels similar to that in early infection. The kinetics of this response were unique to the H5N1 group and likely due to a resurgence in viral replication in the animal still alive on day 7 and to the unremitting influx of immune cells, suggesting that the host response was unable to control the H5N1 infection.
Fig. 3.
Stronger IFN, inflammatory, and innate immune transcriptional induction in the H5N1 group. Heat maps were generated with genes of interest after performing a 1-way ANOVA (P ≤ 0.01, Benjamini-Hochberg FDR) with statistical cut-off criteria of a 5-fold change in at least 2 experiments (P ≤ 0.01). Up-regulation relative to the reference pool is in red, down-regulation in green, and lack or statistically nonsignificant regulation in the darker shades of these colors. *, this transcriptional profile is from a non-lesion area of the animal euthanized on day 7, since tissues from H5N1 day 6 death were not suitable for RNA isolation.
Taken together, these data show that the H5N1 virus resulted in a more productive infection than the other viruses in both the lower and upper respiratory tracts of macaques, with evidence of limited viremic dissemination and concurrently strong and protracted induction of genes relevant to IFN, inflammatory, and innate immune responses.
H5N1 Virus in Macaques Resulted in More Severe Clinical Disease and Pathology than H1N1 Viruses Containing 1918 HA, NA and/or NS Genes.
To closely monitor differences between experimental groups, animals were clinically scored twice daily (Table S1). Animals in the H5N1 group exhibited more severe clinical signs compared to those in the other groups within 24 h PI. Symptoms included anorexia, depression, coughing, diarrhea, a stress leukogram on day 1, and a death on day 6. A complete blood count revealed a thrombocytopenia from day 2 PI, worsening through day 7 when the platelet count reached a low of 139 × 109/L, a value comparable to that found in humans infected with H5N1 viruses (29). This thrombocytopenia is considered idiopathic despite a possible connection with viremia-induced damage to the vascular endothelium (30) and independent from occasional disseminated intravascular coagulation at the end-stage of the disease in humans. Animals in the H5N1 group also had dramatically higher levels of IL-6 in both serum and lung tissue and of TNF-α in lung tissue on days 1 and 2. While the H5N1 and 1918HANA virus groups had comparable lung levels of IFN-γ on day 1, the H5N1 group had much higher levels on days 2 and 7 (Fig. S2), the latter again corresponding to the resurgence of viral replication and the up-regulation of innate immunity genes. IL-2, -4, and -5 were also measured in serum and lung tissue by cytokine bead arrays, but failed to reveal significant differences among experimental groups. Trends with measured cytokines were corroborated by results obtained by microarray (Fig. 3) and real-time PCR analysis performed on lung samples with representative infection and pathology.
In both the H5N1 and 1918 reassortant virus groups, pathology findings showed that all lung lobes were affected by bronchopneumonia seen both grossly (consolidation and edema) and microscopically (bronchiolitis and alveolitis). However, pathology scores in the H5N1 group averaged 5.18 (SE 0.13) on a scale of 0 to 6, versus 3.2 (SE 0.17), 3.0 (SE 0.19), and 1.2 (SE 0.13) in the 1918HANA, 1918HANANS, and Texas group, respectively, over the 7 days of the study (Fig. S1B). A Mac387 stain, specific for myeloid lineage cells, revealed extensive neutrophil and macrophage infiltration—a consistent feature with HPAI viruses in all models studied to date (31–33)—in the lungs of both the H5N1 and 1918 recombinant virus-infected animals (Fig. 4A). However, the degree of infiltration in the H5N1 animals far surpassed the others on day 7. The 1918HANANS infection resulted in more intense infiltration than the 1918HANA virus, but only on day 2. Consistent with human disease, H5N1 infection resulted in severe necrotizing bronchiolitis and alveolitis evident within 24 h PI (34). A similar but less severe inflammation was noted in the animals infected with the 1918HANA recombinant virus (Fig. 4B). No gross or histological lesions ascribable to the experimental infections were observed in any tissues outside the respiratory tract and draining lymph nodes. In summary, both H5N1 and 1918 recombinant viruses produced severe pathology, but in the case of the H5N1 virus, the degree of local inflammation, assessed by transcriptional induction, tissue cytokine secretion, and granulocytic infiltration, was substantially greater and more prolonged. The H5N1 virus also induced a more severe systemic reaction, suggested by high serum cytokine levels and clinical signs.
Fig. 4.
Sustained inflammatory response in H5N1 infected macaques. (A) Mac 387 staining (10×) of lung sections with representative pathology and infection from each experimental group on days 1, 2, 4, and 7. This image illustrates the prolonged and more severe degree of granulocyte and macrophage infiltration in the H5N1 group and the intermediate phenotype of the 1918HANANS group for Mac387. (B) H&E staining (20×) of terminal bronchioles in the H5N1 and 1918HANA group on day 1, showing the severe damage to bronchiolar epithelium and severe infiltration of immune cells in the lumen of H5N1 animals.
H5N1 Virus Caused Margination of Circulating T Lymphocytes and Reduction of Dendritic Cells in Lungs and Draining Lymph Nodes.
To gain a better understanding of how these viruses affected other aspects of the host response, we conducted flow cytometric analysis of circulating lymphocytes and observed a dramatic depletion of CD4+ and CD8+ T cells through day 7, with a progressive and only partial recovery (Fig. S3 A and B). This effect was clearly most evident in the H5N1 group. This transient phenomenon is reportedly mainly due to vascular margination of T lymphocytes, induced by a direct interaction with circulating type I IFNs (35), although H5N1 viruses were shown to also cause T-cell apoptosis in multiple tissues in the mouse model (36).
Another event possibly impacting the adaptive response in the H5N1 group was a notable disappearance of activated (CD83+) dendritic cells in lungs and draining lymphoid tissue over the 7-day infection (Fig. 5A). In contrast, dendritic cells in the peripheral circulation decreased in absolute numbers in the H5N1 group compared to the others (Fig. S4), suggesting that their absence in infected tissues was not the result of impaired recruitment or extravasation. Subsequent immunolabeling for activated caspase-3 suggested that dendritic cells underwent apoptosis in situ early in infection with H5N1 virus. This was determined by morphological examination of apoptotic cells (Fig. 5B), as well as by double CD83/activated caspase-3 staining, which highlighted activated dendritic cells undergoing apoptosis (Fig. 5B).
Fig. 5.
Loss of dendritic cells due to apoptosis in H5N1 infected macaques. (A) CD83 staining in lungs and tracheobronchial lymph nodes was scored according to the following scale: 0 = none apparent; 0.5 = few/rare scattered cells; 1 = small numbers of positive cells, often in clusters; 2 = moderate increase in numbers of positive cells; 3 = moderate increase in numbers of positive cells; 4 = marked increase in interstitiae and/or BALT of the lung or in the paracortical zone of lymph nodes. Note: The entire section (min 7 sections per animal) was examined for CD83+ cells, translating into at least 30–60 high power fields. Day 6 death in H5N1 group is illustrated as day 7 for simplicity. A 2-way ANOVA was performed and revealed that groups were significantly different from one another (P < 0.001) over the entire course of the study, although not on any specific day. (B) CD83, activated caspase-3, and resulting double-labeling (100×) of apoptotic mature dendritic cells in lung tissue of an H5N1 animal on day 4. In most cases, apoptotic cells (caspase-3+) were identified as dendritic cells, mainly through morphology, due to the poor expression of CD83 in apoptotic cells.
In summary, the H5N1 virus perturbed the cell-mediated antiviral response and, through increased or premature apoptosis of dendritic cells normally present in lung and lymph node tissues during acute infection, may also have impacted antigen presentation needed to initiate the adaptive immune response.
Discussion
We performed a systematic comparison of early host responses during infections with highly pathogenic influenza viruses in a nonhuman primate model. This study revealed features unique to HPAI that may help us understand how these characteristics synergize to result in the poor prognosis seen in human patients. Lack of affinity of avian influenza viruses for the most common receptor type (α2–6-bound sialic acids) found in the human upper respiratory tract (8, 37) is believed to have thus far prevented person-to-person transmission and therefore the onset of an H5N1 pandemic. To overcome these affinity differences between avian- and human-adapted viruses, which most likely also exist in nonhuman primates, we inoculated the majority of the viruses directly into the lower respiratory tract. The H5N1 virus resulted in a more widespread lower respiratory tract infection than the other viruses, in particular there appeared to be a greater propensity to infect type II pneumocytes through day 7. Interestingly, the H5N1 virus also replicated better in the upper respiratory tract despite the paucity of cells containing α2–3- bound sialic acids (the optimal receptor for H5N1 influenza virus), substantiating that a high viral inoculation load might overcome suboptimal receptor specificity for the H5N1 virus (38). The H5N1 virus also successfully spread to several extrarespiratory tissues, albeit to a limited extent.
The H5N1 infection resulted in very strong and protracted induction of genes relevant to inflammatory and innate immune responses. While reassortant 1918 influenza viruses also proved much more virulent in the nonhuman primate model than a contemporary human H1N1 virus, overall pathology and clinical course with the 1918HANA virus were less severe compared to the fully reconstructed 1918 virus (12). This was an unexpected finding, given the high virulence of the 1918HANA recombinant virus in mice. Also in contrast to the mouse model, the 1918 reassortant containing the 1918 HA, NA and NS1 genes was of equal virulence to 1918HANA virus in the macaque model, suggesting species-specificity of NS functionality (39).
Virulence of HPAI Viruses May Be Closely Tied to Preferential Infection of Type II Pneumocytes.
Our data, when considered in light of other studies, suggests a connection between the ability of the H5N1 virus to infect type II pneumocytes and the extent of viral replication in the lower respiratory tract. These cells outnumber type I pneumocytes by 2-fold and have a faster metabolism (40) due to their role as stem cells and as source of surfactant with antimicrobial, immunomodulatory, and anti-inflammatory properties (41). These functions are in all likelihood compromised by infection, resulting in disrupted lung repair and increased vulnerability of remaining viable alveolar tissue. Indeed, while the 1918 recombinant viruses resulted in severe pathology, the amount of damage we observed in the H5N1 animals was extreme and unique in the lack of pneumocyte hypertrophy and hyperplasia, which are sign of repair, toward day 7. As early as 24 h PI, the H5N1 virus also destroyed bronchiolar epithelium, Clara cells, which act as stem cells and metabolize toxic substances at that level of the respiratory tract, and mucus-producing cells, which facilitate mechanical clearance.
The targeting of type II pneumocytes has been seen in human patients (34) who died 7 to 10 days after becoming symptomatic. It was also noted in other nonhuman primate studies with the fully reconstructed 1918 pandemic virus (12) and other H5N1 isolates, but only for the first 48 h PI, in contrast to the present study. This suggests that even subtle differences in cell tropism may account for differences in virulence among highly pathogenic influenza viruses in the nonhuman primate model and in humans.
High Virulence of H5N1 Virus Is Associated with Increased Host Responses.
The H5N1 virus caused intense transcriptional induction and secretion of inflammatory cytokines, as evidenced by the lung microarray and bead array data, and as documented in human patients (23, 34). IL-6, recently suggested as a key mediator of acute lung injury in mice (42), was detected at high levels in serum. An increase in circulating serum cytokines is known to damage vascular endothelial cells, resulting in vascular fluid and protein leakage, and ultimately fibrin deposition into the alveolar space (40). This finding is particularly relevant to our H5N1 infection since the fibrin organization we observed constitutes the first step toward pulmonary fibrosis and permanently reduced physiologic reserve. Inflammatory proteins can be produced by almost any infected cells and by immune cells, including alveolar macrophages (43). They can also be produced in response to oxidation of surfactant phospholipids (42), another consequence of infection. However, type II pneumocytes have a marked ability to secrete large amounts of cytokines, such as TNF-α, GM-CSF, MCP, and IL-8, in response to various insults (44, 45) and can be induced to secrete IL-1β, IL-6, RANTES, MIP-2. KC2, and MCP-1, in response to TNF-α (45, 46), the last being produced by alveolar macrophages during H5N1 infection. Therefore, it is possible that the much greater cytokine induction by the H5N1 virus was as much a consequence of the response of individual, infected cells as it was a consequence of the numbers of infected cells. This cytokine and innate response was still very much in effect on day 7 in lung tissue, in contrast to the response in animals infected with other viruses. This finding was consistent with gene expression in H5N1-infected ferrets (28), and in bronchial tissues of 1918-infected macaques (12). This induction did not result in comparable protein production on day 7 for the cytokines we measured, suggesting either a delay between transcription and translation or repressive mechanisms at the translation stage.
H5N1 Infection May Disrupt Immunity Beyond the Innate Response.
We observed a notable disappearance of dendritic cells in the lungs of H5N1-infected animals over the course of the study. During infection, these cells express several maturation markers including CD83, which we readily detected in both lung and draining lymph nodes throughout the infection in groups other than the H5N1 animals. Conventional dendritic cells (cDC), normally present in healthy lung tissue, are key antigen-presenting cells that start migrating to draining lymph nodes within 12 h PI while secreting chemokines that attract, among others, granulocytes, seen in abundance particularly in the H5N1 group. This subset of dendritic cells, as well as its equivalent in peripheral circulation, is susceptible to H5N1 virus infection and subsequent apoptosis (47), which could partially explain our results. Plasmacytoid dendritic cells (pDC), scarce at baseline, normally become the predominant population in lung tissue ≈48 h PI, where they secrete large amounts of type I IFNs and T cell chemokines (48). Therefore, these cells may have significantly contributed to the powerful IFN response that we observed on the arrays 2 days PI. pDCs are presumed to be more resistant to infection with H5N1 viruses (47, 49), yet they failed to express CD83 after day 2 and may have been among the cells undergoing apoptosis as revealed by double-labeling for CD83 and activated caspase-3 (Fig. 5B). pDC may have had limited success attracting lymphocytes to the lungs due to the timing of their disappearance, the overwhelming presence of PMNs in lung tissue, and the likely IFN-induced vascular margination of CD4+ and CD8+ T cells. Thus, apoptosis of dendritic cells, to the extent and at the time it was observed in the H5N1 group, may have compromised either antigen-presenting or other immunoregulatory functions associated with an optimal induction of an adaptive response.
In conclusion, H5N1 virulence is a multipronged mechanism that causes severe lung pathology with potentially permanent tissue damage within 24 h PI, accompanied by excessive and sustained type I IFN, inflammation, and innate immune induction. The intense host response is unsuccessful in controlling the rapidly progressing infection, consistent with the high mortality in humans. Our results suggest that H5N1 viruses may also deregulate the induction of an adaptive response through several mechanisms involving loss of dendritic cells and type I IFN-induced vascular margination of CD4+ and CD8+ T lymphocytes, making them mostly unavailable at critical time points during infection.
Methods
Animals.
Thirty-four cynomolgus macaques (M. fascicularus) (half of each sex), weighing 2.1–2.7 kg, were exported from the Huazheng laboratory animal breeding center, Zuo Village Chengjiao Town, Conghua, Guangzhou, China and received by Covance Research Products. Animals were quarantined for 6 weeks, prescreened, and certified as specific pathogen-free. Husbandry and procedures were approved by the Battelle Institutional Animal Care and Use Committee and by the Battelle Biosafety Committee. Influenza-infected animals were housed in an ABSL-3 enhanced facility.
Viruses.
The H5N1 human influenza isolate A/Vietnam/1203/2004 was obtained from the World Health Organization (WHO) influenza collaborating laboratory at the Centers for Disease Control (CDC), Atlanta, GA. The reassortant A/Texas/36/1991 influenza viruses possessing the A/South Carolina/1/18 HA (GenBank AF117241) and A/Brevig Mission/1/1918 NA (GenBank AF250356) or the A/South Carolina/1/18 HA, A/Brevig Mission/1/1918 NA, and A/Brevig Mission/1/18 NS (GenBank AF333238) were generated as previously described (50, 51). Recombinant viruses were plaque-purified, and the sequences of the virus segments were confirmed by reverse transcription-PCR and sequencing.
Experimental Design.
Animals assigned to 4 experimental groups (n = 8) matched for age, weight, and sex, were inoculated by intratracheal, intranasal, tonsillar, and conjunctival routes with a total of 107 pfu of A/Vietnam/1203/2004 (H5N1) virus, A/Texas/36/91 (H1N1) virus or reassortants of this virus containing either 2 (HA, NA) or 3 (HA, NA, NS) genes from the 1918 virus. Two animals per group were scheduled for sacrifice on days 1, 2, 4, and 7. Two additional animals were used as uninfected control animals and killed on day 7. Blood was collected from all animals 2 weeks before infection and on days 2, 4, and 7 or until sacrifice, for hematology, immunophenotyping, and cytokine analysis. Blood was also collected on day 1 from all animals except those with a day 7 endpoint, due to limitations in the total permissible blood draw. At necropsy, the lungs, tracheobronchial and retropharyngeal lymph nodes, heart, liver, spleen, kidneys, brain, and gastrointestinal tract were examined and sampled either by snap-freezing for viral isolation, cytometric bead array, real-time PCR, or microarray analysis, or by fixation in 10% formalin for histology and immunohistochemistry.
Histology and Immunochemistry.
Histological examination was performed as previously described (13–15). Samples from the lungs, hilar lymph nodes, tonsils, and spleens were immunolabeled for influenza A virus nuclear protein (NP) with a mouse monoclonal antibody (Fitzgerald Industries Intern) (14) or rabbit polyclonal antibody (anti-HK/97; CDC Influenza Branch), for myeloid cells with Mac387 (DAKO), for dendritic cells with anti-CD83 (13), and for activated caspase-3 (Cell Signaling Technology). The protocols used have been described (13, 15, 52). All microscopy was performed on an Olympus BX41 light microscope (Olympus). Photomicrographs were acquired with an Olympus Q-Color 3 camera and associated computer software. Pathology and antigen scores were assigned to each animal according to methods described in the SI. Statistical calculations were performed with SigmaStat for Windows and figures were made with SigmaPlot for Windows (Systat Software).
Viral Titers.
Lung samples were frozen at −80 °C, homogenized, and tested for viable virus by TCID50 assays using a modification of the method reported by the WHO (53). Briefly, 90% confluent MDBK cells in 96-well plates were inoculated with 100 μL of serial 10-fold dilutions of each sample, and positive and negative control samples. Plates were incubated at 37 °C in a humidified incubator (5% CO2) for 96 h. TCID50/mL was calculated using the Spearman–Kärber method (54).
Cytokine Bead Array.
Cytokines tested were: IL-2, IL-4, IL-5, IL-6, TNF-α, IFN-γ. One sample from the left middle lung lobe was analyzed for each animal. Frozen lung tissue was homogenized in 1 mL of T-PER Tissue Extraction Reagent (Pierce) containing protease inhibitor mixture (minitabs, Roche) and centrifuged at 750 × g for 10 min at 4 °C. Cytokine levels in lung and serum were assessed using the Cytometric Bead Array (CBA) Nonhuman Primate Th1/Th2 assay kit (Becton Dickinson) according to the manufacturer's instructions. Assays were read on an LSR II flow cytometer (BD PharMingen) and analyzed using the BD Cytometric Bead Array Software (V 1.4).
Flow Cytometry.
Whole blood was incubated at room temperature for 30 min in the dark with (i) CD3(SP34–2), CD14(M5E2), CD20(2H7), CD56(B159), CD45(H130), CD83(HB15e), CD123(7G3), CD1a (NA13/4-HLK AbD Serotec), and CD16(3G8) monoclonal antibodies to identify plasmacytoid dendritic precursors; and (ii) CD20, CD14, CD4(L200), CD8(RPA-T8), CD56, CD45 (Miltenyi Biotech), and CD3 monoclonal antibodies to define major leukocyte subsets. Lineage-positive leukocyte subsets were defined by using monoclonal antibodies to CD3, 20, 14, and 56 labeled with APC. All antibodies were from BD PharMingen except where noted. Red blood cells were lysed using an ammonium chloride solution and the resultant leukocytes were washed twice with BD PharMingen Stain Buffer, and fixed with 4% paraformaldehyde. The fixed leukocytes were washed an additional time with BD PharMingen Perm/Wash Buffer, and stained with DAPI (Molecular Probes/Invitrogen). Events were initially selected based on DNA content with additional subsets isolated by their CD45 fluorescence and side scatter properties. A total of 30,000 gated events were collected with a FACSAria or LSR II flow cytometer using FACSDiva 5.0.1 software.
Macaque Oligonucleotide Arrays Analysis and Quantitative Real-Time PCR.
Total RNA was extracted from tissues for macaque oligonucleotide arrays and quantitative real-time PCR (viral gene probe sequences available upon request) as previously described (13, 15, 55). Oligoarray analyses consisted of comparing array profiles of individual lung samples (with representative pathology and degree of infection) from infected animals to pooled equal masses of mRNA from all lung lobes of 7 reference cynomolgus macaques obtained through the tissue program of the University of Washington National Primate Research Center.
Supplementary Material
Acknowledgments.
This work was supported by Battelle Internal Research and Development funds and by National Institutes of Health Grants R01AI46954, P01AI58113, and U01AI070469 (to A.G.-S.); R24 RR16354–04, P51 RR00166-45, and R01 AI022646-20A1 (to M.G.K.); K08 AI059106-02 (to Cambridge Research Biochemicals); and R03 AI075019-01 (to H.B.-O.).
Footnotes
Conflict of interest statement: A.G.-S. owns patent positions for reverse genetics of influenza viruses.
Data deposition: The data reported in this paper have been deposited in the Expression Array Manager at http://viromics.washington.edu/informatics/Project/Publications/begin.view.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813234106/DCSupplemental.
References
- 1.Ng WF, To KF. Pathology of human H5N1 infection: New findings. Lancet. 2007;370:1106–1108. doi: 10.1016/S0140-6736(07)61490-1. [DOI] [PubMed] [Google Scholar]
- 2.Gu J, et al. H5N1 infection of the respiratory tract and beyond: A molecular pathology study. Lancet. 2007;370:1137–1145. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 4.Uyeki TM. Global epidemiology of human infections with highly pathogenic avian influenza A (H5N1) viruses. Respirology. 2008;13(s1):S2–S9. doi: 10.1111/j.1440-1843.2008.01246.x. [DOI] [PubMed] [Google Scholar]
- 5.Morens DM, Fauci AS. The 1918 influenza pandemic: Insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 6.Kash JC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 8.Kuiken T, et al. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 9.Tumpey TM, et al. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szretter KJ, et al. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maines TR, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci USA. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 13.Baskin CR, et al. Functional genomic and serological analysis of the protective immune response resulting from vaccination of macaques with an NS1-truncated influenza virus. J Virol. 2007;81:11817–11827. doi: 10.1128/JVI.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskin CR, et al. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina) J Virol. 2004;78:10420–10432. doi: 10.1128/JVI.78.19.10420-10432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baas T, et al. An integrated molecular signature of disease: Analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80:10813–10828. doi: 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, et al. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiss GK, et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci USA. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo SH, Hoffmann E, Webster RG. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 2004;103:107–113. doi: 10.1016/j.virusres.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Solorzano A, et al. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol. 2005;79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, et al. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2006;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 22.Mibayashi M, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beigel JH, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 24.To KF, et al. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J Med Virol. 2001;63:242–246. doi: 10.1002/1096-9071(200103)63:3<242::aid-jmv1007>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 25.Uiprasertkul M, et al. Influenza A H5N1 replication sites in humans. Emerg Infect Dis. 2005;11:1036–1041. doi: 10.3201/eid1107.041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong SS, Yuen KY. Avian influenza virus infections in humans. Chest. 2006;129:156–168. doi: 10.1378/chest.129.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen KY, Wong SS. Human infection by avian influenza A H5N1. Hong Kong Med J. 2005;11:189–199. [PubMed] [Google Scholar]
- 28.Cameron CM, et al. Gene expression analysis of host innate immune responses during lethal H5N1 infection in ferrets. J Virol. 2008;82:11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiwanitkit V. Hemostatic disorders in bird flu infection. Blood Coagul Fibrinolysis. 2008;19:5–6. doi: 10.1097/MBC.0b013e3282f185a6. [DOI] [PubMed] [Google Scholar]
- 30.Sumikoshi M, et al. Human influenza virus infection and apoptosis induction in human vascular endothelial cells. J Med Virol. 2008;80:1072–1078. doi: 10.1002/jmv.21185. [DOI] [PubMed] [Google Scholar]
- 31.Perrone LA, Plowden JK, García-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4(8):e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu T, et al. Acute respiratory distress syndrome induced by avian influenza A (H5N1) virus in mice. Am J Respir Crit Care Med. 2006;174:1011–1017. doi: 10.1164/rccm.200511-1751OC. [DOI] [PubMed] [Google Scholar]
- 33.Zitzow LA, et al. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76:4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korteweg C, Gu J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am J Pathol. 2008;172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- 36.Tumpey TM, Lu X, Morken T, Zaki SR, Katz JM. Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J Virol. 2000;74:6105–6116. doi: 10.1128/jvi.74.13.6105-6116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholls JM, et al. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007;13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- 39.Basler CF, et al. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci USA. 2001;98:2746–2751. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ali J, Summer WG, Levitzky MG. Pulmonary Pathophysiology. 2nd Ed. Atlanta: McGraw-Hill; 2004. p. 250. [Google Scholar]
- 41.Sorensen GL, Husby S, Holmskov U. Surfactant protein A and surfactant protein D variation in pulmonary disease. Immunobiology. 2007;212:381–416. doi: 10.1016/j.imbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Imai Y, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 44.Fehrenbach H. Alveolar epithelial type II cell: Defender of the alveolus revisited. Respir Res. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma AK, Fernandez LG, Awad AS, Kron IL, Laubach VE. Proinflammatory response of alveolar epithelial cells is enhanced by alveolar macrophage-produced TNF-alpha during pulmonary ischemia-reperfusion injury. Am J Physiol. 2007;293:L105–L113. doi: 10.1152/ajplung.00470.2006. [DOI] [PubMed] [Google Scholar]
- 46.Sato K, et al. Type II alveolar cells play roles in macrophage-mediated host innate resistance to pulmonary mycobacterial infections by producing proinflammatory cytokines. J Infect Dis. 2002;185:1139–1147. doi: 10.1086/340040. [DOI] [PubMed] [Google Scholar]
- 47.Thitithanyanont A, et al. High susceptibility of human dendritic cells to avian influenza H5N1 virus infection and protection by IFN-alpha and TLR ligands. J Immunol. 2007;179:5220–5227. doi: 10.4049/jimmunol.179.8.5220. [DOI] [PubMed] [Google Scholar]
- 48.Grayson MH, et al. Controls for lung dendritic cell maturation and migration during respiratory viral infection. J Immunol. 2007;179:1438–1448. doi: 10.4049/jimmunol.179.3.1438. [DOI] [PubMed] [Google Scholar]
- 49.Hao X, Kim TS, Braciale TJ. Differential response of respiratory dendritic cell subsets to influenza virus infection. J Virol. 2008;82:4908–4919. doi: 10.1128/JVI.02367-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tumpey TM, et al. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc Natl Acad Sci USA. 2004;101:3166–3171. doi: 10.1073/pnas.0308391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tumpey TM, et al. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: Functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bielefeldt-Ohmann H, et al. Transplacental infection with non-cytopathic bovine viral diarrhoea virus types 1b and 2: Viral spread and molecular neuropathology. J Comp Pathol. 2008;138:72–85. doi: 10.1016/j.jcpa.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 53.WHO. WHO Manual on Animals Influenza Diagnosis and Surveillance. Geneva: World Health Organization; 2002. [Google Scholar]
- 54.Hamilton MA, Russo RC, Thurston RV. Trimmed Spearman-Karber method for estimating median lethal concentrations in toxicity bioassays. Environ Sci Technol. 1977;11:714–719. [Google Scholar]
- 55.Bielefeldt-Ohmann H, et al. Intestinal stromal tumors in a simian immunodeficiency virus-infected, simian retrovirus-2 negative rhesus macaque (Macaca mulatta) Vet Pathol. 2005;42:391–396. doi: 10.1354/vp.42-3-391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.