Abstract
The genome of Trypanosoma brucei is unusual in being regulated almost entirely at the post-transcriptional level. In terms of regulation, the best-studied genes are procyclins, which encode a family of major surface GPI-anchored glycoproteins (EP1, EP2, EP3, GPEET) that show differential expression in the parasite's tsetse-fly vector. Although procyclin mRNA cis-regulatory sequences have provided the paradigm for post-transcriptional control in kinetoplastid parasites, trans-acting regulators of procyclin mRNAs are unidentified, despite intensive effort over 15 years. Here we identify the developmental regulator, TbZFP3, a CCCH-class predicted RNA binding protein, as an isoform-specific regulator of Procyclin surface coat expression in trypanosomes. We demonstrate (i) that endogenous TbZFP3 shows sequence-specific co-precipitation of EP1 and GPEET, but not EP2 and EP3, procyclin mRNA isoforms, (ii) that ectopic overexpression of TbZFP3 does not perturb the mRNA abundance of procyclin transcripts, but rather that (iii) their protein expression is regulated in an isoform-specific manner, as evidenced by mass spectrometric analysis of the Procyclin expression signature in the transgenic cell lines. The TbZFP3 mRNA–protein complex (TbZFP3mRNP) is identified as a trans-regulator of differential surface protein expression in trypanosomes. Moreover, its sequence-specific interactions with procyclin mRNAs are compatible with long-established predictions for Procyclin regulation. Combined with the known association of TbZFP3 with the translational apparatus, this study provides a long-sought missing link between surface protein cis-regulatory signals and the gene expression machinery in trypanosomes.
Author Summary
Trypanosomes, the tropical parasites that cause African sleeping sickness, show a number of biological peculiarities that distinguish them from other eukaryotes. One is the unusual way in which they regulate gene expression. Unlike most eukaryotes, trypanosomes do not regulate gene expression by controlling the rate of messenger RNA synthesis, but, instead, control the stability of messenger mRNAs (and, hence, their abundance) and also their rate of translation into protein. The best-studied model for this “post-transcriptional” gene expression control in trypanosomes is the procyclin mRNAs, which encode the major surface proteins of the parasite in the tsetse fly. In this study we demonstrate that a small kinetoplastid-specific protein (TbZFP3) co-associates with the mRNAs for some procyclin isoforms (EP1, GPEET procyclin) but not others (EP2, EP3 procyclin). Furthermore, we show that this is dependent upon sequences in the EP1 procyclin 3′untranslated region known to govern its mRNA turnover and protein synthesis. Finally, we demonstrate that limited over-expression of TbZFP3 causes a change in the surface protein expression profile on cultured parasites from GPEET to EP1 Procyclin. Our data identify TbZFP3 as an important post-transcriptional regulator of Procyclin expression, the first such protein factor identified.
Introduction
The pathway of mRNA control in eukaryotes involves regulatory steps at multiple stages. This is reflected by the large investment of eukaryotic genomes in RNA binding proteins, with hundreds of genes in yeasts and mammals being devoted to functions requiring RNA interaction. As the diverse roles of these proteins, and their interactions with specific subsets of mRNAs, are investigated, it is becoming clear that post-transcriptional control represents a regulatory network of a complexity and importance likely greater than transcriptional control [1].
Perhaps the most extreme example of a group of organisms with emphasis on post-transcriptional control is the kinetoplastid parasites. These organisms are of medical, veterinary and economic importance because they are responsible for an enormous burden of disease within the tropics, including a variety of cutaneous and visceral diseases (caused by Leishmania spp.), Chagas disease (caused by Trypanosoma cruzi) and African sleeping sickness (caused by Trypanosoma brucei). Here, an absence of detectable RNA II polymerase promoters for protein coding genes and the general organisation of transcription units into polycistronic arrays necessitates almost complete reliance on post-transcriptional control for regulated gene expression [2]. Supporting this, the genome of these parasites reveals a complexity and composition of encoded RNA binding proteins exceeding, and distinct from, that found in the crown group of eukaryotic organisms [3],[4].
Gene regulation is particularly important in kinetoplastid parasites because their life cycle is complex, involving passage through a mammalian host and within distinct compartments of an arthropod vector [5]. One of the best-characterised life-cycle differentiation events involves exchange of the major surface antigens as African trypanosomes passage from mammalian blood to the midgut of their haematophagous vector, the tsetse fly [6]. In the bloodstream trypanosomes stay ahead of the immune response by expressing, sequentially and hierarchically, thousands of different antigenic surface coats comprised of variant surface glycoprotein (VSG) [7]. However, upon differentiation in the tsetse, the VSG coat is replaced by a family of glycophosphatidyl inositol (GPI)-anchored proteins known as Procyclins. There are two types of Procyclin proteins, which mainly differ by the type of amino acid repeats they contain at their C-termini. One set of proteins, the EP isoforms (encoded by the EP1-1, EP1-2, EP2 and EP3 genes) contain 22–30 [E-P] internal repeat peptides whereas GPEET Procyclins (encoded by one copy of GPEET) contain 6 [G-P-E-E-T] repeats, which can be phosphorylated at the Thr residues. In the tsetse, Procyclins follow a programmed expression and their C-terminal repeat peptides, together with their complex GPI anchors, may provide protection for the parasite from the action of tsetse gut hydrolases [8]–[11].
Sequence-dependent signals in the 3′ untranslated region (3′ UTR) of each procyclin mRNA govern their expression and have been the subject of intense investigation, providing the paradigm for gene expression control in kinetoplastid parasites [2]. Although the 3′UTRs of EP1, 2 and 3 and GPEET procyclin mRNAs are highly similar, the genes are differentially regulated in distinct phases of tsetse infection or in vitro. For example, the GPEET procyclin 3′UTR contains an element, absent in the closely related EP1 3′UTR, that differentially regulates its expression in response to glycerol and the activity of mitochondrial enzyme activities [12],[13]. However, whilst the cis-acting control sequences for procyclin mRNAs are very well characterised [14]–[17], protein factors that recognise these regulatory domains have remained unidentified, despite considerable effort. Here we establish the specific association and regulation of procyclin mRNA isoforms by a kinetoplastid-specific protein factor that associates with polyribosomes, providing the first example in these organisms of surface protein regulation by an mRNA-associated regulatory factor.
Results
TbZFP3 immunoprecipitation differentially selects procyclin isoform mRNAs
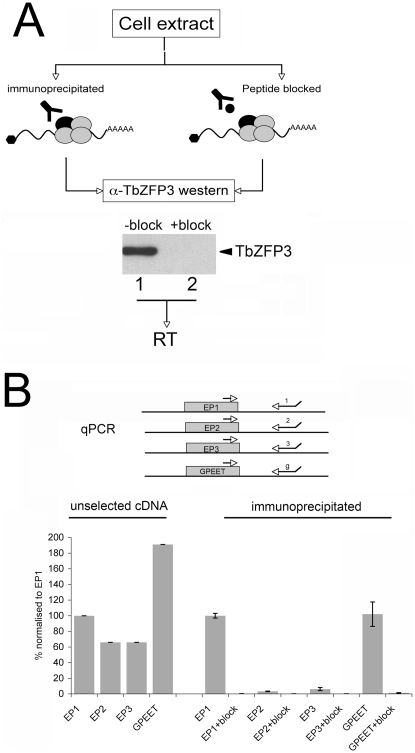
Previous immunoprecipitation experiments using an antibody specific for a small CCCH-protein implicated in developmental control, TbZFP3 (Tb927.3.720), demonstrated co-precipitation of mRNA for the procyclic-form specific surface proteins, Procyclins [18]. Since different Procyclin isoforms exhibit distinct profiles of mRNA and protein expression in the tsetse fly [8],[11],[19] we investigated whether each isoform mRNA was co-precipitated with equivalent efficiency by TbZFP3. Figure 1A shows a typical experiment where immunoprecipitation from cell extracts resulted in a selection for TbZFP3 (lane 1), this being blocked in the presence of the peptide immunogen used to raise the TbZFP3-specific antibody (lane 2). The resulting co-selected mRNAs were then reverse transcribed and subjected to quantitative real-time (qRT) PCR using primers specific for each procyclin transcript isoform [11] (Figure 1B). In parallel reactions, total RNA from the starting cultures was also analysed with each primer set, allowing us to compare the relative level of each procyclin isoform mRNA in unselected and TbZFP3-immunoprecipitated material. In total mRNA of the cell extracts, both EP2 and EP3 were present at 66% of EP1 levels (EP1 is normalised to 100% in Figure 1B), approximating to their observed relative abundance in culture and in the tsetse midgut [11]. As expected in this parasite strain [12],[20],[21], GPEET mRNA was also abundant (191% with respect to EP1) in the unselected material. In contrast to unselected cDNA, TbZFP3-immunoprecipitated material showed a strikingly differential abundance of the isoforms, such that EP1 and GPEET were the dominant selected transcripts, with EP2 and EP3 selected at much lower level (3.3% and 6.2% of immunoprecipitated EP1, respectively). Importantly, use of the peptide block prevented the immunoprecipitation of each procyclin mRNA isoform, demonstrating specificity of the selection. Supporting this qRT-PCR data, non-selective amplification and cloning of procyclin cDNAs derived from TbZFP3-immunoprecipitated material isolated 21/27 (78%) EP1 sequences and 6/27 (22%) GPEET sequences, with no clones containing EP2 or EP3 derived sequences. We conclude that although EP1, EP2, EP3 and GPEET procyclin mRNAs are each abundant in the unselected mRNA pool, TbZFP3 is preferentially associated with EP1 mRNA, but also GPEET procyclin mRNAs.
Figure 1. Co-immunoprecipitation of EP1 and GPEET procyclin mRNA, but not EP2 and EP3 mRNA, by TbZFP3.
(A) Schematic representation of TbZFP3 immunoprecipitation. Cell extracts were subject to a clearing spin and immunoprecipitated in the presence or absence of the peptide immunogen against which the TbZFP3 antibody was raised (“Peptide blocked”). Western blotting with anti-serum to TbZFP3 was used to detect selection in the absence (Lane 1) or presence (Lane 2) of blocking peptide. (B) Three replicate immunoprecipitations from three distinct trypanosome procyclic lines were carried out, with the selected material being subject to reverse transcription and amplification using primers specific for EP1, EP2, EP3, or GPEET mRNAs. Values are normalised in unselected and selected material to the level of EP1 mRNA. Error bars = SD.
Procyclin mRNA association requires integrity of the CCCH domain in TbZFP3
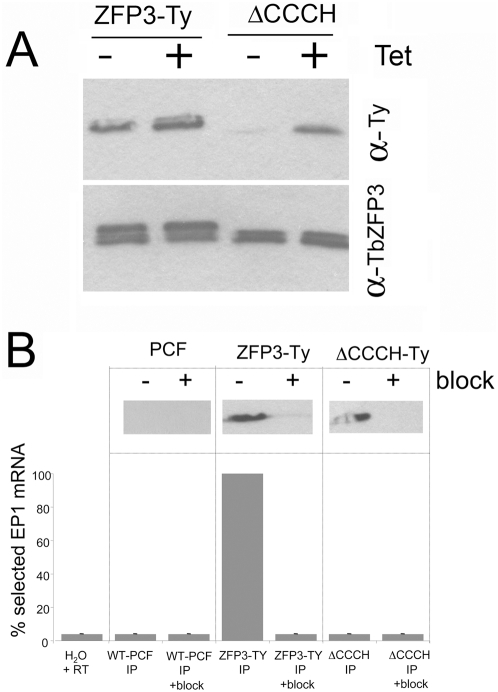
To determine whether the co-immunoprecipitation of procyclin mRNAs with TbZFP3 was dependent on its predicted RNA-binding domain we examined a cell line expressing a mutant form of TbZFP3 lacking the CCCH zinc finger domain (TbZFP3 ΔCCCH; [18]). This mutant incorporated a C-terminal Ty1-epitope tag to allow it to be specifically immunoprecipitated in the context of endogenous TbZFP3 using the BB2 antibody which detects the Ty1 epitope [22]. As a control, wild type TbZFP3 was also expressed with a C-terminal Ty1 tag with the relative expression of each ectopically expressed protein being examined by Western blotting using either the antibody against TbZFP3 (this detecting endogenous and ectopically expressed TbZFP3) or BB2 (detecting only the ectopically expressed protein). Figure 2A shows the relative expression of each ectopically expressed protein in each cell line, confirming their approximately equivalent abundance. Thereafter, cell extracts from each line were used in immunoprecipitation experiments to select the ectopic TbZFP3 using the BB2 antibody, and the co-selection of procyclin EP1 mRNA assayed. This demonstrated selection of EP1 mRNA with TbZFP3-Ty, as expected, whereas deletion of the CCCH domain prevented co-precipitation of EP1 mRNA (Figure 2B). Thus, the integrity of the predicted RNA binding domain in TbZFP3 is necessary for co-immunoprecipitation of EP1 mRNA.
Figure 2. The CCCH domain of TbZFP3 is necessary for EP1 procyclin mRNA co-association.
(A) Western blot of the expression of the Ty-tagged ΔCCCH mutant of TbZFP3 and the Ty-tagged wild type TbZFP3 expressed in transgenic parasites. The ectopically expressed proteins were detected by incorporation of the Ty-1 epitope tag in to the transgenic protein, allowing their detection and immunoprecipitation. The upper panel shows cell extracts reacted with anti-Ty1 epitope tag antibody BB2, the lower panel shows the same cell extracts reacted with anti-TbZFP3 antibody. This detects the endogenous TbZFP3 as well as the ectopically expressed, Ty-tagged copy. Note that the TbZFP3-Ty migrates more slowly than the endogenous protein due to incorporation of the 10 amino acid epitope tag, whereas the ΔCCCH mutant comigrates with the endogenous protein due to deletion of the CCCH domain. (B) RNA immunoprecipitation of EP1 procyclin mRNA using the BB2 antibody to select TbZFP3-Ty or ΔCCCH ZFP3 ectopic protein from cell extracts. The relative selection of EP1 mRNA is normalised to selection with TbZFP3-Ty in each case. The relative immunoprecipitation of each ectopic protein in the presence or absence of a Ty1-epitope specific peptide block is shown at the top of each sample to demonstrate the efficiency and specificity of immunoprecipitation. This represents a Western blot of the immunoprecipitated material reacted with the BB2 antibody, specific for the Ty-1 epitope tag incorporated into the ectopically expressed TbZFP3-Ty or TbZFP3 ΔCCCH.
Sequence-specific affinity selection of EP1 mRNA via TbZFP3 immunoprecipitation
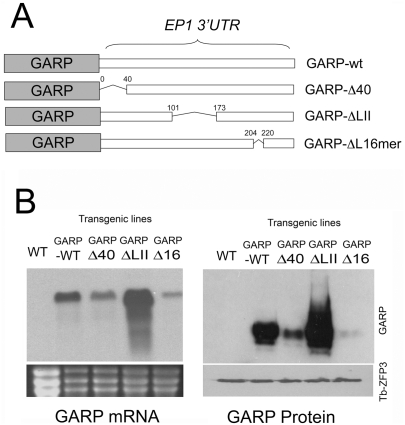
The sequences which regulate procyclin gene expression have been very well characterised in transgenic parasites by use of reporter genes linked to wild type or mutant forms of the EP1 mRNA 3′ UTR. This has identified a number of regulatory regions that act to either positively or negatively control expression [14], [23]–[26]. Minimally, three domains contribute to EP procyclin regulation: a positive control element in the first 40 nt after the stop codon (“Loop I”), a negative element contained within 101–173 nt (‘Loop II’) and a further positive element comprising a highly conserved 16 nt stem loop structure (“Loop III”). To determine whether TbZFP3 RNA–immunoprecipitation generated sequence-specific selection of EP1 procyclin mRNA, we generated a series of cell lines transfected with previously characterised reporter constructs (kindly provided by Professor I. Roditi, University of Bern). These comprised a GARP coding region reporter [27] linked to either the wild type EP1 procyclin 3′UTR or mutants lacking each regulatory domain (Δ40, ΔLII or Δ16mer) (Figure 3A). Initially the anticipated effects on reporter gene expression for each construct were confirmed by analysing the GARP mRNA and protein levels in the resulting transfected cell lines (Figure 3B). Matching previous analyses of these deletions [23], the mRNA abundance of GARP was reduced in the Δ40 (62% of wild type levels) and Δ16mer cell lines (26% of wild type levels), but significantly elevated in the ΔLII cell line (210% of wild type levels). Similarly, Western blotting of protein extracts from these cell lines with a GARP antiserum [28] confirmed that the levels of GARP protein translated from the expression constructs matched previous observations, with abundant GARP generated in the ΔLII cell line and little detectable protein when the 16mer element was deleted.
Figure 3. Construction and in vivo expression analysis of GARP reporters with mutated EP1 3′UTRs.
(A) Schematic representation of the reporter constructs used to assay TbZFP3 selection, highlighting deleted regions in each construct as described in [23]. (B) GARP RNA and protein expression generated in the reporter cell lines transfected with the constructs illustrated in (A). Relative loading is indicated, for Northern blots, by the rRNA levels (revealed by ethidium bromide staining) or, for Western blots, by TbZFP3 levels.
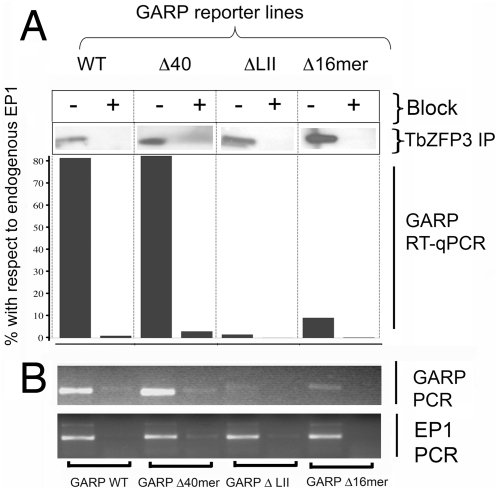
Having generated cell lines stably transfected with each reporter construct, extracts from each were subjected to TbZFP3-immunoprecipitation, either in the presence or absence of blocking peptide and analysed for the selection of TbZFP3, (Figure 4A, “TbZFP3 IP”) or of the reporter GARP mRNA (Figure 4A; “GARP-RT-qPCR”). Importantly, in each case the relative selection of GARP mRNA was compared with, and normalised to, the selection of endogenous EP1 procyclin, ensuring the efficiency of immunoprecipitation from each extract was equivalent (Figure 4A and 4B). In the cell line containing GARP linked to wild type EP procyclin 3′UTR, efficient selection of the reporter mRNA was observed with this being abolished in the presence of the blocking peptide (80% and 1% respectively, normalised to the relative immunoprecipitation of endogenous EP1 mRNA). When the Δ40 cell line was examined efficient selection of GARP transcripts was also observed (81% of endogenous EP1, with 3% of endogenous EP1 in the presence of the peptide block). However, when either the negative control element contained in Loop II of the EP1 procyclin 3′UTR, or the 16mer stem-loop structure were deleted, selection with TbZFP3 was reduced to only 1.5% or 9% of endogenous EP1 mRNA, respectively. This did not represent inefficient immunoprecipitation since endogenous EP1 procyclin mRNA was selected at an equivalent level in all cell lines (Figure 4B and data not shown). Moreover, it was not simply dependent on target mRNA abundance because the ΔLII–derived GARP mRNA was highly expressed (Figure 3B). This demonstrated that TbZFP3 immunoprecipitation showed sequence-specific selection of the EP procyclin 3′UTR, this being individually dependent upon integrity of the Loop II and the 16mer regulatory domains.
Figure 4. Sequence specific co-precipitation of EP1 3′UTR with TbZFP3.
(A) Co-immunoprecipitation of each GARP reporter mRNA with TbZFP3 either in the presence or absence of blocking peptide (“GARP RT-qPCR”). qRT-PCR values are normalised to the value for co-selected endogenous EP1 mRNA in each cell line. Control amplifications lacking input RNA or reverse transcriptase failed to amplify any product. Above each bar chart is shown Western blots for TbZFP3 (“TbZFP3 IP”), using proteins from each immunoprecipitation (plus or minus peptide block) used to isolate TbZFP3 and bound mRNAs from each cell line. These confirm specificity of the selection. (B) Independent confirmation of the real-time PCR data generated in a separate immunoprecipitation, with the products visualised on ethidium bromide stained agarose gels (“GARP-PCR”). The co-selection of endogenous EP1 mRNA is also shown (“EP1-PCR”).
TbZFP3 is a positive regulator of EP1 Procyclin operating at the protein level
Having demonstrated that EP1 procyclin mRNA co-selects with TbZFP3 via known regulatory domains we determined if TbZFP3 could specifically regulate EP procyclin mRNA abundance. Initially, we made use of transgenic procyclic and bloodstream form lines that ectopically over-express TbZFP3 under tetracycline control. Figure 5A (lanes 1–4) shows endogenous and ectopically expressed TbZFP3 mRNA in each cell line, whereas lanes 5–8 shows hybridisation to the same RNAs of a generic EP procyclin riboprobe. This revealed no evidence for a specific enrichment of any EP mRNA in response to TbZFP3 induction in procyclic forms nor appearance of EP mRNA in bloodstream forms (where Procyclin is not normally expressed). Furthermore, quantitative RT-PCR specific for EP1, EP2, EP3 procyclin revealed no specific change of EP1 mRNA with respect to EP2 or EP3 mRNAs, although the expression of all mRNAs increased slightly (∼20%; Figure 5B). Similarly, RNAi directed to TbZFP3 (resulting in 60% reduction of protein expression; Figure 6A) resulted in no specific regulation of any procyclin mRNA isoform, although all procyclin mRNAs as well as several housekeeping genes showed an overall reduction of mRNA abundance. This suggests non-specific or indirect effects or, potentially, a more widescale consequence of TbZFP3 knock-down on mRNA abundance (Figure 6B and data not shown). Nonetheless, the analysis demonstrated that there was no differential change in the abundance of procyclin isoform mRNAs caused by enhanced or reduced TbZFP3 expression.
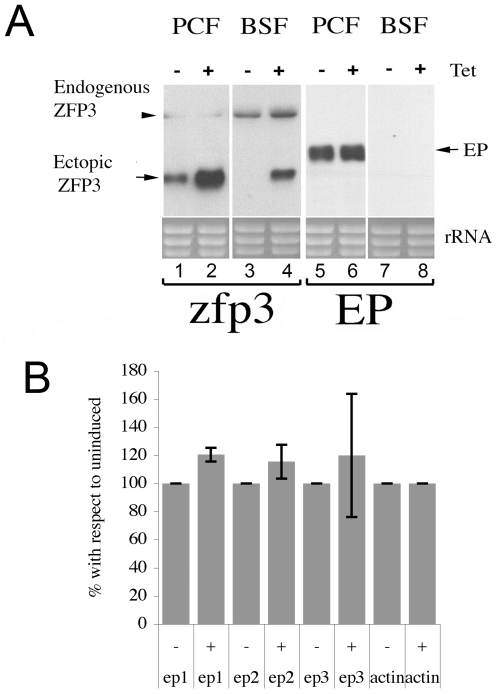
Figure 5. Effect of ectopic over-expression of TbZFP3 on procyclin mRNA levels.
(A) Northern blots of procyclic forms (PCF) and bloodstream form (BSF) transgenic trypanosome lines that induce the ectopic expression of TbZFP3 under a tetracycline regulatable promoter. Lanes 1–4 show the expression of the ectopic TbZFP3 (arrowed) in the absence or presence of induction. Endogenous TbZFP3 mRNA is also shown (arrowhead). Lanes 5–8 (“EP”) show the same RNAs hybridised with a riboprobe detecting all EP procyclin isoforms. The relative loading is indicated by rRNA below each panel. (B) Relative expression of EP1, EP2, and EP3 mRNAs after ectopic expression of TbZFP3 as determined by quantitative RT-PCR. Values are normalised to actin. All procyclin isoform mRNAs showed an approximately 20% increase in abundance, but no specific isoform showed a consistent change with respect to another. GPEET mRNA levels were also not affected (data not shown). Error bars = SD.
Figure 6. RNAi against TbZFP3 generates reduced mRNA abundance for all procyclin mRNA isoforms.
The TbZFP3 coding region was cloned into the RNAi vector p2T7i [41] and transfected into the RNAi competent procyclic cell line, PTT. Cells were induced to undergo RNAi by growth in medium containing tetracycline (1 µg/ml) for 48 h. Protein extracts from the induced and uninduced samples, and from the parental cell line, were then assayed for TbZFP3 expression by Western blotting (A). RNAs isolated from the respective samples were then assayed for relative procyclin mRNA expression by quantitative RT-PCR (B). All isoforms showed an inducible reduction in abundance in the presence of tetracycline (when TbZFP3 RNAi is induced; “+”). Some reduction was also seen in the absence of tetracycline (“−”) due to leaky RNAi.
To monitor the relative protein expression of individual Procyclin isoforms we made use of an established mass spectrometry approach to detect Procyclins. Thus, cell lines induced to ectopically express TbZFP3 for 48 h, 72 h or 1 week were subject to delipidation and butanol extraction followed by aqueous HF treatment to release the Procyclin proteins from their GPI-anchors. The released full length Procyclins were then further subject to mild acid treatment [29], a procedure that cleaves the EP isoforms at the Asp-Pro bonds and partially cleaves GPEET between Asp-Gly. The resulting extracts were analysed by negative ion MALDI-TOF-MS to detect the presence and abundance (based on their comparable ionisations) of the [M-H]− pseudomolecular ions representing C-terminal fragments of GPEET and EP1-1, EP1-2, EP2 and EP3 proteins. Consistent with expectation for this parasite strain, the dominant surface protein was GPEET Procyclin [20],[21], with lower expression of the EP Procyclin isoforms (Figure 7A–7C). However, ectopic expression of TbZFP3 (generating 1.5 and 2.3 fold overexpression at 48 h and 72 h, respectively), progressively elevated EP1-1 and EP1-2 Procyclins (Figure 7D–7F) above either uninduced controls (Figure 7A–7C) or the parental cell line grown in the presence of tetracycline (Figure S1). Furthermore, expression of GPEET was progressively reduced (∼5 fold) from being the dominant surface molecule to being a minor component with respect to EP1 after 7 days of induction (Figure 7F). In contrast to these two proteins, both allelic variants of EP3 procyclin (EP3-1 and EP3-5) remained relatively unchanged, whereas EP2 was not detected in any cell population matching previous studies. Consistent with the specific regulation of Procyclin expression by TbZFP3, examination of the Procyclin protein signature of the cell line expressing the ΔCCCH mutant of TbZFP3, which does not co-select procyclin mRNAs, did not reveal any change in the profile of expressed proteins regardless of whether the ectopic expression of the mutant protein was induced or not (Figure S2).
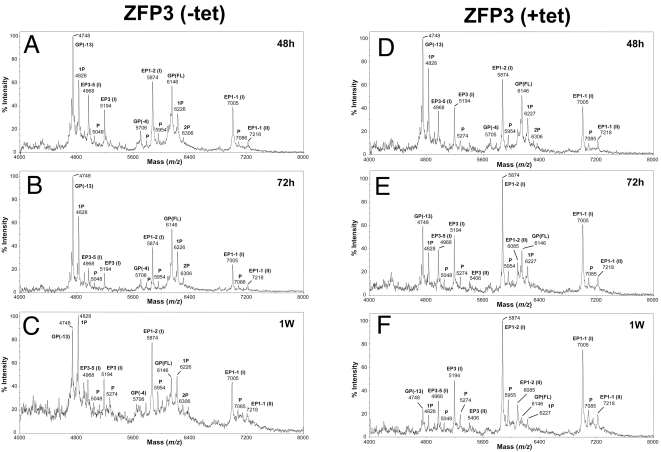
Figure 7. Procyclin isoform regulation by TbZFP3.
Negative ion MALDI-TOF mass spectra of Procyclin isoforms extracted from cells uninduced (A–C) or induced (D–F) to express ectopic TbZFP3 to a level ∼2-fold its endogenous abundance. In response to elevated TbZFP3 levels, the expression of EP1-1 and EP1-2 (which share an identical 3′UTR) is elevated, whereas GPEET expression is reduced. (A) and (D) show uninduced and induced cells after 48 h, (B) and (E) show cells after 72 h, and (C) and (F) show cells after 1 week. EP3-5 is an allelic copy of the EP3 gene in the 427 strain [42]. EP C-terminal polypeptides (I) and (II) represent forms containing the sequence P (EP)nG-EtN and PDP (EP)nG-EtN, respectively. GP (FL), (-4), and (-13) indicate full-length GPEET and its polypeptides lacking four and thirteen N-termini aminoacids, respectively. P, indicates levels of peptide phosphorylation.
To examine the basis of the altered expression of EP1 and GPEET Procyclins after TbZFP3 ectopic expression, we assayed the relative co-immunoprecipitation with TbZFP3 of each procyclin mRNA isoform in the TbZFP3-uninduced population or after 72 h induction. Figure 8 shows a representative semi-quantitative analysis of the relative selection of EP1 and GPEET mRNA in each cell population. This reveals that the relative selection of EP1 mRNA increased upon TbZFP3 ectopic expression, whereas the efficiency of GPEET mRNA selection was diminished. We conclude that moderate elevation of TbZFP3 levels alters the relative association with EP1 and GPEET mRNAs, this resulting in a change of trypanosome surface antigen expression, inducing a change from GPEET to EP1 Procyclin as the dominant surface protein on procyclic forms.
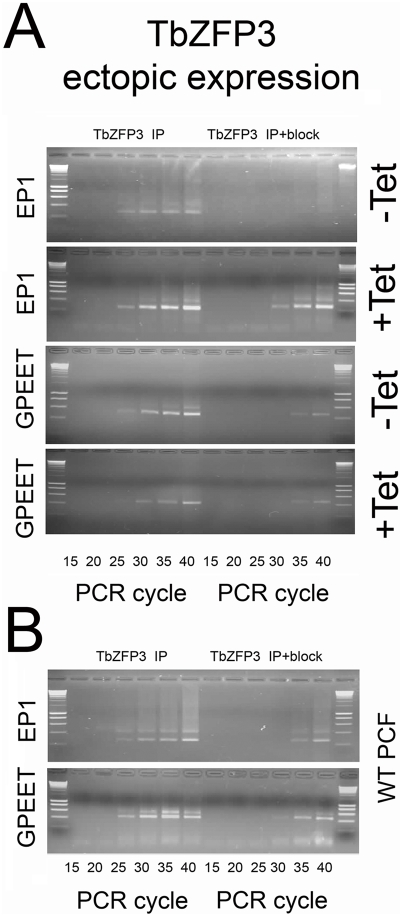
Figure 8. Ectopic expression of TbZFP3 alters its relative association with EP1 and GPEET mRNAs.
(A) RNA-immunoprecipitation of EP1 and GPEET mRNAs by TbZFP3 specific antibody (in the presence or absence of blocking peptide). Immunoprecipitations were carried out using extracts from cells uniniduced (−Tet) or induced (+Tet) to ectopically express TbZFP3. Semi-quantitative assays were performed to directly visualise the amplified products, with samples being isolated after 15, 20, 25, 30, 35, and 40 cycles. The efficiency of EP1 cDNA amplification is enhanced upon TbZFP3 ectopic expression, whereas the efficiency of GPEET cDNA amplification is reduced. All samples were amplified in parallel, resolved on matched gels, and captured at identical exposures. The data are representative of three independent assays. (B) The equivalent assays were also carried out on wild-type procyclic forms as controls. For each reaction, samples generated in the absence of reverse transcriptase failed to amplify any product for EP1 or GPEET, thereby confirming the absence of contaminating genomic DNA (data not shown).
Discussion
The experiments in this paper identify the developmental regulator, TbZFP3, as an isoform-specific regulator of Procyclin surface coat expression in trypanosomes. Specifically, we demonstrate (i) that endogenous TbZFP3 shows sequence-specific co-association with distinct procyclin mRNA isoforms, (ii) that ectopic overexpression of TbZFP3 does not enhance the mRNA abundance of selected transcripts, but rather that (iii) their protein expression is regulated in an isoform-specific manner, as evidenced by mass spectrometric analysis of the Procyclin expression signature in transgenic cell lines. Unlike the wild type TbZFP3 protein, a mutant form of TbZFP3 lacking its C×8C×5C×3H predicted RNA-interaction motif and which cannot co-associate with procyclin mRNAs does not alter Procyclin expression. We have already demonstrated that TbZFP3 promotes differentiation when associated with the translational machinery (this being dependent upon its predicted RNA and protein interaction motifs) and that this occurs only in the parasite life cycle stage at which Procyclin proteins are expressed [18]. Hence, our work provides a long-sought ‘missing link’ between the intensely studied cis-regulatory signals for the procyclin gene family and the general gene expression machinery, this being the translational apparatus.
TbZFP3 shows specific co-association in vivo with EP1 and GPEET procyclin mRNA, whereas the distinctly regulated transcripts EP2 and EP3 are not co-immunoprecipitated. Although deletion of the predicted RNA-binding domain in TbZFP3 prevents the co-association with EP1 mRNA, our studies do not formally distinguish between direct intermolecular contact between TbZFP3 and target mRNAs and indirect contact dependent on other protein factors. Hence, we use the term TbZFP3mRNP to define the composition of the immunoprecipitated material comprising TbZFP3, procyclin mRNAs and, possibly, other identified (e.g. [18]; see below) and unidentified co-operating factors. Nonetheless, by using an immunoprecipitation approach employing an antibody directed to the endogenous TbZFP3 protein in wild type parasites we demonstrate that the observed co-association with procyclin mRNAs is physiological, and directed by TbZFP3 in its normal cellular context. Interestingly, the differential selection of different procyclin mRNA isoforms by the TbZFP3mRNP matches their overall sequence similarity, with EP1 and GPEET 3′UTR sequences being significantly more closely related than EP2 and EP3 (Figure S3). Nonetheless, EP1 and GPEET are differentially regulated in vivo, with GPEET expression being repressed as EP1 is upregulated during differentiation to late procyclic forms in vitro and in the tsetse fly [8],[12]. Significantly, this matches the observed effects of TbZFP3 ectopic overexpression, whereby EP1 expression is elevated to become the dominant surface protein and GPEET expression is repressed, this correlating with enhanced association of the TbZFP3mRNP with EP1 mRNA and diminished association with GPEET mRNA. Although copy number control of Procyclins on the parasite surface could accentuate this switch, it is significant that the inverse control of these surface molecules is regulated by only subtle changes in the abundance of TbZFP3 (∼1.5–2.5 fold). This suggests exquisitely regulated control of Procyclin isoform expression in response to TbZFP3 levels.
In addition to isoform-specific selection, the TbZFP3mRNP exhibits sequence-specific association with the EP1 mRNA 3′UTR, this being dependent upon the integrity of two well-characterised regulatory regions - the ‘Loop II’ and the 16mer stem loop region. Previous analyses have demonstrated that these sequences provide negative and positive control elements for EP1 procyclin expression, respectively. The Loop II region acts as a translational repressor and mRNA destabilisation element in procyclic forms, whereas the 16mer is a translational enhancer, which suppresses the action of the Loop II region [23],[24]. In insect stages, it was predicted that a macromolecular complex would associate with both elements and so shield the ‘Loop II’ element from recognition by a negative regulator, thereby promoting gene expression [23]. Our findings are compatible with this, invoking a model (Figure 9) in which TbZFP3 competes with a negative regulator binding ‘Loop II’, such that TbZFP3 over-expression promotes EP1 Procyclin expression (at the expense of GPEET), whereas RNAi mediated removal of TbZFP3 results in reduced procyclin mRNA abundance. Interestingly, expression of the Loop II deletion reporter construct revealed that TbZFP3mRNP-binding is not necessary for efficient mRNA or protein expression (Figure 3B), suggesting that the TbZFP3mRNP acts primarily as an anti-repressor (Figure 9), matching earlier predictions for the procyclin regulatory machinery [2],[17],[23],[30]. This is analogous to the regulation of nanos RNA during Drosophila embryogenesis, whereby overexpression of Oskar displaces the translational repressor Smaug bound to the nanos 3′UTR [31].
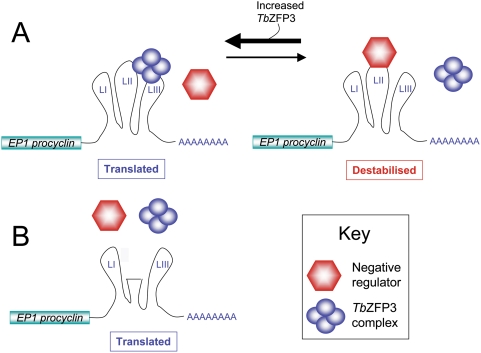
Figure 9. Model for the function of TbZFP3 in EP1 Procyclin expression.
(A) The expression of EP1 procyclin is governed by the balance between a negative regulator (hexagons) and a TbZFP3-containing mRNP complex (circles), each of which competes for binding at the Loop II region. When bound by the negative regulator, EP1 procyclin is poorly translated and its mRNA destabilised. When TbZFP3 expression is increased by ectopic over-expression, the negative regulator is out-competed and expression of EP1 Procyclin increases. (B) When the Loop II region is deleted, neither TbZFP3 nor the negative regulator can bind. In the absence of either factor, EP1 Procyclin expression is enhanced above wild type levels. This demonstrates that TbZFP3 binding is not required for efficient expression, instead suggesting that the TbZFP3 complex acts as an anti-repressor.
CCCH proteins in eukaryotes are involved in all levels of gene expression through RNA recognition, usually this being dependent upon the integrity of at least two juxtaposed CCCH fingers [32]. TbZFP3, however, has only a single CCCH motif and is a member of a family of small CCCH zinc finger proteins (TbZFP1, TbZFP2 and TbZFP3), unique to kinetoplastids, and each <140 amino acids long. These proteins interact in yeast 2-hybrid assays and co-immunoprecipitate in vivo, suggesting that they provide a modular function in order to confer specificity of binding [33]. Whether this is part of a single mRNP complex, or distinct complexes with differential specificities for different genes or in different life cycle stages, remains to be determined. This complexity of interactions may define the specificity of different mRNA classes selected by the TbZFP3mRNP, or moderate their differential efficiency of selection and hence regulation, as observed with EP1 and GPEET mRNA regulation under conditions of TbZFP3 ectopic expression. To be understood in depth, such interactions will need to be analysed on a case-by-case basis for individual RNAs as has been done here for procyclin mRNAs. Nonetheless, analogous combinatorial interaction between RNA binding proteins in kinetoplastid parasites to confer target specificity and regulation has previously been proposed for the small RNA binding proteins TbUBP1 and TbUBP2 [34], homologues of the regulators of mucin gene expression in T.cruzi [35].
TbZFP2 and TbZFP3 are constitutively expressed and associate with the translation apparatus in procyclic forms but not in bloodstream forms [18]. Our demonstration here that the TbZFP3mRNP co-associates with procyclin mRNA regulatory elements that control translation, thus promoting EP1 surface protein expression without enhancing EP1 mRNA abundance, suggests a role for TbZFP3 in translational control. Supporting this, a mutant TbZFP3 lacking the predicted RNA-interaction domain neither co-associates with procyclin mRNAs (Figure 2) nor the translational apparatus [18] and induces no consistent change in Procyclin expression (Figure S2). Temporally-regulated translational control is a key aspect of cell-type development in the Plasmodium parasite, whereby translational repression via the DDX6 RNA helicase family member DOZI regulates gametocyte mRNA expression and life-cycle differentiation [36]. Interestingly, these transcripts share a 47 nt U-rich control element [37], similar to the regulatory U-rich 26mer elements enriched in procyclic form-specific transcripts [38] and comprising part of the Loop II region of EP1 procyclin recognised by TbZFP3. This points to common mechanisms of developmental control among widely divergent eukaryotic protozoan pathogens.
Translational control is believed to be a major mechanism of gene regulation in trypanososmatid parasites [39],[40]. Although the general mRNA degradation and translational machineries are broadly conserved in these evolutionarily ancient eukaryotic organisms [3], it is the kinetoplastid-specific trans-acting regulators that provide the key to understanding their extreme emphasis on post-transcriptional control. Moreover, targeting unique components of the translational machinery in pathogens is a major strategy in antimicrobial therapies. Thus, discovering novel regulators interacting with this apparatus provides both new understanding of gene expression and new possibilities to intervene in the virulence and spread of these devastating parasites.
Materials and Methods
Cell Lines
Procyclic form or bloodstream form T. brucei Lister 427 trypanosomes were used throughout. Cell lines engineered for TbZFP3 ectopic expression in procyclic or bloodstream forms have been described previously and were cultured in SDM-79 or HMI-9, respectively [18].
Immunoprecipitation and qRT-PCR
Immunoprecipitation using TbZFP3-specific antisera, RNA extraction and reverse transcription have been described previously [18]. Blocking peptides used were N-DSSQMQQVGHDVPPMA-C for TbZFP3 and N-EVHTNQDPLD-C for Ty1, each being titrated prior to use. SYBR green qRTPCR reactions were performed using Roche reagents as per specifications for the LightCycler system. The 5′ primers for actin, ep and gpeet and 3′ primers specific to EP1, EP2, EP3, GPEET, or the Anchor sequence were described previously [11],[18]. cDNA was amplified as follows: 10 min, 95°C; 30×[8 s, 95°C; 9 s, 55°C; 12 s, 72°C] with fluorescence acquired at 82°C. The amplification was followed by a melting temperature analysis that measured PCR product fluorescence during a temperature increase from 65°C to 95°C at 0.1°C/s to determine product melting temperature and confirm specificity. Product identities were further verified by gel electrophoresis and DNA sequencing. In all cases, serial dilutions of input cDNAs confirmed the quantitative efficiency of the reactions and “no reverse transcriptase” controls confirmed the absence of contaminating genomic DNA in the RNA preparations.
Northern and Western blotting
Northern blotting involved resolution of 3–5 µg of total trypanosome mRNA on formaldehyde agarose gels resolved in MOPS buffer. Hybridization of blots used digoxigenin labelled riboprobes, detected using anti-DIG alkaline phosphatase-conjugated antibody and visualised using CDP-star as a reaction substrate (Roche). Western blots were detected and quantitated using a Li-COR Odyssey system, using alpha-tubulin as an internal standard.
Mass spectrometry
Mass spectrometry was carried out according to the methodology described in [8],[19],[29]. Briefly, parasite pellets were freeze-dried and then extracted twice with 200 µl of Chloroform/Methanol/Water, 10∶10∶3 (V/V/V), under sonication (10 min). After centrifugation, the delipidated pellets were then extracted 3 times with 150 µl of 9% butanol (ButOH), also under sonication. The ButOH fractions contain the Procyclins. All ButOH fractions were then freeze-dried and submitted to dephosphorylation using 50 µl of 48% aqueous hydrofluoric acid (aq.HF), at 0°C for 24 h. After aq.HF incubation the samples were freeze-dried again and washed twice with water. The samples were then dried and further incubated with 200 µl of 40 mM TFA, 20 min at 100°C (mild acid conditions), in order to assist visualization of the Procyclin C-termini and the identification of each isoform. Under this condition, the Asp-Pro bonds of most of the EP isoforms are cleaved whereas GPEET partially releases 13 amino acids at its N-terminus. Equivalent amounts of each sample were mixed with α-cyano (matrix) and analysed by negative-ion MALDI-TOF-MS using a Voyager-DE STR instrument.
Supporting Information
Procyclin isoform expression in parental procyclic forms.
(0.51 MB DOC)
Procyclin isoform expression in transgenic cells expressing the ΔCCCH mutant of TbZFP3-Ty.
(0.58 MB DOC)
Overall similarity between the 3′UTRs of EP1, EP2, EP3, and GPEET procyclin mRNAs.
(2.22 MB DOC)
Acknowledgments
We thank Douglas Lamont (Proteomics Facility, Dundee University) for access to the MALDI mass spectrometer, Professor Judi Allen (University of Edinburgh) for Lightcycler access, and Dr. Toni Aebischer (University of Edinburgh) for thoughtful comments on the manuscript. Reporter constructs and GARP antiserum were kindly provided by Professor Isabel Roditi, University of Bern.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a Wellcome Trust Programme grant to KRM and a Wellcome Trust Career Development Fellowship and Wellcome Trust Value in People (VIP) award to AAS.
References
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Clayton CE. NEW EMBO MEMBER'S REVIEW Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roditi I, Liniger M. Dressed for success: the surface coats of insect-borne protozoan parasites. Trends Microbiol. 2002;10:128–134. doi: 10.1016/s0966-842x(02)02309-0. [DOI] [PubMed] [Google Scholar]
- 7.Marcello L, Barry JD. From silent genes to noisy populations-dialogue between the genotype and phenotypes of antigenic variation. J Eukaryot Microbiol. 2007;54:14–17. doi: 10.1111/j.1550-7408.2006.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, et al. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci U S A. 2001;98:1513–1518. doi: 10.1073/pnas.041611698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagamune K, Acosta-Serrano A, Uemura H, Brun R, Kunz-Renggli C, et al. Surface sialic acids taken from the host allow trypanosome survival in tsetse fly vectors. J Exp Med. 2004;199:1445–1450. doi: 10.1084/jem.20030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guther ML, Lee S, Tetley L, Acosta-Serrano A, Ferguson MA. GPI-anchored proteins and free GPI glycolipids of procyclic form Trypanosoma brucei are nonessential for growth, are required for colonization of the tsetse fly, and are not the only components of the surface coat. Mol Biol Cell. 2006;17:5265–5274. doi: 10.1091/mbc.E06-08-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urwyler S, Vassella E, Van Den Abbeele J, Renggli CK, Blundell P, et al. Expression of procyclin mRNAs during cyclical transmission of Trypanosoma brucei. PLoS Pathog. 2005;1:e22. doi: 10.1371/journal.ppat.0010022. doi:10.1371/journal.ppat.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassella E, Den Abbeele JV, Butikofer P, Renggli CK, Furger A, et al. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- 13.Vassella E, Probst M, Schneider A, Studer E, Renggli CK, et al. Expression of a major surface protein of Trypanosoma brucei insect forms is controlled by the activity of mitochondrial enzymes. Mol Biol Cell. 2004;15:3986–3993. doi: 10.1091/mbc.E04-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hehl A, Vassella E, Braun R, Roditi I. A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1994;91:370–374. doi: 10.1073/pnas.91.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol Cell Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schurch N, Hehl A, Vassella E, Braun R, Roditi I. Accurate polyadenylation of procyclin mRNAs in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotz HR, Biebinger S, Flaspohler J, Clayton C. PARP gene expression: control at many levels. Mol Biochem Parasitol. 1998;91:131–143. doi: 10.1016/s0166-6851(97)00196-5. [DOI] [PubMed] [Google Scholar]
- 18.Paterou A, Walrad P, Craddy P, Fenn K, Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life cycle development. J Biol Chem. 2006;281:39002–39013. doi: 10.1074/jbc.M604280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassella E, Acosta-Serrano A, Studer E, Lee SH, Englund PT, et al. Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J Mol Biol. 2001;312:597–607. doi: 10.1006/jmbi.2001.5004. [DOI] [PubMed] [Google Scholar]
- 20.Treumann A, Zitzmann N, Hulsmeier A, Prescott AR, Almond A, et al. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J Mol Biol. 1997;269:529–547. doi: 10.1006/jmbi.1997.1066. [DOI] [PubMed] [Google Scholar]
- 21.Butikofer P, Ruepp S, Boschung M, Roditi I. ‘GPEET’ procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem J. 1997;326 (Pt 2):415–423. doi: 10.1042/bj3260415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 23.Furger A, Schurch N, Kurath U, Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotz HR, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3026. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drozdz M, Clayton C. Structure of a regulatory 3′ untranslated region from Trypanosoma brucei. RNA. 1999;5:1632–1644. doi: 10.1017/s1355838299990623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hehl A, Pearson TW, Barry JD, Braun R, Roditi I. Expression of GARP, a major surface glycoprotein of Trypanosoma congolense, on the surface of Trypanosoma brucei: characterization and use as a selectable marker. Mol Biochem Parasitol. 1995;70:45–58. doi: 10.1016/0166-6851(95)00003-j. [DOI] [PubMed] [Google Scholar]
- 28.Bayne RA, Kilbride EA, Lainson FA, Tetley L, Barry JD. A major surface antigen of procyclic stage Trypanosoma congolense. Mol Biochem Parasitol. 1993;61:295–310. doi: 10.1016/0166-6851(93)90075-9. [DOI] [PubMed] [Google Scholar]
- 29.Acosta-Serrano A, Cole RN, Mehlert A, Lee MG, Ferguson MA, et al. The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J Biol Chem. 1999;274:29763–29771. doi: 10.1074/jbc.274.42.29763. [DOI] [PubMed] [Google Scholar]
- 30.Roditi I, Furger A, Ruepp S, Schurch N, Butikofer P. Unravelling the procyclin coat of Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:117–130. doi: 10.1016/s0166-6851(97)00195-3. [DOI] [PubMed] [Google Scholar]
- 31.Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 32.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 33.Hendriks EF, Robinson DR, Hinkins M, Matthews KR. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 2001;20:6700–6711. doi: 10.1093/emboj/20.23.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann C, Benz C, Brems S, Ellis L, Luu VD, et al. Small trypanosome RNA-binding proteins TbUBP1 and TbUBP2 influence expression of F-box protein mRNAs in bloodstream trypanosomes. Eukaryot Cell. 2007;6:1964–1978. doi: 10.1128/EC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Orso I, Frasch AC. TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and Poly(A)-binding proteins forming a ribonucleoprotein complex. J Biol Chem. 2002;277:50520–50528. doi: 10.1074/jbc.M209092200. [DOI] [PubMed] [Google Scholar]
- 36.Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, et al. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braks JA, Mair GR, Franke-Fayard B, Janse CJ, Waters AP. A conserved U-rich RNA region implicated in regulation of translation in Plasmodium female gametocytes. Nucleic Acids Res. 2007;36:1176–1186. doi: 10.1093/nar/gkm1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 2006;34:5312–5324. doi: 10.1093/nar/gkl598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen-Freue G, Holzer TR, Forney JD, McMaster WR. Global gene expression in Leishmania. Int J Parasitol. 2007;37:1077–1086. doi: 10.1016/j.ijpara.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Horn D. Codon usage suggests that translational selection has a major impact on protein expression in trypanosomatids. BMC Genomics. 2008;9:2. doi: 10.1186/1471-2164-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaCount DJ, Bruse S, Hill KL, Donelson JE. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol Biochem Parasitol. 2000;111:67–76. doi: 10.1016/s0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- 42.Hall BS, Pal A, Goulding D, Acosta-Serrano A, Field MC. Trypanosoma brucei: TbRAB4 regulates membrane recycling and expression of surface proteins in procyclic forms. Exp Parasitol. 2005;111:160–171. doi: 10.1016/j.exppara.2005.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Procyclin isoform expression in parental procyclic forms.
(0.51 MB DOC)
Procyclin isoform expression in transgenic cells expressing the ΔCCCH mutant of TbZFP3-Ty.
(0.58 MB DOC)
Overall similarity between the 3′UTRs of EP1, EP2, EP3, and GPEET procyclin mRNAs.
(2.22 MB DOC)