Abstract
The transcription factor Yin Yang 1 (YY1) is a multifunctional protein that can activate or repress gene expression depending on the cellular context. YY1 is ubiquitously expressed and highly conserved between species. However its role varies in diverse cell types and includes proliferation, differentiation and apoptosis. This review will focus on the function of YY1 in the nervous system including its role in neural development, neuronal function, developmental myelination and neurological disease. The multiple functions of YY1 in distinct cell types are reviewed and the possible mechanisms underlying the cell specificity for these functions are discussed.
Keywords: brain, neuron, oligodendrocyte, chromatin, HDAC
1. Introduction
Yin Yang 1 (YY1) is a multifunctional nuclear protein that can act as transcriptional repressor or activator. It was identified in 1991 as a protein that represses the activity of the adeno-associated viral (AAV) P5 promoter in the absence of the oncoprotein E1A, while it activates the promoter in the presence of E1A (Shi et al. 1991). Inspired by its dual transcriptional activity, Shi and colleagues named the protein "Yin Yang 1" from the Chinese "Yin", for repression and "Yang" for activation (Shi et al. 1997).
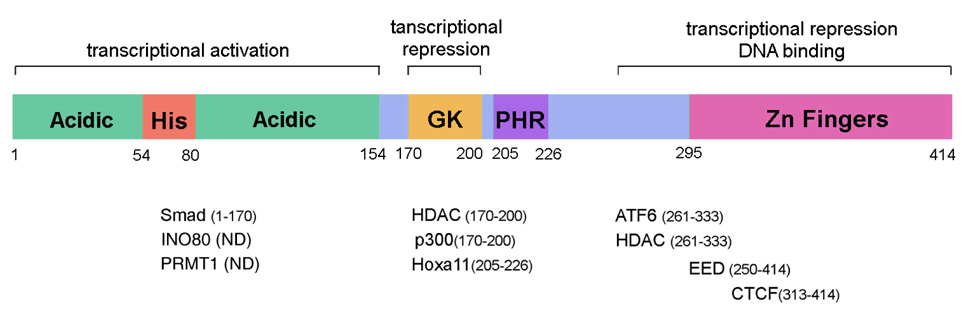
Human YY1 is a protein composed of 414 amino acids with a calculated molecular weight of 44kDa (Figure 1). YY1 contains four C2H2-type zinc fingers at the C-terminus (aa 295–414), which are responsible for sequence-specific binding to the consensus DNA recognition sequence: 5’-(C/g/a)(G/t)(C/t/a)CATN(T/a)(T/g/c)-3’ (Hyde-DeRuyscher et al. 1995; Yant et al. 1995). The N-terminus contains a histidine-rich region (His) flanked by acidic aminoacids, and that serves as transcriptional activation domain (Fig.1). The center of the molecule is glycine and lysine-rich (aa 170–200), and corresponds to the interaction domain with HDAC and together with the C terminus, forms the transcriptional repression domain (Shi et al. 1997; Yao et al. 2001). An additional region within the central domain (PHR, aa 205–226) mediates the interaction with the homeobox Hox proteins (Brown et al. 1998; Wang et al. 2004; Luke et al. 2007). YY1 is highly conserved between species (aa identity: human vs. mouse 98.8%, human vs. Xenopus 90.3%) and is ubiquitously expressed in different tissues including brain, heart, limb and immune system (Pisaneschi et al. 1994; Shi et al. 1997; Donohoe et al. 1999; Affar et al. 2006; Liu et al. 2007b; He et al. 2007). The promoter of yy1 does not contain a conventional TATA box, but has multiple Sp1 binding sites and is GC rich, thereby resembling the promoter of a large subset of housekeeping and growth regulated genes (Safrany and Perry 1993; Yao et al. 1998). These features suggested that YY1 might have an important role in development. Consistent with this prediction, genetic ablation of yy1 in mice resulted in lethality shortly after implantation (Donohoe et al. 1999).
Figure 1.
Schematic diagram of human YY1 protein and its binding partners. The transcriptional activation and repression domains and the DNA binding domain are indicated. His, histidine-rich domain; GK, glycine-lysine-rich domain; PHR, PHO homology region. The protein/complex associated with YY1 are shown below each region of interaction. In parenthesis the aminoacid residues involved in binding are indicated. ND means not determined.
2. Role of YY1 in cell proliferation, differentiation and survival
The role of YY1 in proliferation is quite controversial. Studies on lymphoid cell lines with genetic ablation of yy1 suggested that this molecule might favor proliferation, since yy1 deficient cells were characterized by slow growth (Sui et al. 2004; Vega et al. 2005). This interpretation was supported by the detection of high levels of YY1 protein in human prostate cancer tissue (Seligson et al. 2005; Gordon et al. 2006). Very different conclusions were reached by studies in human breast cancer cells, where YY1 was identified as proliferation inhibitor due to its role as transcriptional repressor of cyclin D1, a critical factor for cell cycle entry (Cicatiello et al. 2004). To render the interpretation of YY1 function even more complex, recent studies conducted in vivo, in mice with conditional deletion of yy1 in the oligodendrocyte lineage suggested that the role of YY1 in progenitor cells is independent of the cell cycle (He et al. 2007).
The role of YY1 in differentiation is also dependent on the specific cell type. In the C2C12 cells, a model system for myoblastic differentiation, down-regulation of YY1 is a prerequisite for the expression of muscle-specific genes such as skeletal alpha actin (Lee et al. 1994). Additional studies further confirmed the inhibitory role of YY1 in muscle differentiation, due to its effect on differentiation signaling pathways (Kurisaki et al. 2003). A similar role for YY1 was reported in keratinocytes, since YY1 negatively regulates differentiation by repressing the keratinocyte-specific gene loricrin (Xu et al. 2004). However, opposite role of YY1 in differentiation was proposed by in vivo studies in yy1 conditional knockout mouse, in the lymphoid system (Liu et al. 2007a) and in the oligodendrocyte lineage (He et al. 2007). In both cases, ablation of yy1 retained the cells at an immature stage, despite the dramatic diversity between the transcriptional program required for B cell differentiation and for oligodendrocyte differentiation. This raises the question of the exact role played by YY1 in cellular differentiation. It will also be interesting to determine whether the role of YY1 as pro-differentiation factor will also be detected in other lineages, by analyzing the phenotype of yy1 conditional mutant mice in other systems.
A protective role of YY1 from apoptosis was suggested by studies using siRNA or genetic targeted mutation in lymphoid cells (Sui et al. 2004). It was suggested that YY1 exerts anti-apoptotic functions by negatively regulating Hdm2-mediated p53 degradation. However, knock-down experiments of yy1 in embryonic carcinoma cell line did not induce apoptosis, despite increased p53 levels (Bain and Sinclair, 2005). In addition, studies in mouse embryonic fibroblasts (Affar el et al. 2006) or in oligodendrocyte lineage cells (He et al. 2007) indicated that decreased levels of YY1 did not affect p53 levels nor they increased apoptosis.
In addition to the described biological functions, new roles for YY1 include cancer biology (reviewed in Gordon et al. 2006) and DNA imprinting (Hendrich et al. 1997; Kim et al. 2006; Donohoe et al. 2007). A thorough discussion of these functions will not be included in this review due to space restriction. In contrast, we shall focus on the function of YY1 in the nervous system.
3. Role of YY1 in the nervous system
3.1 YY1 in the developing nervous system
The potential function of YY1 in the developing nervous system was first suggested by the phenotypic analysis of yy1+/− mice. Complete ablation of yy1 in mice resulted in early embryonic lethality which precluded the analysis of later developmental stages. However, a small subset (16.7% – 24%) of yy1 heterozygotes displayed growth retardation and neurulation defects. The brains of the abnormal embryos demonstrated exencephaly, asymmetric structure and the presence of pseudo-ventricles (Donohoe et al. 1999).
Studies in Xenopus after knock-down of the Xenopus homologue of YY1 (XYY1) using anti-sense morpholino oligonucleotides (MO) revealed similar neurulation defects (Morgan et al. 2004). While a dramatic reduction of XYY1 protein levels (below 20% of the controls) resulted in early embryonic lethality (Morgan et al.2004), a partial depletion of XYY1 resulted in antero-posterior patterning defects and reduction of head structures (Kwon and Chung 2003; Morgan et al. 2004). Together, these studies in mouse and Xenopus supported a pro-differentiative role of YY1 in the developing nervous system. Consistent with this concept, over-expression studies by injecting XYY1 mRNA in Xenopus embryos, was sufficient to induce neural tissue, although it had no effect on the induction of mesodermal tissue (Satijn et al. 2001).
To begin elucidating the molecular mechanisms underlying the role of YY1 in nervous system development, gene expression profiling was conducted in XYY1 depleted Xenopus embryos (Table 1). The genes whose expression was decreased upon yy1 deletion could be grouped into two main categories: transcription factors involved in patterning (i.e. homeobox genes, engrailed2, Otx2 and Krox20) and those involved in neural crest cell specification and migration (i.e. slug, snail).
Table 1. List of genes regulated by YY1 in the nervous system.
The function of YY1 (activation or repression of the target genes), is indicated. Note that the region or specific cell type of expression, the general function of the gene and the specific reference for each gene are also described
| Gene | Function of YY1 | Region or cell type | Function | System used | References |
|---|---|---|---|---|---|
| Bace1 | activation | brain | β-secretase of APP and NRG-1 | PC12 cell, rat neuron and astrocyte | Nowak et al. 2006 |
| DBH | activation | neuron | Dopamine β-hydroxylase | Human neuroblastoma SK-N-BE(2)C | Seo et al. 1996 |
| Dynamin I | repression | brain | Receptor-mediated endocytosis in neuron | Mouse neuroblastoma cell NS20Y | Yoo et al. 2000 |
| Engrailed 2 | activation | Midbrain-hindbrain junction | Midbrain-hindbrain patterning, autism | Xenopus | Kwon and Chung 2003 |
| FE65 | activation | neuron | APP interacting protein, Alzheimer disease | Rat brain extracts and Chinese hamster ovary cell | Zambrano et al. 1997 |
| Glast | repression | astrocyte | Glutamate transporter | Chicken Bergmann glial cells | Rosas et al. 2007 |
| Id4 | Repression | oligodendrocyte | Myelination inhibitor | Mouse | He et al. 2007 |
| Krox20 | activation | Hindbrain, Schwann cell | Hindbrain development, Schwann cell myelination | Xenopus | Kwon and Chung 2003 |
| Nrp1 | activation | neuron | Neuronal marker | Xenopus | Satijn et al. 2001; Morgan et al. 2004 |
| Otx2 | activation | neural crest, forebrain and midbrain | Anterior neuroectoderm development, eye development | Xenopus; F9 embryonal carcinoma cells | Kwon and Chung 2003; Takasaki et al. 2007 |
| plp | activation | oligodendrocyte | Myelin protein | Rat oligodendrocyte CG4 cell | Berndt et al. 2001 |
| REST | activation | Non-neuronal cells | repress neuronal genes expression | mouse embryonic fibroblast; SH-SY5Y cells | Affar et al. 2006; Jiang et al. 2007 |
| Slug | activation | neural crest | Neural crest migration | Xenopus | Morgan et al. 2004 |
| Snail | activation | neural crest | Neural crest migration | Xenopus | Morgan et al. 2004 |
| Tcf4 | repression | brain | Wnt signal downstream effecter | Mouse | He et al. 2007 |
The pattern of YY1 expression in the developing midbrain, hindbrain, and cerebellar primordia at midgestation further validates the importance of this molecule in neural development (Donohoe, et al., 1999). Indeed several YY1-dependent genes with decreased expression after XYY1 knockdown (i.e.Otx2, engrailed 2 and Krox20) play an important role in the development of these brain regions (Kwon and Chung, 2003) and their concomitant decrease of expression may explain the defective anterior-posterior axial patterning and reduction of head structures observed in both mice and Xenopus with reduced YY1 expression. Engrailed 2, for instance marks the boundary between midbrain and hindbrain (Hemmati-Brivanlou et al. 1991), while Krox20 regulates hindbrain patterning since Krox20 null mice lack rhombomeres 3 and 5 (Schneider-Maunoury et al. 1993). Otx2 is expressed in the anterior neuroectoderm, including forebrain and midbrain (Blitz and Cho, 1995), and this expression pattern is dependent on the presence of an enhancer containing YY1 consensus binding sequence (Takasaki et al. 2007). YY1 directly binds to two cis-sites in the promoter and the enhancer region of the Otx2 gene (Takasaki et al. 2007) and mutations of this binding site within the enhancer region, abolished the expression of Otx2 in the anterior neuroectoderm, suggesting a critical role of YY1 in determining the regional patterning of expression (Takasaki et al. 2007).
The additional group of genes with decreased expression upon deletion of Xyy1 included the neural marker Nrp1 and the neural crest markers slug and snail (Morgan et al., 2004). Slug is a member of the Snail family of zinc finger proteins that directly bind to the DNA motif called E-box (Nieto et al. 1994). Both Slug and Snail are able to promote the epithelial to mesenchymal transition (Bolos et al. 2003). Inhibition of slug gene in Xenopus and chick embryos decreases the number and motility of neural crest (NC) cells, indicating its role in specification and migration of neural crest cells (Carl et al. 1999; LaBonne and Bronner-Fraser, 2000). Additional evidence demonstrating YY1 direct regulation of slug expression include electrophoretic mobility shift assay, chromatin immunoprecipitation and slug promoter-driven GFP reporter assays (Morgan et al 2004). It is worth mentioning that the other YY1-regulated gene Otx2, similar to slug and snail is also expressed in neural crest cells at the migratory phase (Kimura et al. 1997). Together these data support an important role for YY1 in regulating anterior patterning in the central nervous system and possibly specification and migration of neural crest cells in the peripheral nervous system.
We have previously mentioned that YY1 has been shown to positively regulate the expression of the homeobox gene Hoxb4 (Gilthorpe et al. 2002). This is a very interesting gene whose forced expression in hematopoietic stem cells has been correlated with trans-differentiation into oligodendrocytes (Miyake et al., 2006). The YY1-dependent regulation of homeobox genes is consistent with its function as Polycomb Group (PcG) protein during early development. PcG proteins are a group of chromatin modulators that were originally identified and characterized in Drosophila as critical factors to maintain the transcriptionally inactive state of homeobox genes in appropriate regions of the embryo (Pirrotta, 1998). Indeed, YY1 shows high degree of homology with the Drosophila PcG protein Pho (Brown et al. 1998&2003). Mutation of pho in Drosophila resulted in a typical homeotic transformation and abnormal development of the central nervous system, and the transient expression of human yy1 was sufficient to rescue the phenotype (Atchison et al. 2003; Srinivasan et al. 2005). The mechanism of PcG repression has been related to the ability of specific enzymatic activities to catalyze repressive histone methylation. These enzymatic activities include EED (embryonic ectoderm development), a vertebrate homologue of the Drosophila PcG protein Extra Sex Combs (ESC) (Satijn et al. 2001; Atchison et al. 2003) and EZH2, related to Drosophila Enhancer of Zeste (E(Z)) (van Lohuizen et al. 1998). The eed−/− mice show defects in very early development, including gastrulation and neural defects (Schumacher et al. 1996). Over-expression of XEED (the Xenopus homologue of EED) and of XYY1, but not of any other PcG protein, induced an ectopic neural axis in Xenopus embryo without any effect on the induction of mesodermal tissue (Satijn et al. 2001).
3.2. YY1 in the neuronal differentiation and function
Neuronal differentiation is characterized by the expression of genes including SCG10, sodium channel type II, synapsin, glutamate receptor, and acetylcholine receptor. These differentiation genes are the main targets of the transcription factor called REST (i.e. repressor element 1 (RE1)-silencing transcription factor). REST plays essential roles in restricting the expression of neuronal genes to neurons by mediating active repression as well as long-term epigenetic silencing of neuronal genes in differentiated non-neuronal cells (Ballas et al. 2005). Interestingly, microarray analysis of yy1 hypomorphic (yy1flox/−) mouse embryonic fibroblasts revealed a significant decrease of REST expression levels (Affar et al. 2006). These results were further validated by the recent characterization of conserved YY1 binding sites in the promoter of the mouse REST gene and by the YY1-dependent positive regulation of the REST promoter in SH-SY5Y cells (Jiang et al. 2007). These results suggested that lack of YY1 might induce neuronal differentiation, a conclusion that is quite different from the one drawn from the studies in Xenopus and yy1 heterozygotes, as discussed in the previous subchapter. In vitro differentiation studies of multipotential progenitors derived from the subventricular zone (SVZ) of yy1flox/flox mice and infected with adenovirus expressing Cre recombinase to delete yy1 in vitro indicated that the ability of these cells to differentiate into Tuj1+ neurons was not affected in the absence of yy1 (He et al. 2007). Yet, this single observation did not exclude an important role of YY1 in early embryonic development as reviewed above, or in the expression of late neuronal differentiation genes.
The role of YY1 in late neuronal differentiation has been suggested by the evidence that this molecule positively regulates the basal levels of expression of dopamine beta-hydroxylase (DBH) in noradrenergic neurons (which catalyzes dopamine to noradrenaline), by binding to specific sites in the promoter (Seo et al. 1996; Yang et al. 1998). YY1 has also been identified as negative regulator of Dynamin I (Yoo et al. 2001), a protein that is highly expressed in the brain and that plays a critical role in clathrin-mediated endocytosis and synaptic vesicle recycling (Kosaka and Ikeda 1983; Cook et al. 1996). The phenotypic characterization of dynamin I −/− mice revealed failure to thrive and death during the first two neonatal weeks. In the knockout mice synaptic vesicle endocytosis was severely impaired during strong exogenous stimulation but it efficiently resumed upon termination of the stimulus. These data suggested that indicating basal synaptic vesicle endocytosis is a dynamin I-independent mechanisms while recycling of vesicles during high levels of neuronal activity requires dynamin-1 function (Ferguson et al. 2007). It remains to be determined whether YY1 activity is differentially modulated by low or high levels of neuronal activity and is responsible for regulating synaptic vesicles recycling.
3.3. YY1 in astrocytes
YY1 function appears to be dispensable also for astrogliogenesis since deletion of yy1 did not affect the generation of astrocytes from neurospheres generated from SVZ cells (He et al. 2007). In this lineage YY1 might play a different role by affecting the expression of molecules involved in glutamate transport. Glutamate is the major excitatory aminoacid neurotransmitter in the vertebrate brain and its extracellular levels are kept at physiologically low levels by transporter systems that are expressed in neurons and glial cells to prevent excitotoxic effects (Gegelashvili and Schousboe 1997). Among the identified glutamate transporters GLAST/EAAT1 is the major player and is expressed almost exclusively in astrocytes throughout the central nervous system, although it is significantly enriched in the Bergmann glial cells (BGC) of the cerebellum (Danbolt et al. 2001). In chick BGC, treatment with glutamate increases binding of YY1 to glast promoter and results in down-regulation of glast transcripts with consequent decreased glutamate re-uptake. Similar results can be obtained by over-expressing yy1. Together these data suggest a specific function of YY1 as transcriptional repressor for glast in cerebellar astrocytes (Rosas et al. 2007). It should be noted that in rat cortical astrocytes, glutamate has an opposite effect on glast by increasing rather than decreasing its expression (Gegelashvili et al. 1996), for this reason future studies will need to determine whether the function of YY1 in cortical astrocytes could be different from that defined in cerebellar BGCs.
3.4 Role of YY1 in myelination
In the central nervous system, myelination is carried out by oligodendrocytes. The function of YY1 in the oligodendrocyte lineage was first reported by Berndt and colleagues, who identified and characterized three YY1 binding elements in the myelin specific proteolipid protein (plp) promoter. Using CAT assay, they demonstrated that YY1 activates plp promoter and gene expression in rat oligodendrocyte CG4 cells (Berndt et al. 2001).
A more direct evidence for the role of YY1 in regulating myelination was provided by the analysis of yy1 conditional knockout mice study in our laboratory (He et al. 2007). By crossing yy1flox/flox mice with mice expressing the recombinase Cre under the oligodendrocyte-specific cnp1 promoter, the specific ablation of yy1 in the oligodendrocytic lineage was achieved. These mice survived until 2 months of age, thereby allowing the detailed analysis of developmental myelination. The yy1 cko mice displayed tremor, ataxia and head wobbling from the second postnatal week and phenotypically resembled dysmyelination mutants. Ultrastructural studies confirmed the decreased number of myelinated axons in white matter tracks of the central nervous system, with the most severe defects in the spinal cord. This phenotype was caused by the arrested development of oligodendrocytes at the progenitor stage, in the absence of changes in proliferation or apoptosis. At a molecular level, this block of differentiation was associated with high levels of expression of transcriptional inhibitors, including Id4 and Tcf4, due to defective recruitment of repressive complexes containing YY1 and the histone deacetylase HDAC1 to their promoters (He et al. 2007). Together, the above mentioned experimental results identified YY1 as a critical player in developmental myelination of the central nervous system.
The function of YY1 in the peripheral nervous system has not been reported yet. However, it is likely that YY1 may also modulate this event since multiple YY1-target genes also regulate the activity or development of the myelinating cells of the PNS. Peripheral myelination is carried out by Schwann cells, which derive from neural crest cells (reviewed in Jessen and Mirsky, 2005). We have previously reviewed the evidence that YY1 regulates multiple genes (i.e. slug, snail, Otx2) that are important for neural crest specification and/or migration. We have also discussed the effect of YY1 on the zinc-finger protein Krox20 a molecule that is directly involved in the maturation of myelinating Schwann cell (Topilko et al. 1994; Le et al. 2005). An additional evidence suggesting a potential role of YY1 in PNS myelination was suggested by the study of YY1 positively regulating Bace1. BACE1 is a type I trans-membrane aspartyl protease that has been recently defined as important for the cleavage of neuregulin-1 (NRG-1) into an active form (Willem et al. 2006). NRG-1 is an axonal membrane associated ligand and plays critical roles in the myelination of the peripheral nervous system (Michailov et al., 2004; Nave and Salzer 2006). As predicted, Bace1 knockout mice revealed striking accumulation of full-length type III NRG-1 proteins in peripheral nerves and severe hypomyelination in peripheral nerves, resembling the phenotype of NRG-1 mutant mice (Willem et al. 2006; Hu et al. 2006). Since YY1 has been shown to be an activator for Bace1 expression in neurons (Rosas et al. 2007), it is conceivable that it may indirectly modulate myelination of peripheral nerves.
4. Role of YY1 in neurodegeneration
In addition to its function in development, YY1 might also play a role in the neurological diseases such as Alzheimer disease (AD). One of the major hallmarks of AD is the brain deposition of the amyloid-beta peptide (Aβ). Aβ is proteolytically cleaved from amyloid precursor protein (APP) by β and γ-secretase and BACE1 (beta-site amyloid precursor protein-cleaving enzyme 1) is one of the major β-secretases (reviewed in Rossner et al. 2006). As mentioned before, YY1 acts as an activator of the BACE1 promoter in neurons and astrocytes, and mutations of the YY1 binding site in the Bace1 promoter decrease its activity, while YY1 over-expression increases its transcriptional activity (Nowak et al. 2006). An alternative possibility is that YY1 might regulate the levels of Aβ indirectly, by modulating the expression of other molecules involved in APP processing, such as FE65. It has been reported that YY1 binds to the FE65 minimal promoter and increases its transcription (Zambrano et al. 1997). FE65 is an adaptor protein with the ability to bind the C-terminal domain of APP (Ermekova et al. 1998). FE65 is highly expressed in neurons, it modulates APP processing and trafficking in several cell lines (Guenette et al. 1999; Santiard-Baron et al. 2005; Wiley et al. 2007) and its expression levels in AD patients’ brain correlates with the severity of the disease and with the risk of developing late-onset AD (Lambert et al. 2000; Delatour et al. 2001). Together these studies suggest a potential role for YY1 in the pathogenesis of Alzheimer disease, although its actual function deserves further investigation.
A possible role for YY1 in neurodegeneration was suggested by the effect of glutamate treatment on YY1-containing protein complexes in cultured neurons (Korhonen et al. 2005). Glutamate is the major excitatory aminoacid neurotransmitter in the vertebrate brain. However high levels of this aminoacid are toxic to neurons and will cause neuronal cell death (Ermak and Davies, 2002). Korhonen et al. discovered that treating cerebellar granule cells with glutamate induced the transition from a high molecular weight protein complex containing YY1 to a smaller protein complex (Korhonen et al. 2005). Treatment with other apoptotic stimuli, such as okadaic acid (inhibitor of serine/threonine protein phosphatase 2A), but not with etoposide (topoisomerase II inhibitor) or trichostatin A (HDAC inhibitor), caused a similar re-organization of YY1 protein complexes, although the biological significance of the high and low molecular weight complexes remains to be determined (Korhonen et al. 2005). An additional role of YY1 in modulating the levels of extracellular glutamate has been previously described and concerns the regulation of the astroglial glutamate transporter GLAST/EAAT1 (Danbolt et al. 2001; Rosas et al. 2007).
Finally, because YY1 is able to affect the expression of engrailed 2 and recent human genetics studies have proposed the existence of a genetic linkage of engrailed 2 with autism (Benayed et al. 2005; Brune et al. 2008), it would be interesting to define whether YY1 itself is involved in autism. In support of its new role, engrailed 2 knockout mice displayed decreased social behavior and defective learning and memory tasks together with a cerebellar-specific increase in serotonin, al of which resembles autism spectrum disorder (Cheh et al. 2006)‥
5. Regulation of YY1 activity
After more than a decade since its discovery, and an overwhelming literature on the opposing roles of YY1 in different cell types and at different developmental stages, it is imperative to ask the question: how can a ubiquitous protein, like YY1, be multifunctional? Generally, there are two ways to endow specificity of function. One possibility is that the protein itself is post-translationally modified in a cell-specific fashion. The other possibility is that the function of YY1 is dependent on the bioavailability of co-factors.
5.1. YY1 expression, subcellular localization and post-translational modifications
While several studies focused on YY1-dependent regulation of downstream target genes, very little is known about the mechanisms regulating YY1 expression itself. Although the levels of YY1 transcripts are relatively constant in the organism, there is some variation during development. For example, as previously mentioned, the XYY1 protein is localized in the anterior neural tube in embryonic Xenopus (Kwon and Chung 2003). For keratinocytes, higher levels of YY1 were detected in undifferentiated cells compared to differentiated cells (Xu et al. 2004). However, this was not the case for oligodendrocyte lineage cells, where the levels remain constant throughout development (He et al. 2007). Future studies on the characterization of YY1 promoter and its mechanisms of regulation will need to be addressed.
An additional modulation of YY1 function is determined by changes in the subcellular distribution. Nucleo-cytoplasmic shuttling of YY1, for instance was correlated with different phases of the cell cycle in Chinese hamster ovary and in HeLa cells (Palko et al. 2004). YY1 was mainly cytosolic during the G1 phase, nuclear in early S phase and cytosolic during late S phase (Palko et al. 2004). Also in Xenopus oocytes the subcellular localization of XYY1 changed at distinct developmental stages and correlated with the DNA binding activity, being nuclear in stage I oocytes, cytosolic in embryos until the early blastula stage and nuclear again after the mid-blastula transition (Ficzycz et al. 2001).
YY1 activity can also be modulated by multiple post-translational modifications, including phosphorylation, acetylation and caspase-dependent cleavage. YY1 contains several Ser/Thr potential phosphorylation sites within the zinc finger DNA-binding domain. Since phosphatase treatment abolished binding of YY1 to the murine leukaemia virus LTR, it was suggested that phosphorylation decreased the ability of YY1 to modulate transcription (Becker et al. 1994). However the binding of YY1 to the AAV p5 promoter was not affected by phosphatase treatment (Shi et al. 1997). Acetylation of lysine residues located in the central region by p300 was also proposed to modulate its activity (Yao et al. 2001). Studies on YY1 dependent regulation of the mouse homeobox gene Otx2 demonstrated that the deacetylated form is capable of bind to both the enhancer and promoter of Otx2, while the acetylated form can only bind to the enhancer (Takasaki et al. 2007). Finally, a recent study in Hela cells reported caspase-dependent cleavage of the first 119 amino acid of YY1 in response to apoptotic stimuli. The newly generated N-terminal truncated YY1 fragment is still able to bind to DNA although it is no longer able to stimulate transcription (Krippner-Heidenreich et al. 2005). It will be important for future studies to determine whether the truncated form of YY1 acts as dominant-negative that interferes with the normal function of YY1.
5.2. Co-factors
The cell-type specific function of YY1 in distinct cell types can be also affected by the availability of the co-factors and by the chromatin conformation of the promoters of its target genes that might be affected by the developmental stage and cell context. One of the most frequently adopted strategies for a ubiquitous protein to gain specificity is interacting with additional factors. The combined configuration would exponentially increase the specificity of the action. Indeed, the majority of the reports about YY1 also mention its binding partners (Figure 1). Some of the interacting factors have a restricted pattern of expression such as Smad, the downstream effector of BMP signaling pathway, Hoxa11 and the polycomb group protein EED, while others are also widely expressed, including HDACs, p300, INO80, PRMT1 and CTCF (Lee et al. 1995; Shi et al. 1997; Yao et al. 2001; Rezai-Zadeh et al. 2003; Satjin et al. 2001; Donohoe et al. 2007). The BMP-dependent recruitment of Smad by YY1 would justify its function in cardiac development where the two factors together result in the activation of the Nkx2.5 promoter (Lee et al. 2004). However, the choice of the protein binding partner determines, at least in part, whether YY1 activates or represses a given promoter. INO80 is a chromatin remodeling complex that catalyzes ATP-dependent nucleosome sliding (Jin et al. 2005). The binding with INO80 is required for YY1-dependent activation of CDC6 gene (Cai et al. 2007). Similarly, targeted recruitment of p300, histone acetyltransferase, and PRMT1, histone H4 (Arg3) methyltransferase, endows YY1 transcriptional activation ability, as reported in ER stress induced Grp78 gene expression (Baumeister et al. 2005). In contrast, HDACs are histone deacetylase that are generally involved in transcriptional repression. A number of reports on YY1 as transcriptional repressor, including our work, indicated its association with HDAC1, 2 or 3 (Osborne et al. 2001; Luke et al. 2006; He et al. 2007; Liu et al. 2007b).
5. Concluding remarks
The complexity and cellular diversity of the nervous system requires a sequence of events that start with the formation of the neuro-ectoderm, is followed by the differentiation into multiple cell types (i.e. neurons, astrocytes and oligodendrocytes) and culminates with the organization of a complex network of interactions between different cell types. Each of these steps requires the temporal and spatial regulation of gene expression. Here we have reviewed evidence suggesting multiple roles for YY1 during development of the central and peripheral nervous system. As reviewed, its function is dependent on a multitude of parameters that include sub-cellular localization, post-translational modifications and binding with other proteins. It is anticipated that a more clear definition of YY1 function will be gained from future conditional mutant mice generated in other lineages including the neuronal and astrocytic lineage.
Acknowledgments
HY is supported by Graduate Fellowship from New Jersey Commission on Spinal Cord Research (08B-010-SCR3). PCB is supported by grants from NIH-NINDS (RO1 NS052738 and RO1NS042925). The authors would like to apologize to all those whose relevant work could not be referenced due to the space restriction.
Abbreviations
- AD
Alzheimer disease
- APP
amyloid precursor protein
- BACE1
beta-site amyloid precursor protein-cleaving enzyme 1
- BGC
Bergmann glial cells
- BMP
bone morphogenetic protein
- DBH
dopamine beta-hydroxylase
- EED
embryonic ectoderm development
- HDAC
histone deacetylase
- NRG
neuregulin
- PcG
polycomb group
- PHR
PHO homology region
- REST
RE-1 silencing transcription factor
- Shh
Sonic hedgehog
- SVZ
subventricular zone
- YY1
Yin Yang 1
REFERENCE
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. Embo J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bain M, Sinclair J. Targeted inhibition of the transcription factor YY1 in an embryonal carcinoma cell line results in retarded cell growth, elevated levels of p53 but no increase in apoptotic cell death. Eur J Cell Biol. 2005;84:543–553. doi: 10.1016/j.ejcb.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Jedlicka P, Templeton NS, Liotta L, Ozato K. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene. 1994;150:259–266. doi: 10.1016/0378-1119(94)90435-9. [DOI] [PubMed] [Google Scholar]
- Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfield S, Smith BJ, Zimmerman RA, Dicicco-Bloom E, Brzustowicz LM, Millonig JH. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem. 2001;77:935–942. doi: 10.1046/j.1471-4159.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Cho KW. Anterior neurectoderm is progressively induced during gastrulation: the role of the Xenopus homeobox gene orthodenticle. Development. 1995;121:993–1004. doi: 10.1242/dev.121.4.993. [DOI] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Brune CW, Korvatska E, Allen-Brady K, Cook EH, Jr, Dawson G, Devlin B, Estes A, Hennelly M, Hyman SL, McMahon WM, Munson J, Rodier PM, Schellenberg GD, Stodgell CJ, Coon H. Heterogeneous association between engrailed-2 and autism in the CPEA network. Am J Med Genet B Neuropsychiatr Genet. 2008;147:187–193. doi: 10.1002/ajmg.b.30585. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense slug RNA. Dev Biol. 1999;213:101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- Cheh MA, Millonig JH, Roselli LM, Ming X, Jacobsen E, Kamdar S, Wagner GC. En2 knockout mice display neurobehavioral and neurochemical alterations relevant to autism spectrum disorder. Brain Res. 2006;1116:166–176. doi: 10.1016/j.brainres.2006.07.086. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, Teti D, Bresciani F, Perillo B, Weisz A. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T, Mesa K, Urrutia R. Three dynamin-encoding genes are differentially expressed in developing rat brain. J Neurochem. 1996;67:927–931. doi: 10.1046/j.1471-4159.1996.67030927.x. [DOI] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Delatour B, Mercken L, El Hachimi KH, Colle MA, Pradier L, Duyckaerts C. FE65 in Alzheimer's disease: neuronal distribution and association with neurofibrillary tangles. Am J Pathol. 2001;158:1585–1591. doi: 10.1016/S0002-9440(10)64113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Ermak G, Davies KJ. Calcium and oxidative stress: from cell signaling to cell death. Mol Immunol. 2002;38:713–721. doi: 10.1016/s0161-5890(01)00108-0. [DOI] [PubMed] [Google Scholar]
- Ermekova KS, Chang A, Zambrano N, de Candia P, Russo T, Sudol M. Proteins implicated in Alzheimer disease. The role of FE65, a new adapter which binds to beta-amyloid precursor protein. Adv Exp Med Biol. 1998;446:161–180. [PubMed] [Google Scholar]
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, Raimondi A, Gong LW, Ariel P, Paradise S, O'Toole E, Flavell R, Cremona O, Miesenbock G, Ryan TA, De Camilli P. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316:570–574. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- Ficzycz A, Eskiw C, Meyer D, Marley KE, Hurt M, Ovsenek N. Expression, activity, and subcellular localization of the Yin Yang 1 transcription factor in Xenopus oocytes and embryos. J Biol Chem. 2001;276:22819–22825. doi: 10.1074/jbc.M011188200. [DOI] [PubMed] [Google Scholar]
- Gegelashvili G, Civenni G, Racagni G, Danbolt NC, Schousboe I, Schousboe A. Glutamate receptor agonists up-regulate glutamate transporter GLAST in astrocytes. Neuroreport. 1996;8:261–265. doi: 10.1097/00001756-199612200-00052. [DOI] [PubMed] [Google Scholar]
- Gegelashvili G, Schousboe A. High affinity glutamate transporters: regulation of expression and activity. Mol Pharmacol. 1997;52:6–15. doi: 10.1124/mol.52.1.6. [DOI] [PubMed] [Google Scholar]
- Gilthorpe J, Vandromme M, Brend T, Gutman A, Summerbell D, Totty N, Rigby PW. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development. 2002;129:3887–3899. doi: 10.1242/dev.129.16.3887. [DOI] [PubMed] [Google Scholar]
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- Guenette SY, Chen J, Ferland A, Haass C, Capell A, Tanzi RE. hFE65L influences amyloid precursor protein maturation and secretion. J Neurochem. 1999;73:985–993. doi: 10.1046/j.1471-4159.1999.0730985.x. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, de la Torre JR, Holt C, Harland RM. Cephalic expression and molecular characterization of Xenopus En-2. Development. 1991;111:715–724. doi: 10.1242/dev.111.3.715. [DOI] [PubMed] [Google Scholar]
- Hendrich BD, Plenge RM, Willard HF. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I. Cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development. 1997;124:3929–3941. doi: 10.1242/dev.124.20.3929. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- Krippner-Heidenreich A, Walsemann G, Beyrouthy MJ, Speckgens S, Kraft R, Thole H, Talanian RV, Hurt MM, Luscher B. Caspase-dependent regulation and subcellular redistribution of the transcriptional modulator YY1 during apoptosis. Mol Cell Biol. 2005;25:3704–3714. doi: 10.1128/MCB.25.9.3704-3714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, Ten Dijke P, Heldin CH, Ericsson J, Moustakas A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol Cell Biol. 2003;23:4494–4510. doi: 10.1128/MCB.23.13.4494-4510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Chung HM. Yin Yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochem Biophys Res Commun. 2003;306:1008–1013. doi: 10.1016/s0006-291x(03)01071-4. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Mann D, Goumidi L, Harris J, Pasquier F, Frigard B, Cottel D, Lendon C, Iwatsubo T, Amouyel P, Chartier-Harlin MC. A FE65 polymorphism associated with risk of developing sporadic late-onset alzheimer's disease but not with Abeta loading in brains. Neurosci Lett. 2000;293:29–32. doi: 10.1016/s0304-3940(00)01477-4. [DOI] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TC, Zhang Y, Schwartz RJ. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Lee KH, Evans S, Ruan TY, Lassar AB. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007a;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Merkler KA, Zhang X, McLean MP. Prostaglandin F2alpha suppresses rat steroidogenic acute regulatory protein expression via induction of Yin Yang 1 protein and recruitment of histone deacetylase 1 protein. Endocrinology. 2007b;148:5209–5219. doi: 10.1210/en.2007-0326. [DOI] [PubMed] [Google Scholar]
- Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech Dev. 2006;123:312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Luke MP, Sui G, Liu H, Shi Y. Yin Yang 1 physically interacts with Hoxa11 and represses Hoxa11-dependent transcription. J Biol Chem. 2006;281:33226–33232. doi: 10.1074/jbc.M606584200. [DOI] [PubMed] [Google Scholar]
- Miyake N, Miyake K, Hisayasu S, Karlsson S, Shmada T. Enforced HoxB4 expressed hematopoietic Stem cell transdifferentiate into oligodendrocytes in adult mouse brains. Mol. Therapy. 2006;13:S39–S40. [Google Scholar]
- Morgan MJ, Woltering JM, In der Rieden PM, Durston AJ, Thiery JP. YY1 regulates the neural crest-associated slug gene in Xenopus laevis. J Biol Chem. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
- Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- Nowak K, Lange-Dohna C, Zeitschel U, Gunther A, Luscher B, Robitzki A, Perez-Polo R, Rossner S. The transcription factor Yin Yang 1 is an activator of BACE1 expression. J Neurochem. 2006;96:1696–1707. doi: 10.1111/j.1471-4159.2006.03692.x. [DOI] [PubMed] [Google Scholar]
- Osborne A, Zhang H, Yang WM, Seto E, Blanck G. Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation. Mol Cell Biol. 2001;21:6495–6506. doi: 10.1128/MCB.21.19.6495-6506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palko L, Bass HW, Beyrouthy MJ, Hurt MM. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J Cell Sci. 2004;117:465–476. doi: 10.1242/jcs.00870. [DOI] [PubMed] [Google Scholar]
- Pisaneschi G, Ceccotti S, Falchetti ML, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas S, Vargas MA, Lopez-Bayghen E, Ortega A. Glutamate-dependent transcriptional regulation of GLAST/EAAT1: a role for YY1. J Neurochem. 2007;101:1134–1144. doi: 10.1111/j.1471-4159.2007.04517.x. [DOI] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Safrany G, Perry RP. Characterization of the mouse gene that encodes the delta/YY1/NF-E1/UCRBP transcription factor. Proc Natl Acad Sci U S A. 1993;90:5559–5563. doi: 10.1073/pnas.90.12.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiard-Baron D, Langui D, Delehedde M, Delatour B, Schombert B, Touchet N, Tremp G, Paul MF, Blanchard V, Sergeant N, Delacourte A, Duyckaerts C, Pradier L, Mercken L. Expression of human FE65 in amyloid precursor protein transgenic mice is associated with a reduction in beta-amyloid load. J Neurochem. 2005;93:330–338. doi: 10.1111/j.1471-4159.2005.03026.x. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, Babinet C, Charnay P. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:648. doi: 10.1038/384648a0. [DOI] [PubMed] [Google Scholar]
- Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L, Bonavida B. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol. 2005;27:131–141. [PubMed] [Google Scholar]
- Seo H, Yang C, Kim HS, Kim KS. Multiple protein factors interact with the cis-regulatory elements of the proximal promoter in a cell-specific manner and regulate transcription of the dopamine beta-hydroxylase gene. J Neurosci. 1996;16:4102–4112. doi: 10.1523/JNEUROSCI.16-13-04102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Sinha S, Lieberburg I. Cellular mechanisms of beta-amyloid production and secretion. Proc Natl Acad Sci U S A. 1999;96:11049–11053. doi: 10.1073/pnas.96.20.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Pan X, Atchison ML. Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J Cell Biochem. 2005;96:689–699. doi: 10.1002/jcb.20562. [DOI] [PubMed] [Google Scholar]
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. Embo J. 2007;26:1649–1659. doi: 10.1038/sj.emboj.7601619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Vega MI, Huerta-Yepez S, Jazirehi AR, Garban H, Bonavida B. Rituximab (chimeric anti-CD20) sensitizes B-NHL cell lines to Fas-induced apoptosis. Oncogene. 2005;24:8114–8127. doi: 10.1038/sj.onc.1208954. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Whiting J, Marshall H, Cook M, Krumlauf R, Rigby PW, Stott D, Allemann RK. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991;5:2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- Wiley JC, Smith EA, Hudson MP, Ladiges WC, Bothwell M. Fe65 stimulates proteolytic liberation of the beta-amyloid precursor protein intracellular domain. J Biol Chem. 2007;282:33313–33325. doi: 10.1074/jbc.M706024200. [DOI] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Control of peripheral nerve myelination by the betasecretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- Wu S, Murai S, Kataoka K, Miyagishi M. Yin Yang 1 induces transcriptional activity of p73 through cooperation with E2F1. Biochem Biophys Res Commun. 2008;365:75–81. doi: 10.1016/j.bbrc.2007.10.145. [DOI] [PubMed] [Google Scholar]
- Xu X, Kawachi Y, Nakamura Y, Sakurai H, Hirota A, Banno T, Takahashi T, Roop DR, Otsuka F. Yin-yang 1 negatively regulates the differentiation-specific transcription of mouse loricrin gene in undifferentiated keratinocytes. J Invest Dermatol. 2004;123:1120–1126. doi: 10.1111/j.0022-202X.2004.23492.x. [DOI] [PubMed] [Google Scholar]
- Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem. 1998;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]
- Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Dupont BR, Ghosh S, Fang Y, Leach RJ, Seto E. Cloning, chromosomal localization and promoter analysis of the human transcription factor YY1. Nucleic Acids Res. 1998;26:3776–3783. doi: 10.1093/nar/26.16.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Jeong MJ, Lee SS, Lee KI, Kwon BM, Park YM, Han MY. Negative regulation of YY1 transcription factor on the dynamin I gene promoter. Biochem Biophys Res Commun. 2001;283:340–343. doi: 10.1006/bbrc.2001.4784. [DOI] [PubMed] [Google Scholar]
- Zambrano N, De Renzis S, Minopoli G, Faraonio R, Donini V, Scaloni A, Cimino F, Russo T. DNA-binding protein Pur alpha and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem J. 1997;328(Pt 1):293–300. doi: 10.1042/bj3280293. [DOI] [PMC free article] [PubMed] [Google Scholar]