Abstract
Yeast and animal homotypic fusion and vacuole protein sorting (HOPS) complexes contain conserved subunits, but HOPS-mediated traffic in animals might require additional proteins. Here, we demonstrate that SPE-39 homologues, which are found only in animals, are present in RAB5-, RAB7-, and RAB11-positive endosomes where they play a conserved role in lysosomal delivery and probably function via their interaction with the core HOPS complex. Although Caenorhabditis elegans spe-39 mutants were initially identified as having abnormal vesicular biogenesis during spermatogenesis, we show that these mutants also have disrupted processing of endocytosed proteins in oocytes and coelomocytes. C. elegans SPE-39 interacts in vitro with both VPS33A and VPS33B, whereas RNA interference of VPS33B causes spe-39–like spermatogenesis defects. The human SPE-39 orthologue C14orf133 also interacts with VPS33 homologues and both coimmunoprecipitates and cosediments with other HOPS subunits. SPE-39 knockdown in cultured human cells altered the morphology of syntaxin 7-, syntaxin 8-, and syntaxin 13-positive endosomes. These effects occurred concomitantly with delayed mannose 6-phosphate receptor-mediated cathepsin D delivery and degradation of internalized epidermal growth factor receptors. Our findings establish that SPE-39 proteins are a previously unrecognized regulator of lysosomal delivery and that C. elegans spermatogenesis is an experimental system useful for identifying conserved regulators of metazoan lysosomal biogenesis.
INTRODUCTION
Lysosome biogenesis is mediated by multiple vesicular budding and fusion events that deliver components between subcellular compartments (Luzio et al., 2003). In Saccharomyces cerevisiae, vesicular fusion at the vacuole (the yeast equivalent of lysosomes) requires the homotypic fusion and vacuole protein sorting (HOPS) complex, which regulates vesicle docking through its interaction with soluble N-ethylmaleimide-sensitive factor-attachment protein receptors (SNAREs) and the Rab protein Ypt7p (Price et al., 2000; Sato et al., 2000; Wurmser et al., 2000). The yeast HOPS complex contains class C Vps proteins Vps11p, Vps16p, Vps18p, and Vps33p (Rieder and Emr, 1997) and class B Vps proteins Vps39p and Vps41p (Seals et al., 2000; Wurmser et al., 2000). In addition to vesicular fusion at the vacuole, the class C Vps proteins also function in Golgi-to-endosome transport and other steps in vacuolar delivery pathways (Srivastava et al., 2000; Peterson and Emr, 2001; Subramanian et al., 2004; Peplowska et al., 2007). Involvement of the class C Vps proteins at multiple stages has also been reported in mammalian cells (Kim et al., 2003; Richardson et al., 2004).
Vesicular trafficking to the lysosome has been extensively described in yeast (Bowers and Stevens, 2005), but it is also known that this process is more complex in metazoans (Dell'Angelica, 2004). Yeast only has lysosomes of a single type, whereas animals have lysosomes and lysosome-related organelles that are functionally diverse. This diversity is evident when comparing the lysosomal trafficking machinery in yeast to those of animals. For example, although there is only one form of Vps33p in yeast (Banta et al., 1990; Wada et al., 1990), two VPS33 homologues (termed VPS33A and VPS33B in humans) have been identified in animals, including Caenorhabditis elegans, Drosophila melanogaster, zebrafish, and mammals (Gissen et al., 2005). A missense mutation of the mouse VPS33A gene (buff) causes a mild platelet-storage pool deficiency and hypopigmentation due to defective melanosome biogenesis (Suzuki et al., 2003). Mutations in the human VPS33B gene cause arthrogryposis-renal dysfunction-cholestasis syndrome (Gissen et al., 2004), which is also usually associated with platelet dysfunction (Eastham et al., 2001). Both VPS33A and VPS33B are required during platelet formation; VPS33A is required for dense granule biogenesis, whereas VPS33B is required for α-granule biogenesis (Suzuki et al., 2003; Lo et al., 2005). These data suggest that the two VPS33 homologues participate in different sets of vesicular fusion events. Current thinking is that VPS33B is required for transport to conventional lysosomes, whereas VPS33A is mainly involved in biogenesis of melanosomes and related lysosomal compartments (Suzuki et al., 2003; Gissen et al., 2005).
We have reported that the C. elegans protein SPE-39 is essential for vesicular trafficking during spermatogenesis (Zhu and L'Hernault, 2003). spe-39 mutant spermatocytes accumulate many ∼100-nm vesicles that apparently cannot fuse to form prominent ∼800-nm vesicular structures (membranous organelles; MOs) that are essential for sperm function (reviewed by L'Hernault, 2006; http://www.wormbook.org). Although MOs are unusual in appearance, SPE-39 has orthologues in many animals (but not in yeast or other unicellular organisms), suggesting that the MO defects in C. elegans spe-39 mutants reflect a conserved process. This notion is supported by an ultrastructural comparison of spe-39 mutants to HOPS mutants identified in other experimental systems. Yeast and Drosophila vps18 mutants (Rieder and Emr, 1997; Sevrioukov et al., 1999) accumulate vesicles that contain internal membranes, and these vesicles seem remarkably similar to the vesicles observed within spe-39 mutant spermatocytes (Zhu and L'Hernault, 2003). In all three experimental systems, these vesicles look like either late multivesicular endosomes (Futter et al., 1996; Bowers and Stevens, 2005) or autophagosomes (Klionsky, 2005). The accumulation of ∼100-nm vesicles in spe-39 mutants suggests that SPE-39 has a role in the fusion of these vesicles with a lysosome-like organelle.
Recent data in Drosophila suggested that there is an interaction between VPS33B and SPE-39 orthologues (Giot et al., 2003; Pulipparacharuvil et al., 2005), but the functional significance of this interaction was not determined. In this article, we examine the role of SPE-39 orthologues in C. elegans and cultured human cells and have taken advantage of the different technical approaches that are possible in these two experimental systems. Our data indicate that SPE-39 orthologues have conserved function in lysosomal biogenesis in animals and suggest that the MO is a lysosome-like organelle in C. elegans sperm.
MATERIALS AND METHODS
Antibodies
The following mouse monoclonal antibodies were used in this study: anti-early endosome antigen 1 (EEA1), anti-Golgi matrix protein of 130 kDa (GM130), anti-γ-adaptin, anti-syntaxin 8, anti-RAB5 (BD Biosciences, San Jose, CA), anti-transferrin receptor (Zymed Laboratories, South San Francisco, CA), anti-CD63, anti-human lysosome-associated membrane protein 1 (LAMP1) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), anti-RAB7 (Abnova, Walnut, CA), anti-cation–independent mannose 6-phosphate receptor/insulin-like growth factor-II receptor (CI-M6PR) (Calbiochem, San Diego, CA), anti-γ-tubulin, anti-β-actin (Sigma-Aldrich, St. Louis, MO), anti-hemagglutinin (HA) epitope (clone 12CA5; Roche Applied Science, Indianapolis, IN), and horseradish peroxidase (HRP)-conjugated anti-maltose-binding protein (MBP) monoclonal antibody (mAb) (New England Biolabs, Ipswich, MA). Rat mAb against the HA epitope (clone 3F10; Roche Applied Science) was used for immunostaining. Rabbit mAb against human epidermal growth factor receptor (EGFR) was from Millipore (Billerica, MA). Polyclonal antibodies against c-MYC and the HA epitope were from Bethyl Laboratories (Montgomery, TX); anti-RAB11a polyclonal antibody was from Zymed Laboratories and anti-green fluorescent protein (GFP) polyclonal antibody was from Synaptic Systems (Göttingen, Germany). The anti-cathepsin D polyclonal antibodies from Millipore and Calbiochem were used for immunostaining and metabolic labeling experiments, respectively. Rabbit antibodies against syntaxins 6, -7, and -13 were a gift from Dr. A. Peden (Cambridge Institute for Medical Research, Cambridge, United Kingdom).
Two Schistosoma japonicum glutathione transferase (GST) fusion constructs were made using the expression vector pGEX-3X (Smith and Johnson, 1988). They contain (human) hSPE-39 regions that encode the N-terminal and C-terminal regions, respectively. Primers BHC2L (5′-CGGGATCCTGAATCGGACAAAGGGTGATGAG-3′) and BHC3R (5′-CGGGATCCAAGCTGTTTCGGCTCTTTAGCTG-3′) amplified an N-terminal fragment encoding amino acids 2-93; primers BHC8L (5′-CGGGATCCTTGGCTTCCATCGGGTTGTCG-3′) and BHC9R (5′-CGGGATCCAGGCAGGAGAGGAGGAAATGAGG-3′) were used to amplify a C-terminal fragment encoding amino acids 406-493. These primers included a BamHI site (underlined) at the 5′ end to allow in-frame insertion of the polymerase chain reaction (PCR) fragments into the BamHI site of the pGEX-3X vector. The GST fusion proteins were expressed in BL21-Gold host bacteria (Stratagene, La Jolla, CA), and cells were lysed by passing twice through a French pressure cell. The soluble fusion proteins were purified by their affinity for glutathione agarose beads (Sigma-Aldrich). A mix of the two fusion proteins was used to immunize BALB/c mice for generating hybridomas. One hybridoma clone (64-8A12) was isolated and found to secrete a mAb that recognized an unknown epitope located in the C-terminal fragment of hSPE-39. The culture supernatant was concentrated 12-fold by ammonium sulfate precipitation and dialyzed against phosphate-buffered saline (PBS). Specificity of this mAb for hSPE-39 was confirmed by immunoblotting (Supplemental Figure 1).
An antiserum was prepared against the rat VPS33B peptide DTLTAVENKVSKLVTDKAAGKITDAFSSL [amino acids 450-478 of the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) record NP_071622.1]. Peptide synthesis and rabbit immunization were performed by Alpha Diagnostic International (San Antonio, TX). Immunoblotting analysis indicated that this antiserum recognizes human VPS33B (Supplemental Figure 2).
Trafficking Assays in C. elegans
The strains DH1006 bIs1[vit-2::GFP; rol-6(su1006)] X (Grant and Hirsh, 1999) and GS1912 arIs37[pmyo-3::ssGFP] I; dpy-20(e1282) IV (Fares and Greenwald, 2001) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). These strains were crossed to create spe-39(eb9) V/nT1[unc-?(n754) let-?] (IV;V); bIs1 X and arIs37 I; dpy-20(e1282)/nT1[unc-?(n754) let-?] IV; spe-39(eb9)/nT1 V that were examined to determine how spe-39 loss of function affected vesicular trafficking in oocytes and coelomocytes, respectively. nT1[unc-?(n754) let-?] (IV;V) is a chromosomal translocation (Ferguson and Horvitz, 1985) used to balance the sterile spe-39(eb9) mutation, and the dominance of n754 allowed unambiguous selection of spe-39 homozygotes for these analyses. The control strain spe-9(hc88ts) I; bIs1 X was constructed for oocyte endocytosis assays. The temperature-sensitive spe-9(hc88ts) mutant hermaphrodites (L'Hernault et al., 1988) are fertile at 20°C, but at 25°C they are completely self-sterile and accumulates unfertilized oocytes, which in this respect is similar to spe-39(eb9). For the oocyte assay, worms were grown at 25°C for 2 d before young adult spe-39(eb9) V; bIs1 X and spe-9(hc88ts) I; bIs1 X hermaphrodites were selected for examination. For the coelomocyte assay, strains were grown at 20°C and young arIs37 I; dpy-20(e1282) IV; spe-39(eb9) V and arIs37 I; dpy-20(e1282) IV adults were selected for examination. Levamisole was used at 10 mM as an anesthetic, and images were recorded by confocal microscopy (see below).
Far Western
The coding sequence of the C. elegans VPS33A gene B0303.9 (also called slp-1) was obtained from an open reading frame clone (Reboul et al., 2003) distributed by Open Biosystems, (Huntsville, AL). The full-length coding sequence of the C. elegans VPS33B gene C56C10.1 was obtained by ligating restriction fragments of the cDNA clones yk1031e8 and yk749c4 (obtained from Y. Kohara, National Institute of Genetics, Nishima, Japan), which contain error-free 5′ and 3′ portions of the coding region, respectively. The cloned coding sequences of the C. elegans VPS33A and VPS33B genes were checked against the NCBI RefSeq records NM_066519.2 and NM_062941.3, respectively, for accuracy. The full-length B0303.9 coding sequence was then amplified with primers MBP33AL (5′-ATGGCAGCCAATGAAGACAGAG-3′) and MBP33AR (5′-CGTCTAGATTAAATATTGGTAATATTATTCAGAAGC-3′), which included an XbaI site (underlined). The PCR fragment was inserted between XmnI (blunt end) and XbaI sites of the pMAL-c2X vector (New England Biolabs) to generate the MBP-CeVPS33A fusion construct. Likewise, primers MBP33BL (5′-ATGGCACCGGGTACCGCCGAG-3′) and MBP33BR (5′-GCTCTAGATTATGATCCAAAAACCGAAGATGCG-3′) were used to generate the MBP-CeVPS33B fusion construct. BL21-Gold was used as host bacteria for expressing the MBP fusion proteins, and the fusion proteins were purified with the pMAL Protein Fusion and Purification System (New England Biolabs) according to the manufacturer's instructions.
To generate the GST-SPE-39 fusion construct, primers BAM5 (5′-CGGGATCCGAATGGCCCTTCGAAGGAAATTC-3′) and 3BAM (5′-CGGGATCCTTACTTCCATTTTATCGACGGATTG-3′) were used to amplify the full-length spe-39 coding sequence from the cDNA clone yk504f7 (obtained from Y. Kohara). Both primers had a BamHI site (underlined) to facilitate the cloning of the PCR fragment into the pGEX-3X vector.
For Far Western assays, GST or the GST-SPE-39 fusion protein were resolved by polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membrane, whereas MBP or MBP fusion proteins were used as probes. The GST-SPE-39 fusion protein forms insoluble inclusion bodies when expressed in host bacteria. The inclusion bodies were pelleted from bacterial lysate, and the pellet was resuspended in Laemmli sample buffer. The sample was then heated in a boiling water bath, and any insoluble material was pelleted by centrifugation at 18,000 × g for 15 min to yield clarified GST-SPE-39. The assay was performed at room temperature for 2 h in 140 mM NaCl, 0.1% Tween 20, and 20 mM Tris, pH 7.6. The bound MBP fusion proteins were detected by an HRP-conjugated anti-MBP mAb followed by the HyGlo Chemiluminescent HRP Antibody Detection Reagent (Denville Scientific, Metuchen, NJ) as the substrate for HRP.
RNA Interference (RNAi) in C. elegans
Two RNAi constructs were made for CeVPS33B. Primers VPS33BIL (5′-GCTCTAGACGATCAGCGAGTCTTCTTTCT-3′) and VPS33BIR (5′-GCTCTAGAAGCGCTCTCAGGTCTTTTTC-3′) were used to amplify an 807 base pairs fragment from the 5′ portion of the coding sequence; and primers VPS33BIIL (5′-GCTCTAGATGCGAAATGATGATGAATAAAGTT-3′) and VPS33BIIR (5′-GCTCTAGAGAATCGCCAGCAGCCGTAAAG-3′) were used to amplify a 664-base pair fragment from the 3′ portion of the coding sequence. The cDNA clone yk749c4 was used as the template. One RNAi construct was made for CeVPS33A with primers VPS33AL (5′-GCTCTAGAGTCCGCCTCCACTCCAC-3′) and VPS33AR (5′-GCTCTAGAACTAAAACCGAATATTCAGCAACT-3′), which amplify an 831-base pair fragment from the template cDNA clone yk706e5 (obtained from Y. Kohara). All six primers contain an XbaI site (underlined) at their 5′ ends, which facilitated the insertion of the PCR fragments into the RNAi vector L4440 (Timmons and Fire, 1998). RNAi was performed by feeding rrf-3(pk1426); him-5(e1490) worms the bacterial strain HT115 (DE3) (Timmons and Fire, 1998; Timmons et al., 2001) transformed with the RNAi constructs. The rrf-3 mutation was used to sensitize the worms to RNAi (Simmer et al., 2002), whereas the him-5 mutation (Hodgkin et al., 1979) was used to obtain males, which were more convenient for analyses of spermatogenesis. All worms were raised at 20°C. Hand-picked rrf-3 him-5 embryos (Po) were transferred to plates upon which double-stranded RNA (dsRNA)-producing bacteria were growing. The plates used for growing RNAi bacterial strains were NGM agar (Brenner, 1974) supplemented with 50 μg/ml carbenicillin (for plasmid selection) and 1 mM isopropyl β-d-thiogalactoside (for inducing dsRNA synthesis). The bacteria for feeding were prepared as described previously (Kamath and Ahringer, 2003). We selected a bacterial strain containing the VPS33BIL/VPS33BIR construct for further analysis of CeVPS33B because it induced a stronger Spe phenotype than either the bacteria containing the VPS33BIIL/VPS33BIIR construct or a mix of the two bacterial cultures. After the embryos hatched and the sexes of the larvae could be identified, virgin Po hermaphrodites were selected and allowed to self-fertilize. The Po worms and their F1 self-progeny were fed exclusively on dsRNA-containing bacteria. F1 hermaphrodites were picked as L4 and scored for oocyte-laying self-sterility (the Spe phenotype) the next day, whereas F1 males were dissected and their sperm were examined as described previously (Zhu and L'Hernault, 2003).
To prepare samples for electron microscopy, the Spe phenotype induced by feeding was further enhanced by injection of dsRNA. The inserts of the two above-mentioned CeVPS33B RNAi constructs were templates for synthesizing dsRNA by the MEGAscript RNAi kit (Ambion, Austin, TX). These two dsRNAs were combined so that each was 25 ng/μl, and this mixture was injected into both gonad arms of Po young adult hermaphrodites. Injected Po hermaphrodites and their F1 self-progeny were continuously fed bacteria that were expressing the CeVPS33B RNAi construct described above. Sperm samples from F1 males were prepared for transmission electron microscopy as described previously (Shakes and Ward, 1989), except that dissected worms were embedded in LX112 resin (Ladd Research Industries, Burlington, VT) for thin sectioning.
Cell Culture
HeLa and human embryonic kidney (HEK) 293T cells were grown in DMEM (with 4.5 g/l glucose, 4 mM l-glutamine, and 0.11 g/l sodium pyruvate; Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS). Transfection of plasmids was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols.
Coimmunoprecipitation
Immunoprecipitation was performed according to procedures described previously (Faundez and Kelly, 2000), except that the ProFound coimmunoprecipitation kit (Pierce Chemical, Rockford, IL) was used for coimmunoprecipitation of hSPE-39 and hVPS33B and the associated control experiments. The application of dithiobis(succinimidyl)propionate (DSP; Pierce Chemical) and immunomagnetic isolation of immune complexes were described previously (Craige et al., 2008).
Confocal and Wide-Field Deconvolution Immunofluorescence Microscopy
Immunofluorescence was preformed as described previously (Salazar et al., 2004; Craige et al., 2008). Briefly, cells were seeded onto coverslips coated with Matrigel (BD Biosciences, Franklin Lakes, NJ). Cells were fixed with 4% paraformaldehyde/PBS for 20 min on ice, free aldehyde was quenched with 25 mM glycine/PBS, and the cells were treated with blocking solution for 1 h. The blocking solution contained 2% bovine serum albumin (BSA), 1% fish skin gelatin (Sigma-Aldrich), 0.02% saponin (Sigma-Aldrich), and 15% horse serum (HyClone Laboratories, Logan, UT), and it was also used for diluting primary and secondary antibodies. The Tyramide Signal Amplification kit (Invitrogen) was used to enhance low signals; in this article, this technique was used to generate the images shown in Figure 10, B and C, and Supplemental Figure 5P. Samples were mounted with Gelvatol (Wang et al., 1999). Images were generated with an MRC-1024 confocal system (Bio-Rad, Hercules, CA) through an Axioskop 50 microscope (Carl Zeiss, Thornwood, NY) equipped with Plan-Apochromat 100× 1.4 numerical aperture (NA; oil) and 63× 1.4 NA (oil) objective lenses. Samples were illuminated with a krypton/argon laser. Emission of GFP, EGFP, and Alexa Fluor 488-conjugated secondary antibodies (Invitrogen) passed through a 522DF32 bandpass filter, and emission of Alexa Fluor 568-conjugated secondary antibodies (Invitrogen) passed through a 605DF32 bandpass filter.
Figure 10.
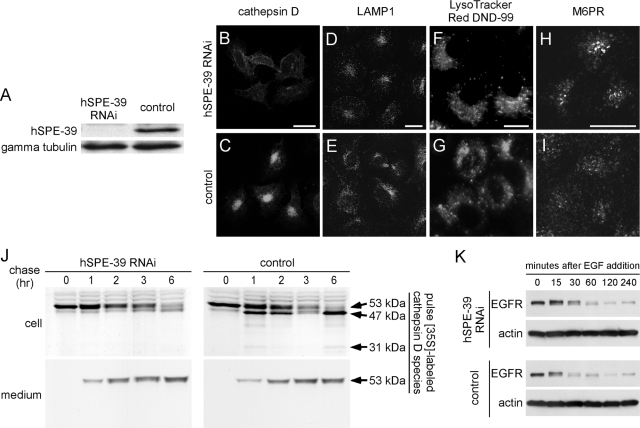
hSPE-39 knockdown in HeLa cells alters M6PR-mediated transport of cathepsin D and causes a delay in the degradation of endocytosed EGFR. (A) Western analysis shows the effectiveness of RNAi by probing hSPE-39 levels in cells treated with hSPE-39 siRNA (left) and control siRNA (right). γ-Tubulin levels were used as a protein loading control. (B–I) HeLa cells transfected with hSPE-39 siRNA (B, D, F, and H) and control siRNA (C, E, G, and I) were stained with anti-cathepsin D antibody (B and C), anti-LAMP1 antibody (D and E), LysoTracker Red DND-99 (F and G), and anti-M6PR antibody (H and I). (J) Metabolic labeling experiment indicates defective cathepsin D delivery and maturation in cells lacking hSPE-39. HeLa cells treated with hSPE-39 RNAi and control RNAi were pulse labeled for 30 min with [35S]methionine and chased for 1, 2, 3, and 6 h. The heavy chain of mature cathepsin D (31 kDa) and its precursor forms (53 and 47 kDa) in both media and cells were immunoprecipitated, resolved by SDS-PAGE, and detected by fluorography. (K) Degradation of EGFR in hSPE-39 siRNA- and control siRNA-treated HeLa cells was induced by EGF and examined with Western analysis. β-Actin levels were used as a protein loading control. Bars, 20 μm.
Wide-field deconvolution microscopy images were acquired with a scientific-grade cooled charge-coupled device (CoolSNAP HQ with ORCA-ERchip) on a multi wavelength, wide-field, three-dimensional microscopy system (Intelligent Imaging Innovations, Denver, CO), by using a 63× numerical aperture 1.4 lens on a 200M inverted microscope (Carl Zeiss). Immunofluorescent samples were imaged at room temperature by using a Sedat filter set (Chroma Technology, Rockingham, UT), in successive 0.20-μm focal planes. Out-of-focus light was removed with a constrained iterative deconvolution algorithm (Swedlow et al., 1997). Images were processed and analyzed using MetaMorph software version 3.0 (Molecular Devices, Sunnyvale, CA). Three consecutive z-series were analyzed per image by thresholding to similar levels and determining colocalization as the percentage of pixel area overlap for the respective channels.
hSPE-39-EGFP Construct
The coding sequence of the hSPE-39 orthologue C14orf133 (also called FLJ12707) was PCR amplified from Marathon-Ready cDNA of whole human brain (Clontech, Mountain View, CA) and inserted into the pGEM-T Easy Vector (Promega, Madison, WI). Randomly chosen clones were sequenced and compared with the NCBI RefSeq record NM_022067.2. An error-free full-length coding sequence was obtained by ligating error-free restriction fragments prepared from these clones. A restriction site was introduced onto each end of the full-length hSPE-39 coding sequence by PCR. The primers used were C2HCDNA (5′-CGGAATTCATGAATCGGACAAAGGGTGATGAG-3′, including an EcoRI site that is underlined) and HCDNAN1 (5′-CGGGATCCTTCTTCCATCGAATTTGCGAGCTG-3′, including a BamHI site that is underlined). The amplified fragment was inserted between the EcoRI and BamHI sites of the pEGFP-N1 vector (Clontech) to make a hSPE-39-EGFP fusion construct for expression in mammalian cells.
HA-tagged hVPS33A, hVPS33B, and hSPE-39 Constructs
A cDNA fragment that contained the coding sequence of the human VPS33A gene (hVPS33A) was amplified from Marathon-Ready cDNA of whole human brain with primers H33AL (5′-GCGGACTCGGTTGGCGTTGTG-3′) and H33AR (5′-TCTATGGGGTTTTACTTCCTTTCA-3′). The hVPS33B coding sequence was obtained from the IMAGE Consortium clone 3449387, which is distributed by American Type Culture Collection (Manassas, VA). The cloned coding sequences of hVPS33A and hVPS33B were checked against NCBI RefSeq record NM_022916.3 and NM_018668.2, respectively, for accuracy. The full-length coding sequence of hVPS33A was amplified with 5′-CACCTGGTGGAGCAAGATGGCG-3′ and 5′-TCAAGCGTAGTCTGGGACGTCGTATGGGTAGAAAGGTTTTTCCATCAGAGCC-3′ and inserted into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen) for expression in mammalian cells. The reverse primer contains antisense coding for the HA tag (underlined) followed by a stop codon. Likewise, primers 5′-CACCGTGATCGCTGCCATGGCT-3′ and 5′-TCAAGCGTAGTCTGGGACGTCGTATGGGTAGGCTTTCACCTCACTCATGGC-3′ were used to make a HA-tagged hVPS33B construct for expression in mammalian cells, whereas primers 5′-CACCATGAATCGGACAAAGGGTGATG-3′ and 5′-TCAAGCGTAGTCTGGGACGTCGTATGGGTAATTCTTCCATCGAATTTGCGAG-3′ were used to make the hSPE-39-HA construct.
Mammalian VPS11, -16, -18, -39, -41, RAB5Q79L, and Hepatocyte Growth Factor-regulated Tyrosine Kinase Substrate (Hrs) cDNA Constructs
Tagged mouse HOPS subunit constructs VPS11-HA, VPS16-HA, and VPS39-GFP were gifts from Robert Piper (Department of Molecular Physiolgy and Biophysics, University of Iowa, Iowa City, IA), and the VPS18-MYC construct was a gift from Liping Wang (United States Department of Agriculture, Agricultural Research Service, University of California, Davis, CA). Full-length human VPS41 coding sequence was amplified from a cDNA clone (catalog no. SC111791; OriGene Technologies, Rockville, MD) with primers 5′-CACCATGGC GGAAGCAGAGGAG-3′ and 5′-CTACAGATCTTCTTCAGAAATAAGTTTTTGTTCT TTTTTCATCTCCAA-3′and inserted into the pcDNA3.1D/V5-His-TOPO vector. The underlined sequence in the reverse primer is the antisense coding of the c-MYC tag. The cloned sequence was checked against NCBI RefSeq record NM_014396.2 and only silent mutations were found. The RAB5Q79L EGFP construct was a gift of Dr. Laura Volpicelli (Emory University, Atlanta, GA), and Hrs-MYC was a gift of Dr. H. Stenmark (Norwegian Radium Hospital, Oslo, Norway).
Small Interfering RNA (siRNA) Transfection and Rescue with siRNA-resistant Construct
One day before transfection, cells were plated in 24-well plates at 2 × 104 cells in 400 μl of growth medium. siGENOME SMARTpool reagent for C14orf133 (Dharmacon RNA Technologies, Lafayette, CO) was tested for hSPE-39 knockdown at a final concentration of 100 nM for transfection and 1 μl of Lipofectamine 2000 (Invitrogen) was used for each well. The growth medium was replaced 6 h after the addition of the transfection reagent and siRNA. Another medium change was done after 2 d. Phenotypic analysis was performed 3–4 d after transfection. Individual duplexes in the SMARTpool were tested with the same procedure and produced similar results. The most efficient duplex was selected for further experiments. Its target sequence on hSPE-39 mRNA is 5′-CAGAAGAGCUUGCGCUAUC-3′. siCONTROL NonTargeting siRNA Pool (Dharmacon RNA Technologies) was used as a negative control.
Silent mutations were made in the hSPE-39-EGFP construct at positions in which perfect matches between the encoded mRNA and the complimentary guide strand of the siRNA are known to be critical for the RNAi effect (Elbashir et al., 2001; Du et al., 2005). In the resulting siRNA-resistant construct, hSPE-39-EGFP(SR), the sense strand sequence corresponding to the target region of the selected siRNA duplex was changed to CAGAAGAATTAGCGCTATC (changed nucleotides are underlined). Site-directed mutagenesis was performed with QuikChange II site-directed mutagenesis kit (Stratagene). Knockdown phenotypes of cells treated with hSPE-39 siRNA were rescued by transfection of the hSPE-39-EGFP(SR) construct 2 d after the initial transfection of siRNA. Cells were inspected one day after the rescuing transfection.
LysoTracker Red DND-99 Staining
Cells were incubated for 45 min at 37°C in a CO2 incubator after the medium was supplemented with 100 nM LysoTracker Red DND-99 (Invitrogen). The cells were then washed twice with PBS and fixed as described above. Stained cells were examined with an Axiovert 200 MOT microscope (Carl Zeiss) equipped with an Achroplan 63× (0.75 NA) LD objective, and images were captured by an ORCA-100 digital camera (Hamamatsu, Bridgewater, NJ).
Metabolic Labeling Analysis
HeLa cells treated with experimental and control siRNA were incubated with methionine-free DMEM (Sigma D0422 supplemented with 4 mM l-glutamine, 0.2 mM l-cystine, 10% dialyzed FBS, and 20 mM HEPES) for 1 h before they were pulse labeled for 30 min with the addition of 200 μCi/ml l-[35S]methionine (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Cells were then washed twice with 37°C Dulbecco's PBS with calcium and magnesium (Mediatech) and chased with normal DMEM supplemented with 5 mM l-methionine, 1 mM l-cystine, 10% FBS, and 20 mM HEPES. At each time point, chase medium was removed and saved on ice, and cells were washed twice with ice-cold PBS. Cells were lysed for 30 min in ice-cold lysis buffer that consisted of 1% NP-40, 0.1% SDS, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 10 mM Tris, pH 7.5, and protease inhibitor cocktail tablet (Roche Applied Science) dissolved according to the manufacturer's instructions. The lysates were cleared by centrifugation at 13,000 × g for 15 min. The saved chase media and lysates were pretreated by incubation with protein A-Sepharose (Sigma-Aldrich) for 1 h before they were incubated with anti-cathepsin D polyclonal antibody-coated protein A-Sepharose for 12 h at 4°C. The Sepharose beads were treated with 0.2% BSA to reduce nonspecific protein binding. After immunoprecipitation, the beads were washed with ice-cold washing buffer consisting of 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, and 50 mM Tris, pH 7.5. Proteins were eluted from the beads with SDS-PAGE sample buffer for electrophoresis. Fluorography was performed with the EN3HANCE reagent (PerkinElmer Life and Analytical Sciences, Boston, MA).
Ligand-induced EGFR Degradation Assay
Cells were incubated in medium supplemented with 0.1% FBS overnight before they were treated with 100 ng/ml epidermal growth factor (EGF) (Sigma-Aldrich) in serum-free medium. The EGF treatment was terminated by subjecting cells to lysis in SDS sample buffer. EGFR degradation was examined by Western analysis.
Sucrose Density Gradient Sedimentation
Clarified Triton X-100–soluble supernatants from HEK293 cells either treated with DSP or the solvent dimethyl sulfoxide alone were sedimented in a 5–20% sucrose gradient prepared in buffer A plus 0.5% Triton X-100 for 13 h at 240,000 × g in a SW55 rotor, 1.5 mg/gradient (Salazar et al., 2004). Twenty samples were collected from the bottom (250 μl/each) and analyzed by Western blots. The following standards were used to calibrate the gradients: horse spleen apoferritin (443 kDa; 16.5 S), bovine serum albumin (66 kDa; 4.6 S), sweet potato β-amylase (200 kDa; 9.4 S), and bovine erythrocyte carbonic anhydrase (29 kDa; 2.9 S).
RESULTS
SPE-39 Is Required for Processing of Endocytosed Proteins in C. elegans
In C. elegans, experimental systems have been developed to study intracellular trafficking in oocytes (Grant and Hirsh, 1999) and coelomocytes (Fares and Greenwald, 2001). Because spe-39 expression is not limited to the testes and our previous findings suggest SPE-39 functions in vesicular traffic (Zhu and L'Hernault, 2003), we used both systems to study the function of SPE-39.
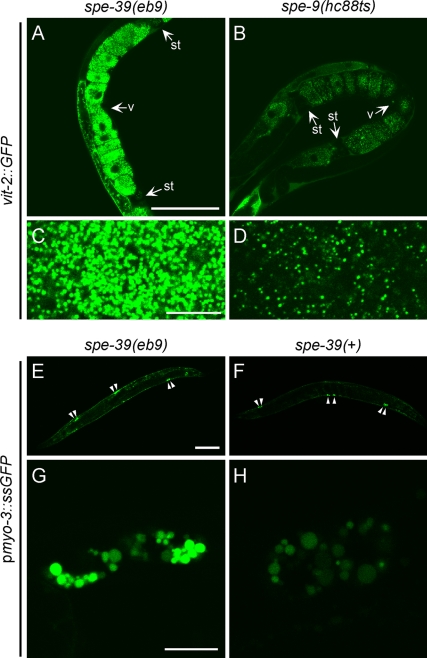
GFP-tagged major yolk protein VIT-2 (also known as YP-170), encoded by the integrated transgene bIs1[vit-2::GFP; rol-6(su1006)], has been used to study the uptake of yolk by oocytes (Grant and Hirsh, 1999). We tested the trafficking of VIT-2-GFP in spe-39(eb9) and spe-9(hc88ts) hermaphrodites. The spe-9(hc88ts) hermaphrodites were selected as a control because at 25°C they are, like spe-39(eb9) mutants, completely self-sterile and have an ovulation rate similar to that of spe-39(eb9) (L'Hernault et al., 1988). This allows comparison between oocytes that have similar exposure to the yolk protein and are not subsequently fertilized to become embryos. In addition, the spe-9 mutation is not expected to affect trafficking in oocytes because SPE-9 is a sperm-specific surface protein (Zannoni et al., 2003) required for sperm–oocyte interaction during fertilization (Singson et al., 1998). In spe-39(eb9) hermaphrodites, the mature oocytes in the uterus contain significantly higher level of VIT-2-GFP (Figure 1A) compared with mature oocytes in spe-9(hc88ts) control worms (Figure 1B). Significantly more yolk granules are found in spe-39(eb9) oocytes (Figure 1C) than in spe-9(hc88ts) control oocytes (Figure 1D), suggesting that processing of internalized yolk protein in oocytes is altered in spe-39 mutants.
Figure 1.
C. elegans SPE-39 is required for yolk protein processing in oocytes and proteolytic degradation in coelomocytes. (A–D) Confocal images of GFP-tagged yolk protein VIT-2, encoded by an integrated transgene, in mature oocytes of young adult spe-39(eb9) (whole uterus in A and magnified yolk granules in C) and control spe-9(hc88ts) (whole uterus in B and magnified yolk granules in D) hermaphrodites. The uterus is located between the two spermathecae (st) and the vulva (v) opens in the middle of the uterus. (E–H) Signal sequence-GFP fusion protein (ssGFP), expressed from an integrated transgene driven by the myo-3 promoter (pmyo-3), was examined in young adult spe-39(eb9) (whole worm in E and a pair of coelomocytes in G) and spe-39(+) (whole worm in F and a pair of coelomocytes in H) hermaphrodites by confocal microscopy. Arrowheads in E and F indicate the location of six coelomocytes in each worm. The two images in each row were recorded with the same settings. Bars, 100 μm (A, B, E, and F) and 10 μm (C, D, G, and H).
The coelomocytes are macrophage-like scavengers in the pseudocoelom and each worm has three pairs of them (Figure 1, E and F). The integrated transgene arIs37[pmyo-3::ssGFP], which encodes GFP fused to a signal sequence (ssGFP) and driven by the promoter of myo-3, has been used to study the endocytic pathway in these cells (Fares and Greenwald, 2001). In worms carrying the transgene, GFP is secreted from body wall muscles into the pseudocoelom, where it is taken up and degraded by the coelomocytes. This transgenic line expresses GFP at a very high level that apparently exceeds the engulfment ability of coelomocytes, and it is also possible that other cell types can internalize this soluble GFP, which would contribute to the noncoelomocyte signal that is always observed. In spe-39(+) worms carrying the transgene, the remaining level of GFP in the pseudocoelom is low (Figure 1F). If ssGFP is expressed in spe-39(eb9) mutants, it shows a slightly stronger labeling in the pseudocoelom (Figure 1E). Coelomocyte vesicular structures accumulated GFP in both spe-39(eb9) (Figure 1G) and spe-39(+) control worms (Figure 1H), but the level of accumulation is dramatically increased in spe-39 mutants. These results suggest that degradation of endocytosed substrates in coelomocytes is disrupted in mutants lacking SPE-39.
SPE-39 Binds C. elegans VPS33A and VPS33B In Vitro
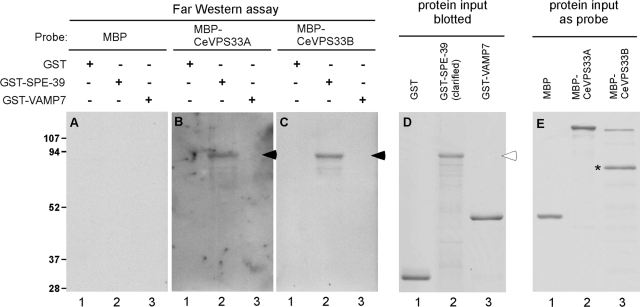
The previously described defective organellar biogenesis in spe-39 mutants suggests that SPE-39 might participate in vesicular fusion (Zhu and L'Hernault, 2003). Prior observations that the Drosophila SPE-39 orthologue interacts with Drosophila VPS33B (Giot et al., 2003; Pulipparacharuvil et al., 2005) suggested an underlying molecular mechanism involving the known features of the class C Vps proteins. To test whether SPE-39 interacts with C. elegans VPS33 homologues, we conducted an in vitro binding assay. Full-length coding sequences of C. elegans spe-39, VPS33A, and VPS33B were used to make GST-SPE-39, MBP-CeVPS33A, and MBP-CeVPS33B fusion proteins, respectively. These proteins were used in a Far Western assay, in which plain GST and GST fusion proteins were resolved by PAGE and blotted to membranes (Figure 2D), whereas plain MBP and MBP fusion proteins were used as probes (Figure 2E). The assay detected interaction between GST-SPE-39 and MBP-CeVPS33A (Figure 2B, lane 2, filled arrowhead) and between GST-SPE-39 and MBP-CeVPS33B (Figure 2C, lane 2, filled arrowhead). In the control experiments, when one of the binding partners was either plain MBP or plain GST (Figure 2A and lane 1 in B and C), interaction was not detected. Because residues 134-166 of SPE-39 are predicted to constitute an α-helix that can potentially form coiled-coil bundles with other α-helices (Lupas et al., 1991), a GST-VAMP7 fusion protein was included as a control to further demonstrate the specificity of this assay. The GST-VAMP7 fusion protein contains the first 190 residues of mouse VAMP7, which include two coiled-coil regions: the regulatory longin domain and the core SNARE motif (reviewed by Rossi et al., 2004). No interactions of GST-VAMP7 with either MBP-CeVPS33A or MBP-CeVPS33B (Figure 2, B and C, lane 3) were detected, suggesting that the observed interaction between SPE-39 and the VPS33 homologues was specific and did not result from nonspecific coiled-coil pairing. These results indicate that the SPE-39 protein interacts with both VPS33 homologues found in C. elegans.
Figure 2.
Far Western assay indicates that SPE-39 interacts with C. elegans VPS33A and VPS33B. (A–C) Blotted GST, GST-SPE-39 fusion protein, and GST-VAMP7 fusion protein were tested for their ability to bind MBP (A), MBP-CeVPS33A fusion protein (B), and MBP-CeVPS33B fusion protein (C). Detected MBP fusion proteins are indicated with filled arrowheads. (D and E) Coomassie Blue staining of input proteins blotted on membrane (D) and used as probes (E). In A–D, lanes 1 and 3 were loaded with 4 μg of GST and GST-VAMP7, respectively, whereas lane 2 was loaded with a sample containing similar amount of full-length GST-SPE-39. The hollow arrowhead marks the full-length GST-SPE-39 fusion protein. Probe proteins were diluted to 1 μg/ml with binding buffer when incubated with blots and 1 μg of each probe protein is shown in E. A major degradation product of MBP-CeVPS33B is indicated with an asterisk (lane 3 in E). Sizes of protein standards (left) are shown in kilodaltons.
VPS33B Is Required for Spermatogenesis in C. elegans
Because SPE-39 binds CeVPS33A and CeVPS33B in vitro, these proteins are likely to interact in vivo and have common biological functions. We tested this potential relationship by reducing the level of each VPS33 homologue and observing whether this caused defective spermatogenesis.
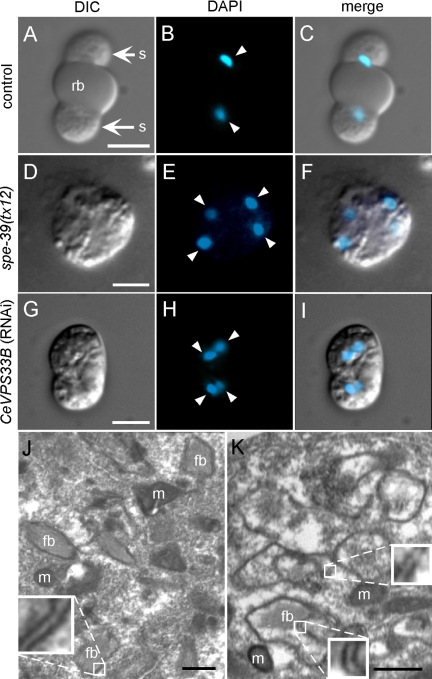
spe-39 mutants are defective in cytokinesis of spermatocytes. In wild-type C. elegans, ∼60% of the primary spermatocytes (4N) divide completely after meiosis I to give rise to two secondary spermatocytes (2N) (Ward et al., 1981). This division is partial for the remaining ∼40% primary spermatocytes, in which both 2N nuclei share a common cytoplasm until meiosis II is completed. In both situations, the spermatocytes undergo a final, asymmetric cytokinesis to give rise to spermatids (1N). The nascent spermatids bud from the spermatocyte periphery, whereas cellular constituents no longer required for sperm function are placed in the central cytoplasmic region. After the spermatids bud, the central part of the cytoplasm is discarded as a residual body (reviewed by L'Hernault, 2006; http://www.wormbook.org). The top row of Figure 3 shows a secondary spermatocyte (2N) undergoing cytokinesis in a control RNAi experiment. This cell has wild-type morphology, which includes the centrally placed residual body and budding spermatids (rb and s in Figure 3A, respectively). The residual body seems smooth when examined by differential interference contrast (DIC), whereas the spermatids have a textured appearance. During spermatid budding, the haploid (1N) nuclei, which are highly condensed at this stage (arrowheads in Figure 3B), are segregated into the nascent spermatids (Figure 3C). In spe-39 mutants, the nuclear events of meiosis can be completed, but cytokinesis of spermatocytes is defective. As a result, spermatocytes usually fail to divide and contain four nuclei (shown for the tx12 mutant in Figure 3, D–F). This phenotype has also been observed in spe-4 (L'Hernault and Arduengo, 1992) and spe-5 (Machaca and L'Hernault, 1997) mutants, and such cells are called terminal spermatocytes.
Figure 3.
RNAi of CeVPS33B induces abnormal FB-MO morphogenesis and defective cytokinesis of spermatocytes. (A–C) A dividing secondary spermatocyte from a male that was fed with HT115 (DE3) bacteria containing the empty RNAi vector L4440. This dividing spermatocyte has normal morphology, which is characterized by two budding spermatids and a centrally placed anucleate cytoplast named the residual body. (D–F) A terminally arrested spermatocyte from a spe-39(tx12) male. (G–I) A terminally arrested spermatocyte from a male fed with HT115 (DE3) bacteria containing the VPS33BIL/VPS33BIR RNAi construct. Each row contains a DIC image (A, D, and G), a corresponding image of nuclei (arrowheads) stained with 4′,6-diamidino-2-phenylindole (DAPI) (B, E, and H) and a merged image (C, F, and I). (J and K) Transmission electron microscopy images of spermatocytes from males treated with CeVPS33B RNAi. J shows a spermatocyte that contains wild-type–looking FBs with closely associated, double-layered membranes. K shows a spermatocyte with defective FB-MO morphogenesis, in which vacuolar spaces are found inside double-layered membranous envelopes. Examples of double-layered membranes are magnified in insets in J and K. fb, fibrous body; m, mitochondrion; rb, residual body; s, spermatid. Bars, 5 μm (A–I) and 0.5 μm (J and K).
We conducted RNAi experiments to examine whether either of the two C. elegans VPS33 homologues plays a role during spermatogenesis. The RNAi experiments were performed by feeding rrf-3(pk1426); him-5(e1490) worms with dsRNA-producing bacteria (see Materials and Methods for explanation). Po worms were fed exclusively with the dsRNA-producing bacteria, and before they became adults, hermaphroditic Po worms were selected and raised individually on the same bacteria. Each Po hermaphrodite produced a brood containing F1 hermaphrodites and males. From each of 10 randomly selected broods treated with CeVPS33B RNAi, we examined five to 12 F1 hermaphrodites for the oocyte-laying self-sterile (Spe) phenotype. Spe F1 hermaphrodites were scored from two of the 10 selected broods, and in the two broods, the incidence of the Spe phenotype was 17% and 60%, respectively. The low incidences of the Spe phenotype in most broods are consistent with previous observations that RNAi is difficult to perform in the testis (Fraser et al., 2000; Gonczy et al., 2000). When the same RNAi method was used for CeVPS33A and the RNAi vector-only control (see Materials and Methods for explanation), Spe worms were not observed. F1 males from CeVPS33B RNAi-treated broods that included many Spe hermaphrodites were dissected so that their sperm could be examined. These males produced spermatocytes that contained four condensed nuclei (Figure 3, G–I), and such terminal spermatocytes were not observed in control experiments (Figure 3, A–C). This indicates that depleting CeVPS33B can produce similar spermatogenesis defects to those found in spe-39 mutants.
The presence of terminal spermatocytes, such as in Figure 3, F or I, is usually associated with defects in formation of the MO and its associated structures (reviewed by L'Hernault, 2006; http://www.wormbook.org). In spermatocytes, the MO develops in intimate association with a bundle of proteinaceous fibers named the fibrous body (FB), and wild type FB-MO ultrastructure is quite uniform and well described (Wolf et al., 1978; Ward et al., 1981). One characteristic feature of the wild-type FB is its envelopment by a double-layered membrane that extends from the MO and intimately surrounds the periphery of the enclosed fiber bundle (Figure 3J and its associated insert box); this essentially wild-type image is of a CeVPS33B RNAi treated spermatocytes that had escaped the effects of RNAi because of its low efficiency in the testis (Fraser et al., 2000; Gonczy et al., 2000). When CeVPS33B RNAi is effective (Figure 3K), the double-layered membrane envelope either did not contain an enclosed FB (upper small box and its associated magnified inset in Figure 3K), or there was a gap between the FB and its surrounding membranes (lower small box and its associated magnified inset in Figure 3K). These results indicate that FB-MO morphogenesis is defective as a result of the CeVPS33B knockdown.
Human SPE-39 Interacts with Both Human VPS33A and VPS33B
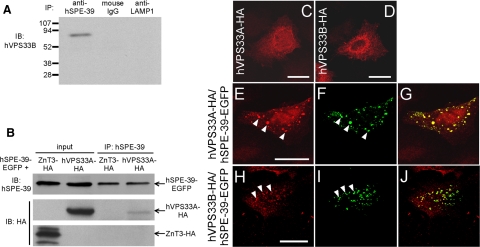
We tested whether human SPE-39 interacts with human VPS33A and VPS33B orthologues to determine whether the interaction between SPE-39 and VPS33 homologues is like that described above for C. elegans and therefore evolutionarily conserved. Immunoprecipitation with an anti-hSPE-39 mAb revealed that endogenous hVPS33B and hSPE-39 coimmunoprecipitated from HeLa cell lysate, whereas in control experiments using either normal mouse immunoglobulin G (IgG) or an anti-LAMP1 mAb, precipitation of hVPS33B was not detected (Figure 4A). Coimmunoprecipitation of hSPE-39 and hVPS33B was also observed in HEK293 cells (Supplemental Figure 3). Coimmunoprecipitation of hVPS33A-HA and hSPE-39-EGFP was detected when lysate of cells coexpressing these two fusion proteins was used (Figure 4B). HA-tagged zinc transporter 3 (ZnT3) (Salazar et al., 2004) was used as a control, which showed no interaction with hSPE-39-EGFP (Figure 4B). We also examined the localization of hSPE-39 and human VPS33 homologues when they were expressed as recombinant proteins in HeLa cells. When expressed singly, either HA-tagged hVPS33A or hVPS33B each had a diffuse cytoplasmic distribution with an elevated concentration in the perinuclear region (Figure 4, C and D). When hVPS33A-HA was coexpressed with hSPE-39-EGFP, its localization in the cytoplasm changed dramatically and these two proteins partially colocalized in a punctate pattern (Figure 4, E–G). Such redistribution was also observed for hVPS33B-HA when hSPE-39-EGFP was coexpressed (Figure 4, H–J). Control experiments showed that plain EGFP did not affect the staining patterns of hVPS33A-HA or hVPS33B-HA (data not shown). These coimmunoprecipitation and immunostaining results indicate that hSPE-39 interacts with both hVPS33A and hVPS33B, although the size and number of puncta observed when hSPE-39-EGFP is overexpressed might not represent what is physiologically normal (see below).
Figure 4.
hSPE-39 interacts with hVPS33A and hVPS33B. (A) Endogenous hSPE-39 bound to the anti-hSPE-39 mAb 64-8A12 coimmunoprecipitates endogenous hVPS33B in HeLa cells (left lane). The normal mouse IgG and anti-LAMP1 mAb H4A3 serve as negative controls (right two lanes). Sizes of protein standards are shown in kilodaltons. (B) hVPS33A-HA and hSPE-39-EGFP were coexpressed in HEK293 cells and were detected with anti-HA and anti-hSPE-39 antibodies, respectively, in the input sample. Immunoprecipitation with anti-hSPE-39 antibody resulted in coimmunoprecipitation of both hVPS33A-HA and hSPE-39-EGFP from the cell lysate. Lysate of cells coexpressing ZnT3-HA and hSPE-39-EGFP was used in a parallel control experiment that detected no coimmunoprecipitation. (C–J) hSPE-39-EGFP changed distribution patterns of hVPS33A-HA and hVPS33B-HA and colocalized with them. (C and D) HeLa cells expressing hVPS33A-HA and hVPS33B-HA, respectively. (E–G) A HeLa cell expressing both hVPS33A-HA and hSPE-39-EGFP. (H–J) A HeLa cell expressing both hVPS33B-HA and hSPE-39-EGFP. All red signals are staining of HA-tagged proteins (C, D, E, and H), whereas all green signals are EGFP-tagged hSPE-39 (F and I). G and J are merged images of the left (red) and middle (green) panels of their respective rows. Corresponding arrowheads in E and F, and in H and I, denote examples of colocalization. Bars, 20 μm.
hSPE-39 Is Associated with the HOPS Complex
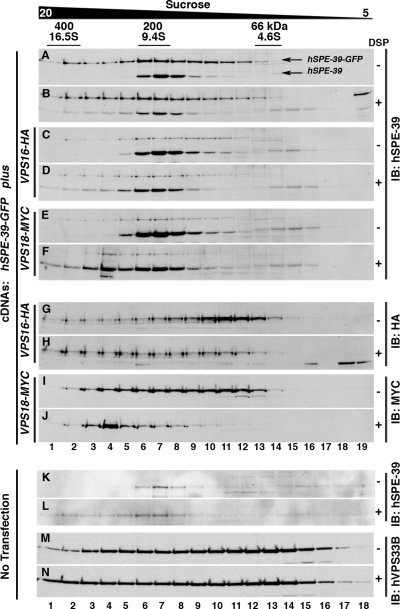
We next examined whether hSPE-39 interacts with other subunits of the HOPS complex. We created HEK cell lines that expressed tagged mammalian VPS11, VPS16, VPS18, VPS39, and VPS41 fusion proteins and transiently transfected them with hSPE-39-EGFP. HEK cells expressing hSPE-39-EGFP alone or coexpressing either VPS16-HA or VPS18-MYC were treated in the absence and presence of 1 mM DSP, a cell-permeant reversible cross-linking agent used for stabilizing low-affinity protein interactions (Craige et al., 2008). Clarified Triton X-100–soluble extracts were resolved by sucrose sedimentation and migration of hSPE-39 and HOPS subunits assessed by immunoblot of sucrose gradient fractions. Both endogenous and EGFP-tagged hSPE-39 migrated as a complex sedimenting at 9.4S (Figure 5A). SPE-39 sucrose sedimentation in hSPE-39-GFP–expressing cells was identical to its sedimentation in nontransfected HEK293 cells (Figure 5K). Addition of DSP increased the proportion of recombinant and endogenous hSPE-39 migrating between 16.5 and 9.4S (Figure 5, B and L). The expression of either VPS16-HA or VPS18-MYC did not changed the sedimentation pattern of endogenous or EGFP tagged hSPE-39 (Figure 5, C–F), which argues against competition between hSPE-39 and these two HOPS subunits. Importantly, in DSP-treated cells, VPS16-HA and VPS18-MYC cosedimented with hSPE-39 between 16.5 and 9.4S, suggesting that these proteins form a complex (Figure 5, G–J). The broad sucrose sedimentation patterns for VPS16-HA and VPS18-MYC are unlikely an artifact of HOPS subunits overexpression because endogenous VPS33B displayed an even broader sedimentation in either the absence or presence of DSP (Figure 5, M and N). These results suggest that, contrary to yeast, mammalian HOPS subunits can be found in multiple stoichiometries, some of which would include hSPE-39.
Figure 5.
hSPE-39 and HOPS subunits cosediment during sucrose gradient fractionation. HEK293 cells stably expressing hSPE-39 EGFP were either mock transfected (A and B) or transfected with plasmids encoding VPS16-HA (C, D, G, and H) or VPS18-MYC (E, F, I, and J). Cells were treated with vehicle (−) or 1 mM of the cell-permeant cross-linker 1 mM DSP (+). Clarified detergent soluble extracts from DSP treated and untreated cells were sedimented in 5–20% sucrose gradients, and fractions were probed with antibodies against hSPE-39 (A–F), HA (G and H), or MYC (I and J) epitopes. Nontransfected HEK293 cells (K–N) were probed for endogenous hSPE-39 (K and L) or hVPS33B (M and N) as controls for sedimentation experiments. hSPE-39 antibodies detect both endogenous and the EGFP-tagged versions of hSPE-39. Markers for Svedberg sedimentation coefficient (S) and molecular weight in kilodaltons are shown above A.
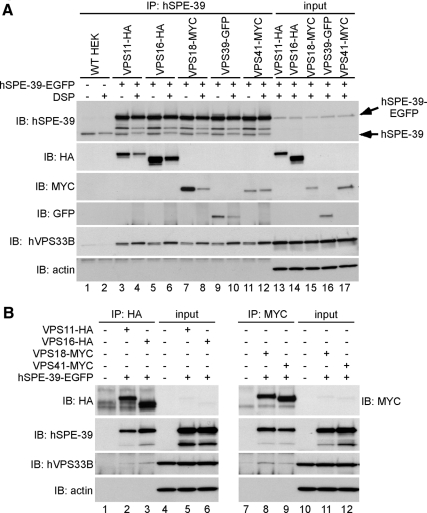
The anti-hSPE-39 antibody was then used to treat lysates from these cells to test whether hSPE-39 immunoprecipitates as a complex containing other HOPS subunits. Both the tagged HOPS subunits and endogenous hVPS33B coprecipitated with hSPE-39-EGFP, irrespective of the presence or absence of 1 mM DSP (Figure 6A). When antibodies against the HA or MYC tags were used for immunoprecipitation, HA- or MYC-tagged HOPS subunits were also found to be associated with hSPE-39-EGFP and endogenous hVPS33B (Figure 6B). In similar experiments, HA-tagged hSPE-39 also coimmunoprecipitated with tagged VPS11, VPS16, VPS18, and VPS41 (Supplemental Figure 4), suggesting that neither the EGFP nor the HA tag contributed to the binding of the hSPE-39 recombinant protein to the HOPS complex. These results are further confirmation that hSPE-39 and the HOPS subunits form a complex and suggest that they are functionally related.
Figure 6.
hSPE-39 is associated with the HOPS complex. (A and B) HEK cell lines expressing tagged HOPS complex subunits were transiently transfected with hSPE-39-EGFP and then used in immunoprecipitation experiments. (A) Cells treated with (even lanes) or without 1 mM DSP (odd lanes) were lysed, and clarified Triton X-100–soluble extracts were immunoprecipitated with monoclonal antibodies against hSPE-39, which recognized both endogenous and EGFP-tagged forms of hSPE-39 (arrows). The presence of HOPS subunits in the immunocomplexes was examined by Western analysis with polyclonal antibodies against HA, MYC, and GFP. hSPE-39 coprecipitated with VPS11-HA (lanes 3 and 4), VPS16-HA (lanes 5 and 6), VPS18-MYC (lanes 7 and 8), VPS39-GFP (lanes 9 and 10), VPS41-MYC (lanes 11 and 12), and endogenous hVPS33B. (B) Wild-type HEK cells (lanes 1 and 7) and HEK cell lines expressing VPS11-HA (lane 2), VPS16-HA (lane 3), VPS18-MYC (lane 8), or VPS41-MYC (lane 9) transiently transfected with hSPE-39-EGFP were lysed and immunoprecipitated with antibodies against HA (left) or MYC (right). Coimmunoprecipitated protein complexes contained hSPE-39-EGFP and endogenous hVPS33B. Each input was loaded at 2.5% of the total amount of cell lysate used for corresponding immunoprecipitation experiments.
hSPE-39 Localizes to Specific Endosomal Compartments
The interaction between hSPE-39 and the HOPS complex suggests that hSPE-39 functions in endosomal/lysosomal trafficking pathways (Kim et al., 2003; Richardson et al., 2004). A series of immunofluorescence experiments was next performed to obtain additional information on the location of hSPE-39 with respect to various cellular compartments. Overexpressed hSPE-39-EGFP does not colocalize with Golgi marker GM130 (Nakamura et al., 1997), early endosome marker EEA1 (Mu et al., 1995), recycling endosome marker transferrin receptor (Hopkins, 1983), late endosome/lysosome membrane proteins LAMP1 (Carlsson et al., 1988) and CD63 (Metzelaar et al., 1991), and lysosomal protease cathepsin D (reviewed by Liaudet-Coopman et al., 2006; Supplemental Figure 5). Although we showed that GFP tagged hSPE-39 could functionally replace the wild-type version (Supplemental Figure 6), we ultimately defined the subcellular localization of hSPE-39 in HEK293 cells because, in contrast to HeLa cells, this cell line possesses endogenous levels of hSPE-39 that are readily detectable by indirect immunofluorescence.
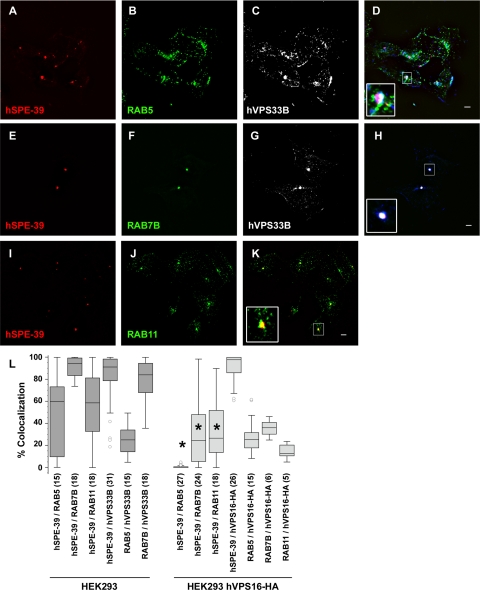
HEK293 cells have a discrete organelle that is revealed by immunofluorescence with hSPE-39 antibodies. Typically, cells contained one to three hSPE-39 perinuclear puncta per cell (Figure 7, A, E, and I, and Supplemental Figure 7, A, C, D, F, G, I, J, L, and M), and all hSPE-39 puncta were positive for either endogenous hVPS33B or VPS16-HA (Figure 7, D and H, and Supplemental Figure 7, A, D, G, and J). In contrast, except for one to three puncta, most of the hVPS33B- or VPS16-HA-positive organelles were devoid of hSPE-39 (Figure 7, D, E, and H; and Supplemental Figure 7, A, D, G, and J). We identified endosomes in these cells with antibodies against the GTPases RAB5, -7, and -11, which label sorting, late, and recycling endosomes, respectively (Grosshans et al., 2006). We found that, in addition to VPS33B and VPS16, hSPE-39 positive organelles were also immunoreactive for either RAB5 (Figure 7D), RAB7 (Figure 7, H and L, and Supplemental Figure 7I), or RAB11 (Figure 7, K and L, and Supplemental Figure 7L). In cells with two hSPE-39 positive puncta only one of them was positive for a RAB (Figure 7, D and K, and Supplemental Figure 7, I and L). Expression of VPS16-HA reduced the colocalization of endogenous hSPE-39 with these three endosomal RABs, but these effects were more pronounced in RAB5-positive endosomes (Figure 7L, compare dark and light gray bars). In fact, RAB5 was not detected in any hSPE-39-containing puncta when cells expressed VPS16-HA (Supplemental Figure 7C). However, quantitative colocalization analysis revealed that VPS16-HA was evenly distributed among RAB5, -7, and -11 endosomes (Figure 7L). Transfection of a plasmid encoding RAB5Q79L was used to assess whether this GTPase defective, constitutively active RAB5 mutant (Li et al., 1994; Stenmark et al., 1994) could drive hSPE-39 into sorting endosomes in VPS16-HA–expressing HEK293 cells. Expression of this mutant induced enlarged sorting endosomes (Raiborg et al., 2002, 2006) that characteristically contained VPS16-HA yet hSPE-39 was undetectable in these organelles (Supplemental Figure 7, E and F). Similarly, sorting endosomes enlarged by overexpression of Hrs-MYC (Raiborg et al., 2002, 2006) were also devoid of hSPE-39 signal (Supplemental Figure 7M). These findings indicate that hSPE-39 is expressed in early RAB5-positive, recycling RAB11-positive, and late RAB7-positive endosomes. However, in the presence of overexpressed VPS16-HA, hSPE-39 is undetectable in early RAB5-positive endosomes.
Figure 7.
hSPE-39 colocalizes with RAB5, RAB11, and RAB7 compartments. HEK293 cells were costained with antibodies against endogenous hSPE-39, RAB5, and hVPS33B (A–D); hSPE-39, RAB7B, and hVPS33B (E–H); and hSPE-39 and RAB11 (I–K). All images were acquired by wide-field deconvolution microscopy. The extent of colocalization of markers was quantitated using MetaMorph software, and the results are depicted in L as a box plot. The asterisk denotes p < 0.0001 by analysis of variance followed by the Student–Newman–Keuls multiple comparison test. The extent of hSPE-39 colocalization with specific RABs was compared in both HEK293 and HEK293 cells expressing VPS16-HA, images of which are shown in Supplemental Figure 7. Dark gray bars represent nontransfected HEK293 cells, and light gray bars depict HEK293 cells stably expressing VPS16-HA. The numbers in parentheses represent the number of analyzed cells obtained from at least two independent experiments. Images of hSPE-39 (A, D, E, H, I, and K) were pseudocolored red, and those of RABs (B, D, F, H, J, and K) were pseudocolored green. Bars, 5 μm.
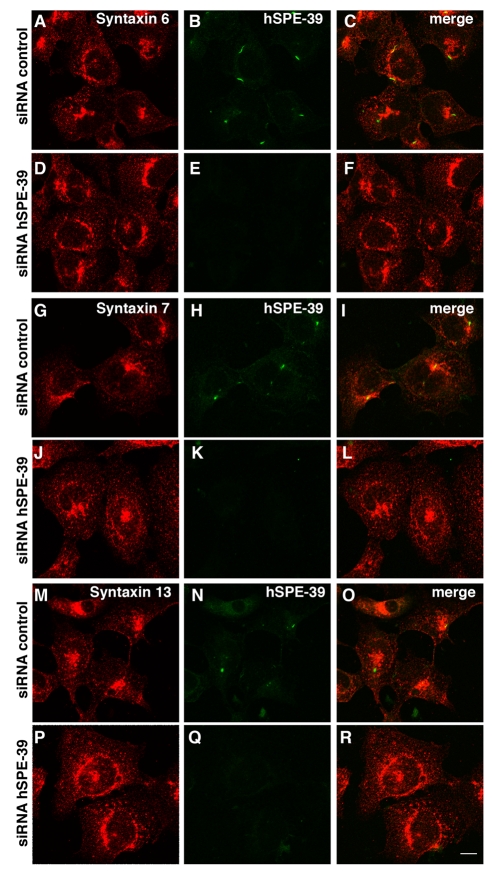
hSPE-39 Knockdown Affects the Morphology of Recycling and Late Endosomal Compartments
We assessed the involvement of hSPE-39 in recycling and late endosomes by examining HEK293 cells treated with hSPE-39 RNAi. Endosome architecture was assessed by confocal light microscopy of endosome-specific SNAREs. Recycling endosomes were identified with syntaxin 13, late endosomes with syntaxins 7 and -8, and syntaxin 6 was used to highlight the Golgi to sorting endosome compartments (Jahn and Scheller, 2006). Syntaxins 13, -7, and -8 are concentrated in the perinuclear area of control siRNA-treated cells (Figure 8, G–I, M–O, and Figure 9B). In contrast, hSPE-39 RNAi down-regulation induced a dispersion of the recycling endosome syntaxin 13 and late endosome syntaxins 7 and -8 (Figure 8, J–L, P–R, and Figure 9, E–H). Moreover, down-regulation of hSPE-39 triggered the formation of occasional enlarged syntaxin 8 late endosomes (Figure 9, F and G, arrow). Analysis of syntaxin 6 in hSPE-39 siRNA down-regulated cells revealed no changes in subcellular distribution (Figures 8, D–F, and 9, E and M). These results are consistent with the preferential localization of hSPE-39 to RAB11-positive recycling endosomes and RAB7-positive late endosomes.
Figure 8.
hSPE-39 siRNA down-regulation alters the distribution of recycling and late endosome markers. HEK293 cells were treated with control or hSPE-39 siRNA oligonucleotides, fixed, and double stained with antibodies against hSPE-39 with either syntaxin 6 (A–F), syntaxin 7 (G–L), or syntaxin 13 (M–R). All images were acquired by confocal microscopy. Bar, 10 μm.
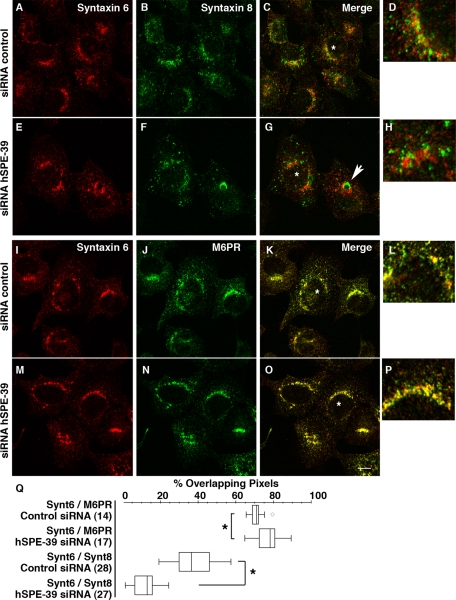
Figure 9.
hSPE-39 siRNA down-regulation alters the distribution of late endosome markers. HEK293 cells were treated with control or hSPE-39 siRNA oligonucleotides, fixed, and double stained with antibodies against syntaxin 6 with either syntaxin 8 (A–H) or M6PR (I–P). hSPE-39 RNAi occasionally induces enlargement of syntaxin 8-positive late endosomes, and an example is indicated by the arrow in G. Redistribution of syntaxin 8 and M6PR was measured relative to syntaxin 6 as overlapping pixels by using MetaMorph software and it is depicted in Q as a box plot. The asterisks in Q denote p < 0.0001 Mann–Whitney U test. The number of analyzed cells (in parentheses) was obtained from at least two independent experiments. All images were acquired by confocal microscopy. Bar, 10 μm.
hSPE-39 Is Required for M6PR-mediated Transport
The above-discussed results suggest that hSPE-39 down-regulation should impair the delivery of cargo transiting through recycling or late endosomes. Prior work has revealed that M6PR moves between the Golgi and endosomes along diverse recycling pathways (Ghosh et al., 2003; Bonifacino and Rojas, 2006). Recycling endosomes and late endosomes are among the endocytic compartments from which M6PR returns back to the Golgi complex (Klumperman et al., 1993; Hirst et al., 1998; Lin et al., 2004; Tortorella et al., 2007). Consequently, we analyzed the subcellular distribution of the mannose-6-phosphate receptor (M6PR) in control and hSPE-39 down-regulated cells. Down-regulation of hSPE-39 in HEK293 cells caused M6PR to move away from the cell periphery and concentrate in the perinuclear area (compare Figure 9J with N). This redistribution was expressed as a significant increase in the percentage of overlapping pixels between syntaxin 6 and M6PR (compare Figure 9L with P), which we quantitated (Figure 9Q).
The hSPE-39 siRNAi experiment was also performed in HeLa cells (Figure 10). After siRNA treatment, no expression of hSPE-39 could be detected by Western analysis (Figure 10A). Cells that lack hSPE-39 show M6PR signals that are high in the perinuclear region (Figure 10H), whereas control cells have a more scattered M6PR distribution (Figure 10I). The M6PR redistribution induced by hSPE-39 knockdown in HeLa cells is more dramatic than what we observed in HEK293 cells, which is likely due to the lower expression of hSPE-39 in HeLa cells. Concomitantly with abnormal M6PR distribution, the immunoreactivity of cathepsin D, a ligand of the M6PR (Ghosh et al., 2003), was significantly decreased by hSPE-39 RNAi. We established the specificity of the hSPE-39 RNAi-induced effects on M6PR and cathepsin D by showing that these phenotypes can be rescued by siRNA-resistant recombinant hSPE-39 expression (Supplemental Figure 6). Similar effects on cathepsin D were obtained by overexpression of hSPE-39-EGFP in HeLa cells (Supplemental Figure 5, P–R). Although cathepsin D levels were affected by the hSPE-39 knockdown, levels of the lysosome marker LAMP1 did not show a significant change (Figure 10, D and E). We also stained the RNAi-treated cells with the fluorescent acidotropic reagent LysoTracker Red DND-99, which labels acidic compartments including lysosomes (Figure 10, F and G). The result showed that the distribution of acidic vesicular compartments is not perturbed. These observations indicate that hSPE-39 knockdown disrupts the M6PR-mediated lysosomal delivery, whereas they also suggest that other lysosomal pathways are either not or less affected.
Pulse-chase experiments were performed to trace the trafficking and maturation of cathepsin D in HeLa cells treated with hSPE-39 RNAi and control RNAi (Figure 10J). Cells were pulse-labeled with radioactive methionine for 30 min and chased for 1, 2, 3, and 6 h. Three species of polypeptides, which correspond to the heavy chain of mature cathepsin D (31 kDa), procathepsin D (53 kDa), and a single-chain active intermediate (47 kDa) (Hasilik and Neufeld, 1980; Gieselmann et al., 1983), respectively, were immunoprecipitated by an anti-cathepsin D polyclonal antibody. Levels of these three cathepsin D species were analyzed in both cell lysates and culture media. In control siRNA-treated cells, proteolytic processing of 53-kDa procathepsin D into the 47-kDa form was detected after only 1 h of chase conditions. By 6 h, most of the 53-kDa procathepsin D was processed into the 47-kDa and the mature 31-kDa species. During the first 3 h of chase conditions, procathepsin D was continuously secreted into the medium. The 47-kDa intermediate species was prominent intracellularly at ∼6 h after labeling, when a slight decrease in the level of extracellular procathepsin D was also detected. This may reflect internalization of extracellular procathepsin D by surface mannose 6-phosphate receptors (reviewed by Ghosh et al., 2003; Liaudet-Coopman et al., 2006). In hSPE-39 RNAi-treated cells, although the level of 53-kDa procathepsin D at the start of the chase was similar to that in control RNAi-treated cells, maturation of cathepsin D was severely retarded. This was evident in the conversion of the 53 kDa into the 47-kDa species. Minimal levels of the 47-kDa cathepsin D species were found in cells after 1–6 h of chase conditions. Moreover, no detectable 31-kDa mature form was observed in hSPE-39 siRNA-treated cells. The extracellular level of procathepsin D continuously increased during the course of the chase, indicating its secretion was not blocked by hSPE-39 knockdown. Thus, loss of hSPE-39 apparently disrupted trafficking of cathepsin D and the disruption occurred after procathepsin D was glycosylated in the Golgi and acquired its normal molecular weight.
We also tested degradation of EGFR as a functional readout to determine whether lysosomal proteolysis is affected as a result of defective lysosomal protease delivery in cells lacking hSPE-39 (Figure 10K). After internalization of EGFR was induced by EGF, the molecular size of EGFR was slightly increased, which has been shown to indicate polyubiquitination of EGFR induced by EGF (Stang et al., 2000). Compared with control cells, cells lacking hSPE-39 show delayed degradation of EGFR at 15 and 30 min after addition of EGF. After 60 min of EGF induction, the EGFR levels in hSPE-39 siRNA- and control RNAi-treated cells became similar. These results suggest that hSPE-39 knockdown compromises, but does not abolish, proteolytic functions of lysosomes.
DISCUSSION
Previously, we showed that vesicular biogenesis during C. elegans spermatogenesis required SPE-39, but the underlying mechanism was unknown. Its widespread expression in both C. elegans and humans suggested that SPE-39 functioned in many tissues (Zhu and L'Hernault, 2003). The present study shows that SPE-39 orthologues play an evolutionarily conserved role in lysosomal delivery, probably via association with the core HOPS complex in a discrete population of endosomes. Consequently, the subcellular distribution of various endosomal SNAREs, as well as M6PR trafficking, requires hSPE-39. These observations are consistent with cathepsin D delivery and lysosomal proteolysis defects observed in hSPE-39 siRNA-treated cells. Additionally, C. elegans spe-39 mutants had defects consistent with decreased degradation of internalized proteins in oocytes and the macrophage-like coelomocytes.
We assessed endogenous hSPE-39 in HEK293 cells to determine its subcellular distribution relative to known HOPS and endocytic markers. HOPS subunits usually reside within numerous structures dispersed along the endocytic route (Figure 7; Kim et al., 2001). Depending on the vesicle, VPS16 or VPS33B colocalized with RAB5 (sorting endosomes), RAB7 (late endosomes), and RAB11 (recycling endosomes). In contrast, hSPE-39 localized in one to three VPS16- or VPS33B-positive dots. In nontransfected cells, these hSPE-39–positive dots colocalized with RAB11, RAB7, or RAB5. In cells where two hSPE-39–positive dots were present, only one of them was positive for a tested RAB (RAB5, RAB11, or RAB7). Perhaps hSPE-39 protein levels were too low in most vesicles to allow their detection by immunofluorescence. Cytosolic RAB effectors can be driven into sorting endosomes by overexpressing a RAB5Q79L GTPase mutation (Raiborg et al., 2002, 2006), but we found that this mutation failed to drive hSPE-39 into additional puncta in VPS16-HA–expressing cells. Therefore, we conclude that hSPE-39 resides within a discrete set of sorting, recycling, and late endosomal compartments. This interpretation was strengthened when we showed that hSPE-39 down-regulation by siRNA altered SNAREs syntaxin 13 (recycling endosomes; Prekeris et al., 1998; Jahn and Scheller, 2006) and syntaxin 7 and syntaxin 8 (late endosomes; Antonin et al., 2000; Jahn and Scheller, 2006; Raiborg et al., 2006) distribution but did not affect syntaxin 6 distribution (Golgi to sorting endosome traffic; Simonsen et al., 1999; Jahn and Scheller, 2006). An attractive hypothesis is that certain vesicles have SNAREs that require hSPE-39 to localize properly, and this might be mediated by hSPE-39 interaction with hVPS33A-B. Mammalian VPS33A-B are Sec1/Munc18 family members present in the HOPS complex, and they are homologous to yeast Vps33p, which binds target membrane-SNAREs (Kim et al., 2001; Wickner, 2002; Gissen et al., 2005). Cells in which hSPE-39 expression is lowered by siRNA show altered M6PR distribution, cathepsin D maturation, and EGF receptor degradation, and perhaps this occurs through HOPS effects on recycling and late endosomal SNAREs. M6PR moves between the Golgi and endosomes along diverse pathways (Ghosh et al., 2003; Bonifacino and Rojas, 2006) that include recycling and late endosomes, which are locations from which M6PR returns back to the Golgi complex (Klumperman et al., 1993; Hirst et al., 1998; Gissen et al., 2004; Tortorella et al., 2007).
HeLa cells treated with hSPE-39 siRNA showed disrupted trafficking of both M6PR and its cathepsin D cargo. However, LAMP1, a lysosomal membrane protein that shows M6PR-independent transport (Karlsson and Carlsson, 1998), is unaffected in hSPE-39 siRNA-treated cells. Similar to hSPE-39–depleted cells, the LAMP1 distribution was unaffected whereas M6PR accumulated in large vesicles within cells expressing dominant-negative mutant forms of RAB7; wild-type RAB7 facilitates membrane fusion at a late stage in the endosome-lysosome pathway (Press et al., 1998). These results suggest that SPE-39 regulates the delivery of a subset of lysosomal resident proteins.
The reduced cathepsin D levels observed when hSPE-39 is overexpressed might be caused by a dominant-negative effect on a multiprotein complex. Because cathepsin D misregulation is implicated in cancer progression (reviewed by Fusek and Vetvicka, 2005; Liaudet-Coopman et al., 2006), altering hSPE-39 levels in vivo may have important consequences. Cathepsin D expression in estrogen receptor-positive breast cancer cells is regulated by estrogen (Cavailles et al., 1993), and a recent study indicates that the hSPE-39 promoter binds estrogen receptor α in a hormone-dependent manner (Laganiere et al., 2005). This suggests that hSPE-39 plays an important role in cathepsin D regulation in vivo.
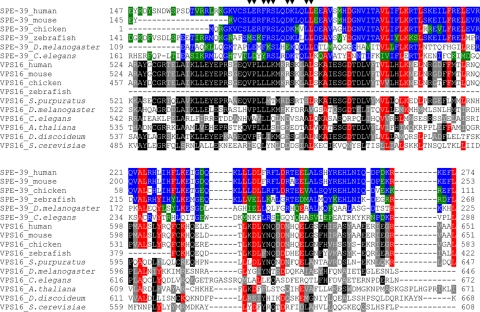
Prior studies suggested that Drosophila has one HOPS complex containing Car (dVPS33A) and dVPS16A and a second complex containing dVPS33B and the Drosophila SPE-39 orthologue, which was considered an alternative VPS16 homologue (Pulipparacharuvil et al., 2005). This differs from our data in both C. elegans and two human cell lines in which SPE-39 orthologues clearly interact with both VPS33 homologues. In addition, we found that mammalian SPE-39 and VPS16 coimmunoprecipitate, cosediment, and colocalize, which suggests they are subunits of a single protein complex. The aligned SPE-39 orthologues and VPS16 orthologues show homology over only a limited region, which suggests they are two distinct protein types. Although the VPS16 C-terminal region (pfam04840) shows homology to SPE-39 orthologues (NCBI Conserved Domain Search, CDD v2.05), there is no detectable homology between the VPS16 N-terminal region (pfam04841) and any SPE-39 orthologue. Furthermore, all SPE-39 orthologues, including Drosophila, have a unique motif (Zhu and L'Hernault, 2003; filled triangles in Figure 11) near the N-terminal end of the VPS16-homologous region (Figure 11). This motif, which is close to the VPS16-homologous region, might have an important, SPE-39–specific function. These observations indicate that SPE-39 is a HOPS complex-interacting protein with a role that differs from VPS16.
Figure 11.
SPE-39 orthologues share homology with the VPS16 C-terminal region and the VPS16-homologous region is adjacent to a SPE-39–specific motif. Sequence alignments for SPE-39 and VPS16 orthologues were generated separately by the ClustalW program version 1.82 (Chenna et al., 2003) by using full-length protein sequences. The homologous regions shared by the two alignments were then aligned manually for a comparison, and the section that contains the VPS16-homologous region and the adjacent SPE-39–specific motif is shown. For C. elegans SPE-39, this section includes residues 161–288 of the full-length protein of 522 amino acids. Identical and similar residues in SPE-39 orthologues are shaded with blue and green, respectively. Identical and similar residues in VPS16 orthologues are shaded with black and gray, respectively. Residues conserved in both SPE-39 and VPS-16 homologues are shaded with red. The following NCBI RefSeq records were used for SPE-39 orthologues: human (NP_071350.2), mouse (NP_598805.2), chicken (XP_421291.1), zebrafish (NP_001001836.1), D. melanogaster (NP_651731.1), and C. elegans (NP_504718.1). The following NCBI RefSeq records were used for VPS16 orthologues: human (NP_072097.2), mouse (NP_085036.2), chicken (XP_424209.1), zebrafish (XP_697990.1), Strongylocentrotus purpuratus (XP_781126.1), D. melanogaster (NP_649877.1), C. elegans (NP_498411.1), Arabidopsis thaliana (NP_565879.1), Dictyostelium discoideum (XP_646623.1), and S. cerevisiae (NP_015280.1). Filled triangles (▼) over the top row of the sequences denote conserved residues of SPE-39 orthologues that constitute a characteristic motif [(LM)-(ED)-x-(FY)-(RK)-S-x-x-(DE)-K-x-x-L-L].
MO precursor vesicles initially bud from the Golgi during C. elegans spermatogenesis, but subsequent events were not understood (Wolf et al., 1978). CeVPS33B RNAi-treated animals produce spermatocytes that share cell division and MO defects with spe-39 mutants (Zhu and L'Hernault, 2003). VPS33B, which interacts with SPE-39, facilitates vesicular fusion to lysosomal compartments in other species (Rieder and Emr, 1997; Sevrioukov et al., 1999; Sato et al., 2000), suggesting that MO assembly uses lysosomal delivery pathways. Mature MOs share several characteristics with bona fide lysosomes. Like lysosomes (reviewed by Sun-Wada et al., 2004), MOs are acidified by V-ATPase activity (our unpublished data). Furthermore, the MO defects shown by spe-5 mutants (Machaca and L'Hernault, 1997) are due to mutations in a V-ATPase B subunit (our unpublished data). Like conventional lysosomes after plasma membrane damage (reviewed by Andrews, 2002) and secretory lysosomes (reviewed by Blott and Griffiths, 2002), MOs fuse with the plasma membrane and play a secretory role (reviewed by L'Hernault, 2006; http://www.wormbook.org). Interestingly, dysferlin facilitates mammalian sarcolemmal repair (Bansal et al., 2003), whereas its C. elegans homologue FER-1 is required for MO fusion with the plasma membrane during sperm maturation (Achanzar and Ward, 1997; Washington and Ward, 2006).
Lysosomal trafficking is morphologically more diverse and requires more components in metazoans than in unicellular organisms. For example, Hermansky–Pudlak syndrome causes pathology that includes defective melanosome and platelet dense granule biogenesis (reviewed by Wei, 2006; Raposo and Marks, 2007; Raposo et al., 2007). Many proteins implicated in this disease and its mouse models are not encoded by the yeast genome (reviewed by Di Pietro and Dell'Angelica, 2005). Our data indicate that SPE-39 is a novel protein required for lysosomal trafficking only in metazoans. This suggests that MO biogenesis during C. elegans spermatogenesis is a useful system for identifying new lysosomal trafficking components unique to animals.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Drs. Melanie L. Styers, James J. Lah, Edwin R. Smith, and Ms. Tamara Lindsay for assistance with mammalian cell culture, immunostaining, and immunoblotting techniques. Nancy L'Hernault provided assistance with electron microscopy. We thank Dr. Andrew Peden for antibodies and Karen A. Newell-Litwa, Dr. Laura Volpicelli, Dr. H. Stenmark, and Dr. Yuji Kohara for various DNA clones used in this study. The Caenorhabditis Genetics Center provided several nematode strains and is funded by the National Institutes of Health National Center for Research Resources. B. F. was supported by the Emory University SURE summer program, which is funded by Howard Hughes Medical Institute award 52003727 to the Center for Science Education, Emory University. This work was also supported by National Institutes of Health grants GM-40697 and GM-082932 (to S.W.L.) and NS-42599 and GM-077569 (to V. F.), funds from Emory College, and an Emory University Research Committee award.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0728) on December 30, 2008.
REFERENCES
- Achanzar W. E., Ward S. A nematode gene required for sperm vesicle fusion. J. Cell Sci. 1997;110:1073–1081. doi: 10.1242/jcs.110.9.1073. [DOI] [PubMed] [Google Scholar]
- Andrews N. W. Lysosomes and the plasma membrane: trypanosomes reveal a secret relationship. J. Cell Biol. 2002;158:389–394. doi: 10.1083/jcb.200205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Holroyd C., Fasshauer D., Pabst S., Von Mollard G. F., Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- Banta L. M., Vida T. A., Herman P. K., Emr S. D. Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol. Cell. Biol. 1990;10:4638–4649. doi: 10.1128/mcb.10.9.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blott E. J., Griffiths G. M. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 2002;3:122–131. doi: 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S. R., Roth J., Piller F., Fukuda M. Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Major sialoglycoproteins carrying polylactosaminoglycan. J. Biol. Chem. 1988;263:18911–18919. [PubMed] [Google Scholar]
- Cavailles V., Augereau P., Rochefort H. Cathepsin D gene is controlled by a mixed promoter, and estrogens stimulate only TATA-dependent transcription in breast cancer cells. Proc. Natl. Acad. Sci. USA. 1993;90:203–207. doi: 10.1073/pnas.90.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Salazar G., Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol. Biol. Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica E. C. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Di Pietro S. M., Dell'Angelica E. C. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- Du Q., Thonberg H., Wang J., Wahlestedt C., Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastham K. M., McKiernan P. J., Milford D. V., Ramani P., Wyllie J., van't Hoff W., Lynch S. A., Morris A. A. ARC syndrome: an expanding range of phenotypes. Arch Dis. Child. 2001;85:415–420. doi: 10.1136/adc.85.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faundez V. V., Kelly R. B. The AP-3 complex required for endosomal synaptic vesicle biogenesis is associated with a casein kinase Ialpha-like isoform. Mol. Biol. Cell. 2000;11:2591–2604. doi: 10.1091/mbc.11.8.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fusek M., Vetvicka V. Dual role of cathepsin D: ligand and protease. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2005;149:43–50. doi: 10.5507/bp.2005.003. [DOI] [PubMed] [Google Scholar]
- Futter C. E., Pearse A., Hewlett L. J., Hopkins C. R. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Dahms N. M., Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Gieselmann V., Pohlmann R., Hasilik A., Von Figureura K. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J. Cell Biol. 1983;97:1–5. doi: 10.1083/jcb.97.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gissen P., Johnson C. A., Gentle D., Hurst L. D., Doherty A. J., O'Kane C. J., Kelly D. A., Maher E. R. Comparative evolutionary analysis of VPS33 homologues: genetic and functional insights. Hum. Mol. Genet. 2005;14:1261–1270. doi: 10.1093/hmg/ddi137. [DOI] [PubMed] [Google Scholar]
- Gissen P., et al. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat. Genet. 2004;36:400–404. doi: 10.1038/ng1325. [DOI] [PubMed] [Google Scholar]
- Gonczy P., et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Grant B., Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B. L., Ortiz D., Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl Acad. Sci. USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasilik A., Neufeld E. F. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J. Biol. Chem. 1980;255:4937–4945. [PubMed] [Google Scholar]
- Hirst J., Futter C. E., Hopkins C. R. The kinetics of mannose 6-phosphate receptor trafficking in the endocytic pathway in HEp-2 cells: the receptor enters and rapidly leaves multivesicular endosomes without accumulating in a prelysosomal compartment. Mol. Biol. Cell. 1998;9:809–816. doi: 10.1091/mbc.9.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. A., Horvitz H. R., Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C. R. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell. 1983;35:321–330. doi: 10.1016/0092-8674(83)90235-0. [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Karlsson K., Carlsson S. R. Sorting of lysosomal membrane glycoproteins lamp-1 and lamp-2 into vesicles distinct from mannose 6-phosphate receptor/gamma-adaptin vesicles at the trans-Golgi network. J. Biol. Chem. 1998;273:18966–18973. doi: 10.1074/jbc.273.30.18966. [DOI] [PubMed] [Google Scholar]
- Kim B. Y., Kramer H., Yamamoto A., Kominami E., Kohsaka S., Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- Kim B. Y., Ueda M., Kominami E., Akagawa K., Kohsaka S., Akazawa C. Identification of mouse Vps16 and biochemical characterization of mammalian class C Vps complex. Biochem. Biophys. Res. Commun. 2003;311:577–582. doi: 10.1016/j.bbrc.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Hille A., Veenendaal T., Oorschot V., Stoorvogel W., von Figureura K., Geuze H. J. Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J. Cell Biol. 1993;121:997–1010. doi: 10.1083/jcb.121.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault S. W. WormBook, editor. The C. elegans Research Community, WormBook. 2006. Spermatogenesis. doi/10.1895/wormbook. 1.7.1. [Google Scholar]
- L'Hernault S. W., Arduengo P. M. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J. Cell Biol. 1992;119:55–68. doi: 10.1083/jcb.119.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault S. W., Shakes D. C., Ward S. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Lefebvre C., Bataille A. R., Robert F., Giguere V. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Barbieri M. A., Colombo M. I., Stahl P. D. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J. Biol. Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- Liaudet-Coopman E., Beaujouin M., Derocq D., Garcia M., Glondu-Lassis M., Laurent-Matha V., Prebois C., Rochefort H., Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Lin S. X., Mallet W. G., Huang A. Y., Maxfield F. R. Endocytosed cation-independent mannose 6-phosphate receptor traffics via the endocytic recycling compartment en route to the trans-Golgi network and a subpopulation of late endosomes. Mol. Biol. Cell. 2004;15:721–733. doi: 10.1091/mbc.E03-07-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo B., Li L., Gissen P., Christensen H., McKiernan P. J., Ye C., Abdelhaleem M., Hayes J. A., Williams M. D., Chitayat D., Kahr W. H. Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2005;106:4159–4166. doi: 10.1182/blood-2005-04-1356. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Poupon V., Lindsay M. R., Mullock B. M., Piper R. C., Pryor P. R. Membrane dynamics and the biogenesis of lysosomes. Mol. Membr. Biol. 2003;20:141–154. doi: 10.1080/0968768031000089546. [DOI] [PubMed] [Google Scholar]
- Machaca K., L'Hernault S. W. The Caenorhabditis elegans spe-5 gene is required for morphogenesis of a sperm-specific organelle and is associated with an inherent cold-sensitive phenotype. Genetics. 1997;146:567–581. doi: 10.1093/genetics/146.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzelaar M. J., Wijngaard P. L., Peters P. J., Sixma J. J., Nieuwenhuis H. K., Clevers H. C. CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem. 1991;266:3239–3245. [PubMed] [Google Scholar]
- Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T.P., Rabouille C., Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Peplowska K., Markgraf D. F., Ostrowicz C. W., Bange G., Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Peterson M. R., Emr S. D. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y. A., Scheller R. H. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B., Feng Y., Hoflack B., Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A., Seals D., Wickner W., Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M. A., Ray S., Sevrioukov E. A., Haberman A. S., Rohrer J., Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Wesche J., Malerod L., Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J. Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- Raposo G., Marks M. S. Melanosomes–dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Marks M. S., Cutler D. F. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr. Opin. Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul J., et al. C. elegans ORFeome version 1. 1, Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Richardson S. C., Winistorfer S. C., Poupon V., Luzio J. P., Piper R. C. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell. 2004;15:1197–1210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. E., Emr S. D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Banfield D. K., Vacca M., Dietrich L. E., Ungermann C., D'Esposito M., Galli T., Filippini F. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Salazar G., Love R., Werner E., Doucette M. M., Cheng S., Levey A., Faundez V. The zinc transporter ZnT3 interacts with AP-3 and it is preferentially targeted to a distinct synaptic vesicle subpopulation. Mol. Biol. Cell. 2004;15:575–587. doi: 10.1091/mbc.E03-06-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. K., Rehling P., Peterson M. R., Emr S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov E. A., He J. P., Moghrabi N., Sunio A., Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Shakes D. C., Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev. Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., Plasterk R. H. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Gaullier J. M., D'Arrigo A., Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- Singson A., Mercer K. B., L'Hernault S. W. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Woolford C. A., Jones E. W. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics. 2000;156:105–122. doi: 10.1093/genetics/156.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang E., Johannessen L. E., Knardal S. L., Madshus I. H. Polyubiquitination of the epidermal growth factor receptor occurs at the plasma membrane upon ligand-induced activation. J. Biol. Chem. 2000;275:13940–13947. doi: 10.1074/jbc.275.18.13940. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Woolford C. A., Jones E. W. The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:2593–2605. doi: 10.1091/mbc.E03-10-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada G. H., Wada Y., Futai M. Diverse and essential roles of mammalian vacuolar-type proton pump ATPase: toward the physiological understanding of inside acidic compartments. Biochim. Biophys. Acta. 2004;1658:106–114. doi: 10.1016/j.bbabio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Oiso N., Gautam R., Novak E. K., Panthier J. J., Suprabha P. G., Vida T., Swank R. T., Spritz R. A. The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc. Natl. Acad. Sci. USA. 2003;100:1146–1150. doi: 10.1073/pnas.0237292100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J. R., Sedat J. W., Agard D. A. Deconvolution in optical microscopy. In: Jansson P. A., editor. Deconvolution of Images and Spectra. San Diego, CA: Academic Press; 1997. pp. 284–307. [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tortorella L. L., Schapiro F. B., Maxfield F. R. Role of an acidic cluster/dileucine motif in cation-independent mannose 6-phosphate receptor traffic. Traffic. 2007;8:402–413. doi: 10.1111/j.1600-0854.2007.00541.x. [DOI] [PubMed] [Google Scholar]
- Wada Y., Kitamoto K., Kanbe T., Tanaka K., Anraku Y. The SLP1 gene of Saccharomyces cerevisiae is essential for vacuolar morphogenesis and function. Mol. Cell. Biol. 1990;10:2214–2223. doi: 10.1128/mcb.10.5.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Achim C. L., Hamilton R. L., Wiley C. A., Soontornniyomkij V. Tyramide signal amplification method in multiple-label immunofluorescence confocal microscopy. Methods. 1999;18:459–464. doi: 10.1006/meth.1999.0813. [DOI] [PubMed] [Google Scholar]
- Ward S., Argon Y., Nelson G. A. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 1981;91:26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington N. L., Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J. Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- Wei M. L. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J. 2002;21:1241–1247. doi: 10.1093/emboj/21.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N., Hirsh D., McIntosh J. R. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J. Ultrastruct. Res. 1978;63:155–169. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoni S., L'Hernault S. W., Singson A. W. Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev. Biol. 2003;3:10. doi: 10.1186/1471-213X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G. D., L'Hernault S. W. The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics. 2003;165:145–157. doi: 10.1093/genetics/165.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.