Abstract
Many Saccharomyces cerevisiae mutants defective in the SUMO pathway accumulate elevated levels of the native 2 μm circle plasmid (2 μm). Here we show that accumulation of 2 μm in the SUMO pathway mutants siz1Δ siz2Δ, slx5Δ, and slx8Δ is associated with formation of an aberrant high-molecular-weight (HMW) form of 2 μm. Characterization of this species from siz1Δ siz2Δ showed that it contains tandem copies of the 2 μm sequence as well as single-stranded DNA. Accumulation of this species requires both the 2 μm–encoded Flp recombinase and the cellular homologous recombination repair (HRR) pathway. Importantly, reduced SUMO attachment to Flp is sufficient to induce formation of this species. Our data suggest a model in which Flp that cannot be sumoylated causes DNA damage, whose repair via HRR produces an intermediate that generates tandem copies of the 2 μm sequence. This intermediate may be a rolling circle formed via break-induced replication (BIR), because mutants defective in BIR contain reduced levels of the HMW form. This work also illustrates the importance of using cir° strains when studying mutants that affect the yeast SUMO pathway, to avoid confusing direct functions of the SUMO pathway with secondary effects of 2 μm amplification.
INTRODUCTION
The 2 μm circle is a naturally occurring 6318-base pair plasmid (2 μm) present in ∼40–60 copies in virtually all strains of Saccharomyces (Broach and Volkert, 1991). 2 μm's only known activity is self-propagation, which is accomplished via two distinct plasmid maintenance mechanisms (Jayaram et al., 2004). One of these allows the plasmid to be amplified if its copy number drops and is mediated by the plasmid-encoded Flp recombinase and two DNA target sites for Flp (FRT) that are present on opposite sides of the plasmid. Flp catalyzes an intramolecular recombination reaction between the two FRT sites, thereby inverting one side of the 2 μm with respect to the other. Futcher (1986) has proposed a model that explains how this activity could allow multiple copies of 2 μm to be made from a single origin firing: if recombination occurs during replication between the converging replication forks (RFs), it would generate a double rolling circle where both forks chase each other around the template in the same direction. The 2 μm sequence would be rereplicated until a second Flp-dependent recombination event permits convergence of the RFs.
Native levels of 2 μm have little effect on the growth rate of wild-type (wt) yeast (Broach and Volkert, 1991). However, in several mutants defective in the SUMO (Smt3) pathway, accumulation of high copy numbers of 2 μm is associated with slow, cold-sensitive growth, and cell cycle delays caused by activation of the DNA damage checkpoint (Zhao et al., 2004; Chen et al., 2005; Dobson et al., 2005; Burgess et al., 2007; Ii et al., 2007b). These strains are also heterogeneous and form irregularly shaped colonies, presumably because different lineages contain different levels of 2 μm, leading to different degrees of growth inhibition. Removal of 2 μm from these strains eliminates these conspicuous growth defects.
SUMO is a ubiquitin-like protein that functions by being attached to other proteins as a posttranslational modification (Johnson, 2004; Hay, 2005; Geiss-Friedlander and Melchior, 2007). SUMO conjugation is essential for viability of Saccharomyces cerevisiae, and SUMO is attached to hundreds of yeast proteins. SUMO attachment is catalyzed by a three-step pathway analogous to the ubiquitin pathway. In yeast, the SUMO pathway consists of the heterodimeric E1 Uba2-Aos1, the E2 Ubc9, and several SUMO ligases (E3s), which include Siz1, Siz2/Nfi1, Mms21, and the meiotic E3 Zip3 (Geiss-Friedlander and Melchior, 2007). SUMO can be cleaved off substrate proteins by SUMO-specific proteases, including the yeast Ulp1 and Ulp2. Ulp1 is also required for maturation of the SUMO precursor and so participates in both SUMO conjugation and deconjugation. Mutants in certain nuclear pore complex (NPC) and nuclear envelope proteins also have sumoylation defects because they mislocalize Ulp1 (Zhao et al., 2004; Chen et al., 2007; Lewis et al., 2007; Palancade et al., 2007). Recently, a factor that acts downstream of sumoylation has been identified: a heterodimer consisting of Slx5/Hex3, and Slx8 acts as a ubiquitin ligase that targets sumoylated proteins for ubiquitin-dependent proteolysis (Ii et al., 2007a; Prudden et al., 2007; Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007). The natural substrates of Slx5·Slx8-dependent ubiquitylation are unknown.
The siz1Δ siz2Δ double mutant, slx5Δ, slx8Δ, ulp1, and certain NPC mutants accumulate as much as 10-fold higher than wt levels of 2 μm DNA (Zhao et al., 2004; Chen et al., 2005; Dobson et al., 2005; Burgess et al., 2007; Ii et al., 2007b). A potential mechanism for SUMO's role in 2 μm stability is suggested by the fact that Flp recombinase is modified by SUMO at a fairly high level (∼10%; Chen et al., 2005). When Flp sumoylation is reduced, either in the siz1Δ siz2Δ mutant or in a strain where FLP is mutated to eliminate its major SUMO attachment site, Flp protein accumulates to higher than wt levels (Chen et al., 2005). These higher levels of Flp could be partially responsible for hyperamplification of 2 μm, because overexpression of Flp also increases 2 μm DNA levels (Murray et al., 1987; Reynolds et al., 1987).
Here we found that the 2 μm hyperamplification in SUMO pathway mutants requires the homologous recombination repair (HRR) pathway. During vegetative yeast growth, the primary role of HRR is to repair double-strand breaks (DSBs) and to resolve aberrant structures generated during DNA replication (Paques and Haber, 1999; Krogh and Symington, 2004). The RAD52 epistasis group is responsible for carrying out HRR and includes genes involved in several distinct processes. The RAD51 subgroup: RAD51, RAD52, RAD54, RAD55, and RAD57, is required for gene conversion events, which require participation of both sides of the DSB in the repair reaction. The RAD51 group can also carry out break-induced replication (BIR), where one end of a DSB invades an intact homologous sequence and establishes a RF that performs both leading and lagging strand synthesis and can copy many kilobases of DNA (McEachern and Haber, 2006; Llorente et al., 2008). This process involves only one side of the DSB. BIR can also take place by a RAD51-independent mechanism that involves RAD52, RAD59, RDH54/TID1, and the MRE11-RAD50-XRS2 complex (MRX). Mutants lacking both known BIR pathways dramatically reduce, but do not entirely eliminate, BIR (McEachern and Haber, 2006; Llorente et al., 2008), suggesting that there are additional BIR mechanisms.
There is no evidence that HR participates in the normal Flp-dependent amplification of 2 μm in wt cells. In vitro, Flp is sufficient to carry out recombination between two FRT sequences (Grainge and Jayaram, 1999). However, experiments using engineered constructs have shown that Flp activity can lead to Rad52-dependent recombination in yeast (Prado et al., 2000; Storici and Bruschi, 2000). Furthermore, Flp's catalytic mechanism suggests how it could form a DSB. Flp and related recombinases initiate their catalytic reaction by forming a single-strand break in the DNA target site, where the active site Tyr of the recombinase is covalently linked to the 3′ phosphate of the broken strand (Grainge and Jayaram, 1999). This feature of recombinase activity is similar to the mechanism of type IB topoisomerases such as eukaryotic topoisomerase I (Top1), which form similar covalent intermediates and cause DSBs when these covalent intermediates are encountered by RFs (Li and Liu, 2001; Pommier, 2006; Koster et al., 2007).
Here we have investigated the mechanism of 2 μm hyperamplification in SUMO pathway mutants and have found that it requires both Flp and HRR. Our results suggest a model in which deficient sumoylation of Flp results in Flp-dependent DNA damage, whose repair via HRR leads to formation of a species that generates multiple tandem copies of the 2 μm DNA sequence.
MATERIALS AND METHODS
Media and Genetic Techniques
Standard techniques were used (Ausubel et al., 2000). Rich yeast medium containing 2% glucose (YPD) and synthetic yeast medium were prepared as previously described (Sherman et al., 1986). Galactose inductions were done by adding 2% galactose to cells pregrown in YP containing 2% raffinose. Cell cycle arrests were done using 10 μM α factor or 15 μg/ml nocodazole.
Plasmid and Yeast Strain Constructions
S. cerevisiae strains used are listed in Supplemental Table S1. Mutations and C-terminal green fluorescent protein (GFP) or epitope tags were introduced into native loci in chromosomal sequences or native 2 μm by transforming strains with appropriate PCR products, as described (Johnson and Blobel, 1999). For modifications of native 2 μm, once a strain containing the correct plasmid was obtained, whole genomic DNA from this strain was transformed into appropriate cir° strains to obtain strains containing only the modified 2 μm plasmid (Chen et al., 2005). The sequence of the hemagglutinin (HA)-His8 tag on Flp and mutant derivatives was GYPYDVPDYAAFLHHHHHHHH. Oligonucleotide sequences are available on request. A CEN-based plasmid expressing untagged Flp from a GAL1 promoter was constructed by inserting the Flp coding region into p415GAL1 (Mumberg et al., 1994).
Analysis of 2 μm Species
Total levels of 2 μm DNA were measured using quantitative PCR (qPCR) as described (Chen et al., 2005), using DNA prepared by glass-bead lysis (Hoffman and Winston, 1987). DNA for Southern blotting was prepared either by a different glass-bead lysis protocol (Adkins and Tyler, 2004) where DNA was treated with 50 μg/ml proteinase K (Figure 3) or by spheroplasting and lysing cells in denaturing conditions as described (Holm et al., 1986; Figures 4, 5, B and C, 6, and 8) with the following modifications: the preparation was treated with RNase at the end of the protocol rather than before the proteinase K step, and the suspension was brought to a concentration of 0.5% SDS before proteinase K treatment. These protocols give somewhat different relative levels of 2 μm DNA between wt and SUMO mutant cells, with glass-bead methods showing a greater apparent difference. This is because the Holm et al. (1986) method produces greater yields of the 2 μm DNA from wt cells (not shown). Chloroquine gels (Figure 3) contained 1.2% agarose and 0.75 μg/ml chloroquine and were run in 1× Tris-borate EDTA (TBE) at 2 V/cm for 24 h at RT (Adkins and Tyler, 2004). Ethidium bromide (EtBr) gels (Figures 4, 5, B and C, 6, A, C and D, and 8) contained 0.6% agarose and 0.3 μg/ml EtBr and were run in 1× TBE at 1–2 V/cm at 4°C for 16–20 h. The gel in Figure 6B contained 1.5% agarose and was run in 1× Tris-acetate EDTA (TAE) at 6 V/cm for 3 h. Lanes in Southern blots were normalized to contain equal amounts of 2 μm DNA, except in Figure 4D, where equal amounts of total DNA were loaded. Individual 2 μm species (Figure 5C) were isolated from EtBr gels made from low-melting point agarose by extracting melted gel slices with phenol and then 25:24:1 phenol/chloroform/isoamyl alcohol, followed by ethanol precipitation. Pulsed-field gel electrophoresis (Figure 5A) was performed using DNA prepared embedded in low-melting point agarose as described (Schwartz and Cantor, 1984). Approximately 20 μl per lane of agarose-embedded DNA was treated with PMSF in Tris-EDTA (TE; pH 8.0), washed with TE supplemented with New England Biolabs (Beverly, MA) buffer 3, melted, and digested to completion with BglII, which does not cleave in 2 μm, to reduce the chromosomal DNA to mostly small (<10 kb) fragments. This preparation was either loaded directly or digested with EagI, which cleaves once in 2 μm. To give partial or complete digests, reactions containing 0.2 or 1.0 U of EagI were incubated at 37°C for 30 min, or reactions containing 20 U were incubated for 2 h. About fivefold more total DNA preparation was loaded in the wt lanes than in siz1Δ siz2Δ lanes (because it was impossible to quantify 2 μm DNA by qPCR in these preparations, which contain unlysed cells and debris). Samples were analyzed by electrophoresis on a 1% agarose gel in 0.5× TBE in a Bio-Rad CHEF-DR II pulsed field electrophoresis system (Hercules, CA) at 200V (6 V/cm) for 19.2 h at 14°C with switch times ramped from 1 to 20 s. Southern transfers were performed as described (Ausubel et al., 2000), and hybridizations (except Figure 6B) were done using a probe containing two PCR products containing base pairs 5953-1270 (including position 1) and 2120-4579 of 2 μm (numbering according to Saccharomyces Genome Database), labeled using the DIG High Prime Labeling Kit (Roche, Indianapolis, IN) according to the manufacturer's instructions. Probes in Figure 6B were prepared by the same method and contained base pairs 1327-1570 of 2 μm (probe A) and base pairs 1712-1946 (probe B). Chemiluminescent signals were quantified using a Bio-Rad Chemidoc gel documentation system. Restriction enzymes were from New England Biolabs.
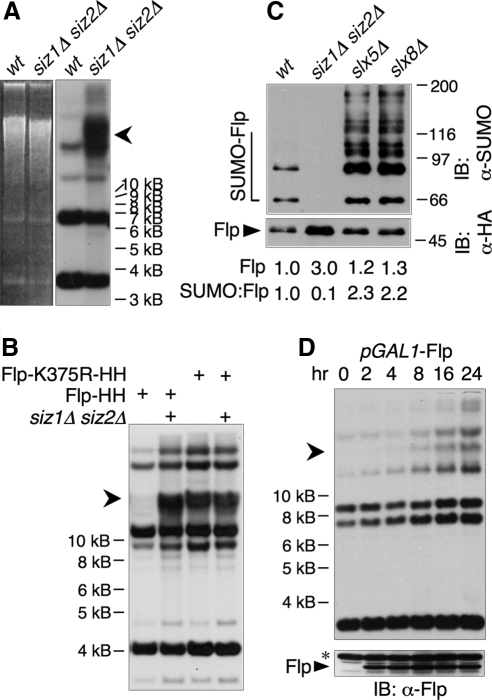
Figure 3.
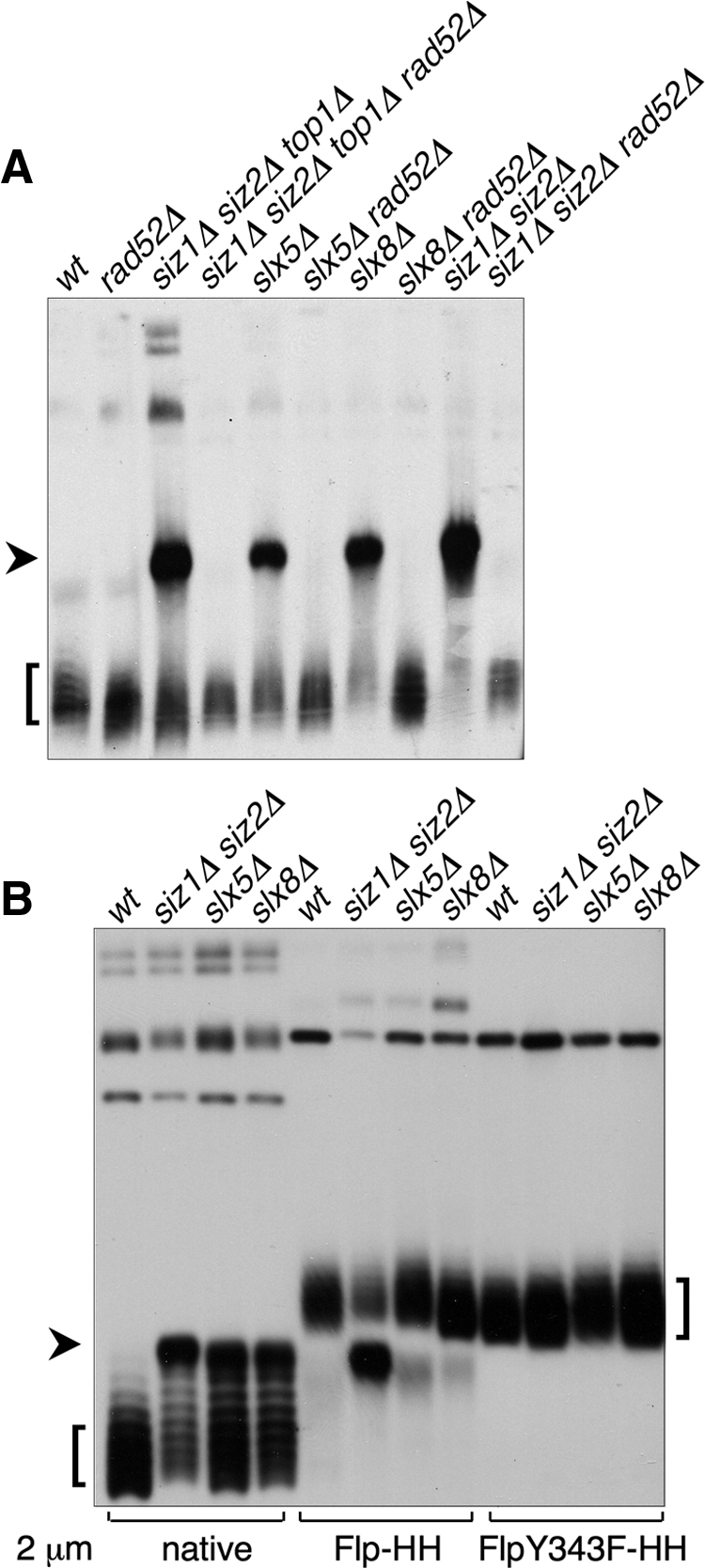
Formation of a HMW 2 μm species in SUMO pathway mutants. (A) Uncut DNA from the indicated strains was analyzed by electrophoresis in an agarose gel containing chloroquine, followed by Southern blotting with a probe against 2 μm. Lanes were normalized to contain equal amounts of 2 μm DNA, as measured by qPCR. Arrowhead indicates the aberrant HMW species. Square bracket indicates supercoiled monomeric 2 μm. (B) DNA from strains of the indicated genotypes containing the indicated versions of 2 μm were analyzed as in A. Both the Flp-Y343F 2 μm variant and the corresponding wt control contained a marker gene, and consequently were larger than native 2 μm. Thus, the supercoiled monomer forms of these plasmids migrated more slowly than native 2 μm, but the HMW form ran at the same position as for native 2 μm.
Figure 4.
Defects in Flp sumoylation lead to formation of the HMW species. (A) Uncut yeast DNA from the indicated strains was analyzed by EtBr-agarose electrophoresis (left) followed by Southern blotting with a probe against 2 μm (right). Lanes contained equal amounts of 2 μm DNA. An arrowhead indicates the HMW form. (B) Uncut DNA from wt or siz1Δ siz2Δ strains containing the indicated versions of 2 μm was analyzed by Southern blotting as in A. (C) Proteins from indicated strains containing 2 μm expressing Flp(Y343F)-HA-His8 (which does not allow 2 μm amplification) were purified by Ni-NTA affinity chromatography and analyzed by SDS-PAGE and immunoblotting with Abs against SUMO (top) and HA (bottom). Arrowhead indicates unmodified Flp, and an open bracket indicates SUMO-modified Flp. Levels of unmodified Flp as well as ratios of total SUMO signal to unmodified Flp are given below the lanes. Quantities are expressed relative to wt. (D) Flp was expressed from the galactose-inducible GAL1 promoter for the indicated times in log phase wt cells containing native 2 μm. A600 was kept ≤2.0. (top) Uncut DNA was analyzed by Southern blotting as in A, except that lanes contained equal amounts of total DNA rather than equal amounts of 2 μm DNA. (bottom) Whole cell lysates from the same samples as in top panel were analyzed by immunoblotting with an Ab against Flp. Asterisk designates a band that cross-reacts with the Ab.
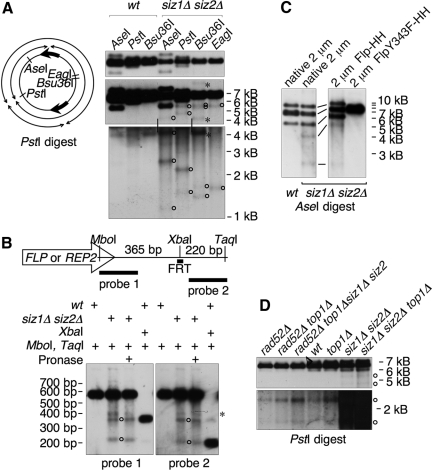
Figure 5.
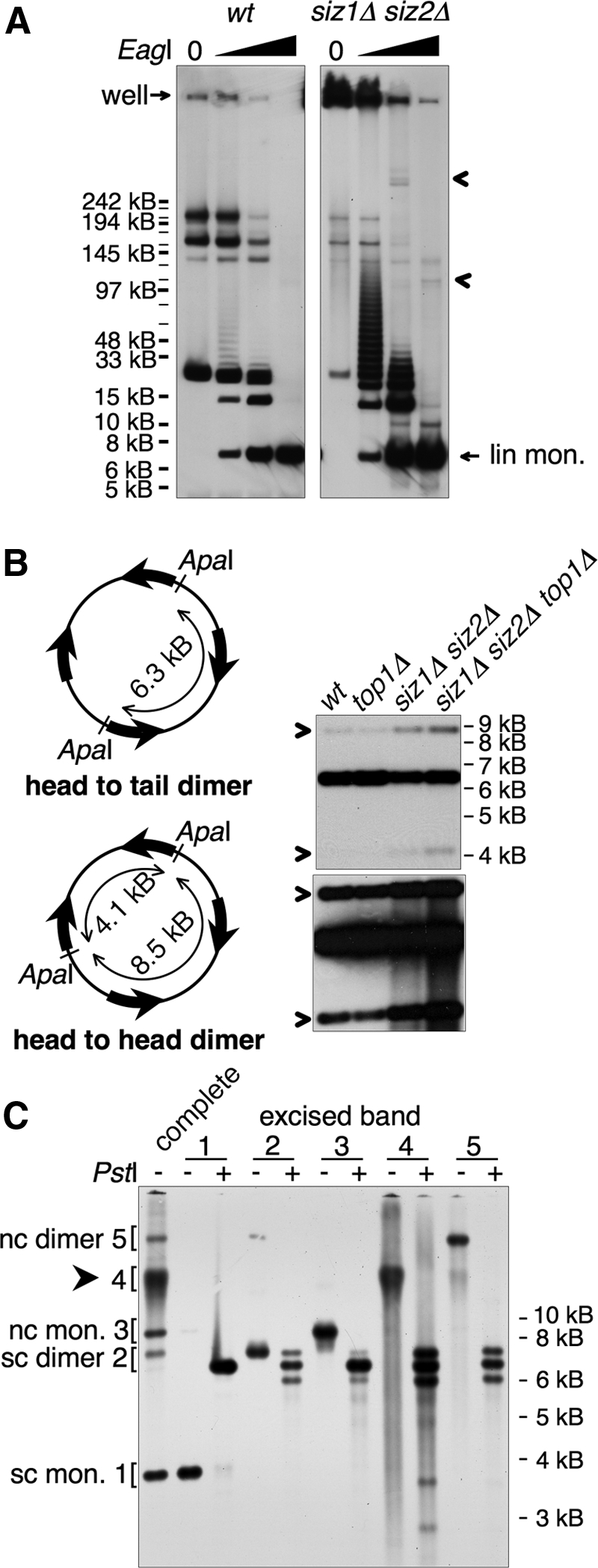
The HMW form contains tandem copies of 2 μm. (A) DNA from indicated strains was left untreated or cleaved with increasing amounts of EagI, as indicated, and analyzed by pulsed-field gel electrophoresis, followed by Southern blotting using a probe against 2 μm. All samples were cleaved to completion with BglII, which does not cleave in 2 μm, to reduce chromosomal DNA to small fragments. wt and siz1Δ siz2Δ panels are from different exposures from the same blot, chosen to normalize the signal from the fully cleaved 2 μm band to the same level, as quantified using the Chemidoc system. The fully cleaved 6.3-kb linear monomer and the position of the well are indicated. Arrowheads designate siz1Δ siz2Δ-specific low-mobility species released by partial restriction digest that may represent nonlinear portions of the HMW species. Markers were NEB MidRange I, which are all indicated, but not all labeled. Major bands in wt uncut sample probably represent the small circular forms of 2 μm, although this has not been shown directly. (B) Left, illustration of head-to-head and head-to-tail dimer linkages and ApaI restriction fragments. Wide arrows denote the 599-base pair internal repeat sequences that contain FRT. Right, DNA from the indicated strains was digested with ApaI and analyzed by Southern blotting with a probe against 2 μm. Bottom, a darker exposure of the top panel. Bands derived from head-to-head dimer linkages are designated with arrows. (C) The five bands indicated on the left were isolated from an EtBr-stained agarose gel and were either analyzed uncut or were digested with PstI, as indicated, followed by Southern analysis as in Figure 4B. Identities of the 2 μm species are indicated. sc, supercoiled; nc, nicked circular; mon., monomer. Arrowhead designates HMW species.
Figure 6.
The HMW form contains apparent DSBs. (A) Left, diagram of 2 μm monomer showing positions of restriction sites and illustrating the four fragments that would be formed by a PstI digest combined with breaks at FRT. Right, yeast DNA from the indicated strains was digested with indicated restriction enzyme and analyzed by Southern blotting with a probe against 2 μm. All panels are different exposures of the same blot. siz1Δ siz2Δ samples were slightly underloaded relative to wt. Vertical lines indicate bands that are the same between the middle and bottom panels. Open circles indicate bands representing apparent DSBs near FRT. Asterisks indicate bands that result from incomplete digests. (B) Top, diagram of 599-base pair internal repeat (IR) sequence, showing positions of restriction sites, FLP/REP2 gene (transcripts from both genes extend into the same side of the IR), FRT, and probes. Flp generates a single-strand break at one of two sites 8 base pairs apart within FRT, and the XbaI site is between these. Bottom, DNA from indicated strains digested with indicated restriction enzymes was analyzed by Southern blotting using the indicated probes (see Materials and Methods). Duplicate samples were run on the same gel and blotted, and the blot was cut in half and analyzed with different probes. Some samples were treated with Pronase in an attempt to test whether part of the Flp protein was still attached to these DNAs, but no effect was seen. Circles designate bands that represent DSBs in the IR. The band marked with an asterisk does not appear to represent a DSB, because no corresponding small band is present. We do not know what this band is. (C) DNA from indicated strains containing indicated versions of 2 μm were analyzed as in A. Only two DSB bands are visible with AseI; the other two predicted bands are not visible because one is the same size as the head-to-head dimer band, whereas the other is small and is obscured by background. Panels are from the same exposure of the same blot. (D) DNA from the indicated strains was analyzed as in A. Panels are from different exposures of the same blot.
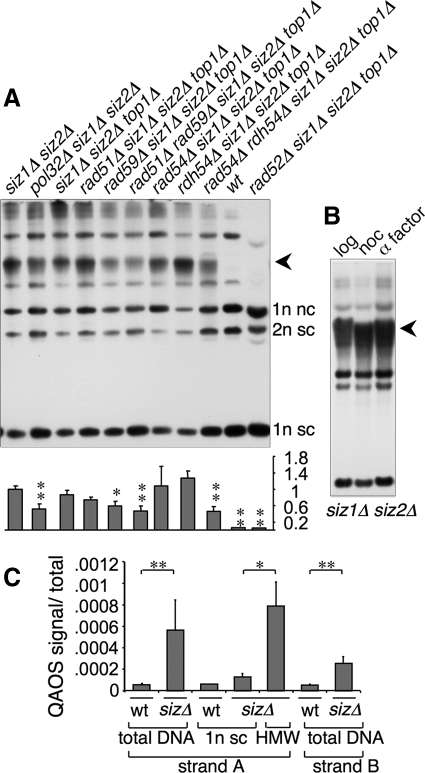
Figure 8.
Genes involved in BIR contribute to formation of the HMW species, which contains ssDNA. (A) Top, uncut DNA from the indicated strains was analyzed by Southern blotting with a probe against 2 μm. Lanes were normalized to contain equal quantities of 2 μm DNA. Arrowhead indicates HMW species. Bottom, graph of the ratio of the HMW species to the sum of the two monomeric species plus the supercoiled dimer species. Ratios are relative to siz1Δ siz2Δ. Three independent experiments were averaged, and SD is given. siz1Δ siz2Δ pol32Δ was compared with siz1Δ siz2Δ; other strains to siz1Δ siz2Δ top1Δ. *p < 0.02; **p < 0.01; calculated using Student's t test. (B) Uncut DNA isolated from siz1Δ siz2Δ cells in log phase (log), after 4-h nocodazole treatment (noc) or after 3-h nocodazole treatment followed by 3-h α factor treatment (α factor) was analyzed as in A. (C) Levels of ssDNA in the indicated DNA preparations was measured using QAOS (see Materials and Methods). Ratio of QAOS signal to total DNA at the same locus is given. “sizΔ” is siz1Δ siz2Δ; “1n sc” is isolated supercoiled 2 μm monomer; “HMW” is isolated DNA from the main chromosomal DNA band, which contains the HMW 2 μm species. Three independent DNA preparations were assayed for each sample, except for the isolated supercoiled monomer from wt cells, where two preparations were used. Error bars, SD, except for wt sc monomer where no error is given. *p < 0.05; **p < 0.01; calculated using Student's t test.
Quantification of Single-stranded DNA in 2 μm by QAOS
The QAOS method (quantitative analysis of single-stranded DNA [ssDNA]) was performed approximately as described (Booth et al., 2001) and is illustrated in Supplemental Figure S2. For each reaction, a DNA sample containing a standard amount of 2 μm DNA (determined by qPCR) was digested with MboI and added to a reaction containing 0.4 pM tagging primer, 200 μM each dNTP, and 0.25 U/10 μl of Taq polymerase in PCR buffer (10 mM Tris, pH 8.85, 25 mM KCl, 5 mM (NH4)2SO4, and 2 mM MgCl2). This was incubated on ice for 5 min, at 25°C for 15 min, and at 37°C for 15 min. Different tagging primers were used to measure ssDNA on each strand (arbitrarily designated strands A and B) at a single locus. Ten microliters of this initial reaction was then included in 30-μl qPCR reactions made using the DyNAmo HS SYBR green qPCR kit (New England Biolabs). Two different qPCR reactions were performed for each initial reaction. One measured QAOS signal and contained the primer specific for the tag plus the appropriate flanking primer. The other measured total DNA at this locus.
Levels of ssDNA are indicated as the QAOS signal/total signal (measured as ΔCt and converted to a fraction). Three independent DNA preparations (made as described for total genomic DNA; Holm et al., 1986) were analyzed for each strain; qPCR reactions were performed in duplicate. We do not believe that our QAOS qPCR reactions measured 100% of the ssDNA in a given sample, or, consequently, that the QAOS/total figure is a precise measurement of the fraction of a given sequence that is single-stranded. The QAOS signal seems likely to be substantially lower than the actual amount of ssDNA. It also appeared that different tagging primers gave somewhat different QAOS signals, and so it would be inappropriate to compare measurements made with different primers. (Thus, we cannot compare the levels of ssDNA on the different strands to each other.) However, this method can still be used to measure relative levels of ssDNA in different samples as long as the same primers are used. The tagging primers were 5′ TGCCCTCGCATCGCTCTCGAATTCCGTTG 3′ at position ∼5690 in 2 μm for strand A and 5′ TGCCCTCGCATCGCTCTCGAAGCAAGT 3′ at position ∼5863 for strand B. The tag primer was 5′ TGCCCTCGCATCGCTCTCGAA 3′ for both strands. The flanking primer for measuring strand A was 5′ AGAAGCAAGTCAGGCTGCCATAGA 3′ at 5872 and for strand B was 5′ ACATCTTCTTCCGTTGGAGCTGGT 3′ at position 5675. The flanking primers were used together to measure total DNA. qPCR was performed in a MJ Research (Waltham, MA) Opticon using cycles of 30 s at 95°C, 45 s at 55°C, and 2 min at 72°C. For analysis of isolated supercoiled and HMW species, bands were isolated from EtBr-stained low-melting agarose gels, and DNA was purified using β-agarase (New England Biolabs) as described by the manufacturer.
Antibodies and Immunoblot Analyses
Yeast whole-cell lysates were prepared by lysis in NaOH (Yaffe and Schatz, 1984). HA and His8-tagged proteins were purified from yeast by Ni-nitrilotriacetic acid (NTA) affinity chromatography (Chen et al., 2005) and subjected to immunoblotting, followed by chemiluminescent detection (Johnson and Blobel, 1999). Antibodies were an affinity-purified rabbit polyclonal antibody (Ab) against Smt3 (SUMO; Johnson and Blobel, 1999), the 16B12 monoclonal Ab against the HA epitope (Covance Research Products, Denver, PA), and a rabbit polyclonal Ab against Flp, generously provided by M. Jayaram (University of Texas, Austin, TX). Photographs of immunoblots were made using chemiluminescent detection (Supersignal; Pierce, Rockford, IL). For quantification of signals, secondary antibodies coupled to fluorescent dyes IRDye 800 (Rockland Immunochemicals, Gilbertsville, PA) and Alexa Fluor 680 (Molecular Probes, Eugene, OR) were used with an Odyssey infrared imaging system (Li-Cor Bioscience, Lincoln, NE).
Microscopy
Fluorescence micrographs were taken using a 63× oil objective on a Leica DM.RXA microscope (Deerfield, IL) with a Cool SNAP fx digital camera (Roper Scientific, Tucson, AZ) and IP Lab software (Scanalytics, Billerica, MA). Photomicrographs of yeast colonies were taken using a tetrad dissecting microscope with a 10× objective and 15× ocular by holding an Olympus Stylus 300 digital camera (Melville, NY) at 3× zoom to the ocular of the microscope.
RESULTS
HRR Is Required for 2 μm Hyperamplification in siz1Δ siz2Δ
We recently reported that a siz1Δ siz2Δ rad52Δ strain is nearly inviable, but that its growth defect is suppressed by deleting the gene encoding topoisomerase I (TOP1; Chen et al., 2007). This phenotype is independent of 2 μm and is observed in strains both containing (cir+) and lacking (cir°) 2 μm. However, this set of mutations also showed a separate effect on 2 μm accumulation. Whereas the siz1Δ siz2Δ cir+ mutant was heterogeneous and contained many arrested cells, the siz1Δ siz2Δ rad52Δ top1Δ cir+ mutant was not only viable but grew uniformly, similar to the cir° version of this strain (Figure 1, A and B), suggesting that 2 μm may not hyperaccumulate in this mutant. To test whether it was the deletion of TOP1 that suppressed 2 μm hyperamplification, we examined a siz1Δ siz2Δ top1Δ cir+ mutant and found that it, like siz1Δ siz2Δ cir+, formed heterogeneous colonies (Figure 1, A and B) and hyperaccumulated 2 μm DNA (Figure 1C). Thus, top1Δ does not affect 2 μm accumulation. This result is important for technical reasons, because elimination of TOP1 suppresses the inviability of siz1Δ siz2Δ mutants lacking genes of the HRR pathway, thereby permitting us to test whether HRR is involved in 2 μm hyperamplification. Deleting RAD52 from siz1Δ siz2Δ top1Δ cir+ eliminated both its colony heterogeneity and its 2 μm DNA accumulation (Figure 1). A similar effect on 2 μm DNA levels was seen comparing siz1Δ siz2Δ cir+ to the barely viable siz1Δ siz2Δ rad52Δ cir+ strain (Figure 1C). This suppression of 2 μm accumulation by rad52Δ was not caused by complete loss of 2 μm in most of the population: 7 of 10 siz1Δ siz2Δ top1Δ rad52Δ single colonies from Figure 1A were cir+, and the DNA used in Figure 1C was prepared from a similar population (not shown). Thus, RAD52-dependent HRR is required for 2 μm hyperamplification in SUMO pathway mutants. Consistent with this conclusion, accumulation of 2 μm in slx5Δ and slx8Δ is also suppressed by deleting RAD52 (Figure 1C; Burgess et al., 2007).
Figure 1.
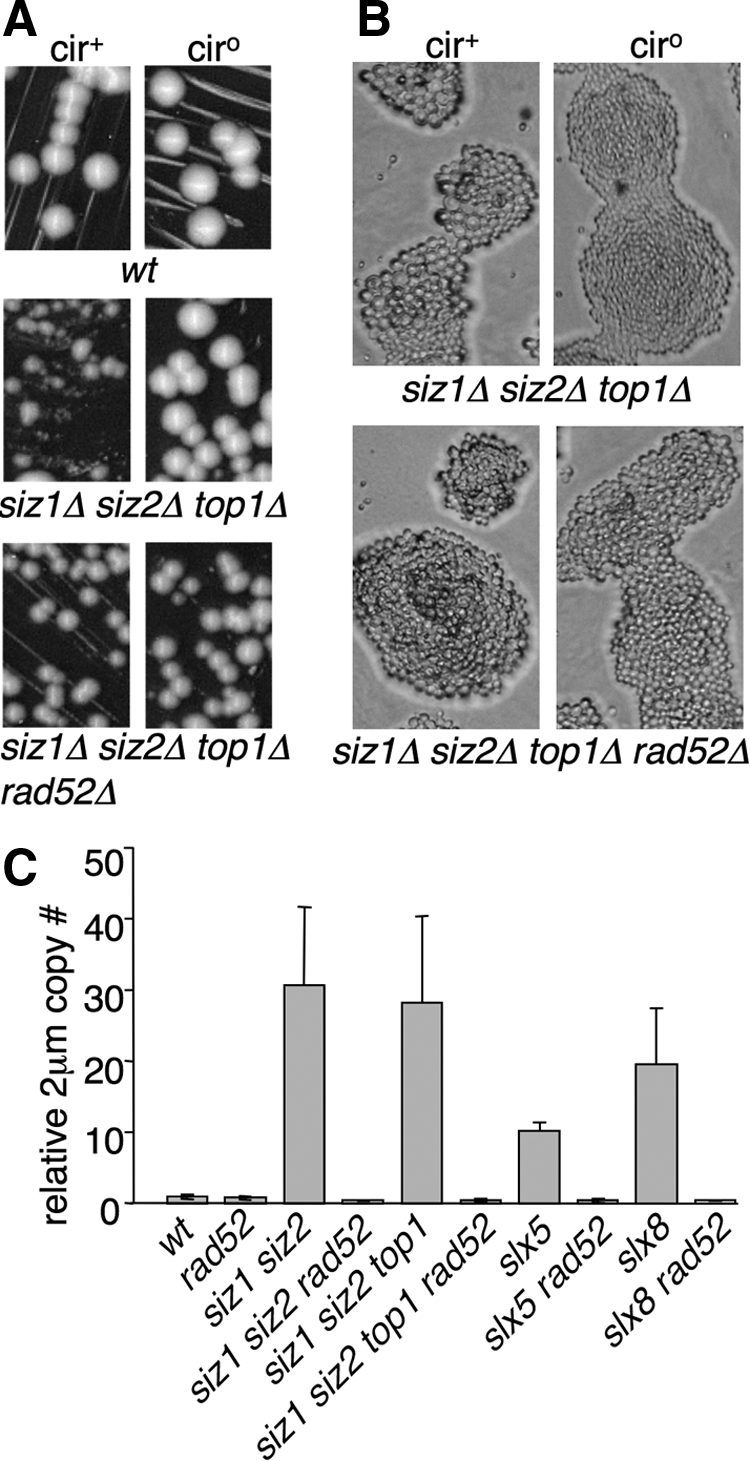
Hyperaccumulation of 2 μm requires homologous recombination. (A and B) Strains of the indicated genotypes were grown at 30°C on YPD plates for 2 d (A) or 1 d (B). Note that differences between cir° and cir+ versions of siz1Δ siz2Δ top1Δ in both colony and cell sizes are not seen in siz1Δ siz2Δ top1Δ rad52Δ. (C) 2 μm DNA levels in the indicated strains were analyzed by qPCR. Quantities are expressed relative to wt. Error bars, SD. Three independent cultures were analyzed for each sample.
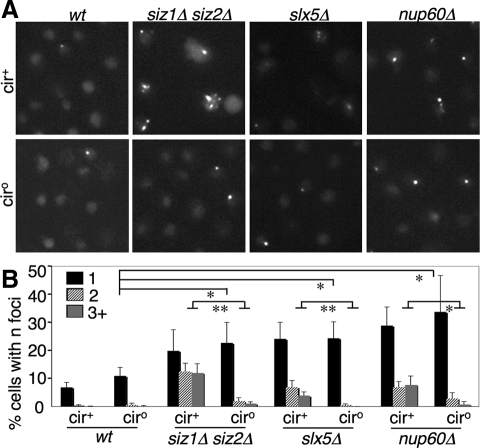
Much of the Increase in Rad52-containing Foci in Sumoylation Mutants Is Related to 2 μm
Other labs have reported that SUMO pathway mutants, including slx5Δ and slx8Δ and the NPC mutants nup60Δ, nup133Δ, and nup84Δ, contain increased levels of Rad52-containing nuclear foci, indicative of elevated ongoing HRR (Burgess et al., 2007; Palancade et al., 2007; Nagai et al., 2008). However, it was not tested to what extent this effect is associated with 2 μm amplification. We counted Rad52-GFP-containing foci in both cir+ and cir° versions of wt, siz1Δ siz2Δ, slx5Δ, and nup60Δ (Figure 2). Mutants containing 2 μm displayed a ∼10-fold increase in the number of cells containing two or more foci, relative to wt. ∼80% of this increase was eliminated by removing the 2 μm from these strains, indicating that most of the increase in cells with multiple foci is due to 2 μm. However, in the absence of 2 μm these mutants still showed a two- to threefold increase in the fraction of cells containing a single Rad52-GFP focus. Thus, although much of the increase in Rad52 foci in SUMO pathway mutants is related to hyperamplification of 2 μm, a significant fraction of the increase involves the chromosomal DNA.
Figure 2.
Part of the increase in Rad52 foci in SUMO pathway mutants is due to 2 μm. (A) Cells of the indicated genotype and containing Rad52-GFP were analyzed by fluorescence microscopy. (B) Rad52-GFP foci were counted in cells from triplicate cultures of indicated genotypes. Percent of cells containing 1, 2, or 3+ foci, with SD, is depicted. Statistical significance is given of comparisons between fractions of various cir° strains having one focus, and fractions of cir° and cir+ versions of the same mutant containing ≥2 foci. *p < 0.05; **p < 0.01; calculated using Student's t test. n = 700-1500 cells per strain.
SUMO Pathway Mutants Accumulate an Aberrant High-Molecular-Weight Form of 2 μm
We next analyzed 2 μm–containing DNA species in SUMO pathway mutants by Southern analysis of uncut DNA. This experiment showed that siz1Δ siz2Δ, siz1Δ siz2Δ top1Δ, slx5Δ, and slx8Δ strains all contained high levels of an aberrant species of 2 μm (Figure 3A). In conventional agarose gel systems, this species always migrated with the uncut chromosomal DNA, suggesting that it is too large to be resolved in these systems (all DNA >∼30 kb migrates at the same rate on these gels; Figure 4A; not shown). Remarkably, this species was not detectable in the corresponding mutant strains lacking RAD52, indicating that its formation requires HRR (Figure 3A). Because 2 μm hyperaccumulation in siz1Δ siz2Δ depends on Flp activity (Chen et al., 2005), we also analyzed DNA from a strain where 2 μm contained the Flp active site mutant Flp-Y343F (Figure 3B). This experiment showed that Flp activity is also required for accumulation of the high-molecular-weight (HMW) species. In all cases, presence of the HMW species correlated with hyperamplification of 2 μm, suggesting that it may be responsible for 2 μm accumulation. However, the cause-effect relationship between formation of the HMW species and hyperamplification cannot be proven directly.
SUMO Attachment to Flp Prevents Accumulation of the HMW Species
Our previous results have suggested that one mechanism by which the SUMO pathway affects 2 μm DNA levels is through sumoylation of Flp (Chen et al., 2005). Next we asked if reducing SUMO attachment to Flp is sufficient to induce formation of the HMW species. Flp contains one major SUMO attachment site, Lys-375, which when mutated eliminates >90% of Flp sumoylation (Chen et al., 2005). Wild-type yeast containing 2 μm expressing Flp-K375R developed high levels of the HMW species (Figure 4B). Thus, this single mutation in the Flp protein is sufficient to induce formation of the HMW species. This result suggests that Flp that cannot be adequately sumoylated carries out the initiating event in generating the HMW species.
We next asked how the SUMO pathway alters Flp function. We have shown previously that reduction of SUMO attachment to Flp, either using the attachment site mutant or siz1Δ siz2Δ, results in accumulation of ∼3–4-fold higher levels of Flp protein (Chen et al., 2005). Here we examined Flp levels in slx5Δ and slx8Δ strains as well. Like siz1Δ siz2Δ, these mutants accumulated somewhat higher levels of Flp protein, but instead of showing reduced sumoylation, these mutants showed dramatically higher levels of SUMO attachment to Flp (Figure 4C). Because these mutants have the same 2 μm–related phenotype as mutants that eliminate Flp sumoylation, this result suggests that Slx5·Slx8 mediates the downstream function of SUMO attachment to Flp (see Discussion).
If a simple increase in Flp levels causes the 2 μm hyperamplification in SUMO pathway mutants, overexpression of Flp in wt cells should cause hyperamplification through a similar mechanism. Induction of Flp expression in log phase cells using a galactose-inducible promoter caused 2 μm DNA levels to increase only slowly, with a gradual increase over 24 h (Figure 4D, top), even though proteins expressed from the GAL1 promoter are induced rapidly, and large amounts of Flp were detected 2 h after induction (Figure 4D, bottom). Amounts of several different 2 μm species increased over this period, including one that comigrated with the chromosomal DNA. However, this form did not accumulate to the levels seen in SUMO pathway mutants, and the other forms increased in similar proportions. While it is difficult to make a direct comparison between this experiment, where Flp levels were increased by orders of magnitude above normal, and SUMO pathway mutants, where Flp levels are increased by a few fold, this result suggests that the effect of SUMO on Flp may be more complex than simply reducing total Flp protein levels.
The 2 μm Sequence Is Present in Tandem Arrays in siz1Δ siz2Δ
We attempted to determine the size of the HMW species using pulsed field gel electrophoresis (see Materials and Methods), but found that much of the 2 μm DNA from siz1Δ siz2Δ cells was trapped in the well (Figure 5A). This trapped DNA probably represents the HMW species. This result most likely suggests that the HMW species contains structures that inhibit migration of large DNAs, such as large circles, branches, or RFs (Hennessy et al., 1991; Christman et al., 1993; Ma et al., 2008). An alternative explanation, that this effect results from large circular structures becoming entrapped in the well by the agarose in which the DNA had been prepared, can be excluded because the HMW species entered the gel when this agarose-embedded sample was analyzed on a standard agarose gel (Supplemental Figure S1A). (This result does not mean that this species does not contain a circular component.) We next performed partial restriction digests on the DNA using an enzyme that cleaves once in the 2 μm sequence (EagI). For siz1Δ siz2Δ DNA, this treatment released large amounts of a ladder of 2 μm species extending to >120 kB (Figure 5A). These forms migrated at the expected positions for linear DNAs containing different numbers of copies of the 2 μm sequence, suggesting that the HMW species contains tandem arrays of 2 μm >20 copies long. The wt sample contained only low levels of the smaller forms. In the siz1Δ siz2Δ samples, restriction digests also released discrete low-mobility 2 μm species (Figure 5A), which may represent portions of the HMW species with nonlinear structures.
When 2 μm DNA isolated from yeast is completely digested with a restriction enzyme that cleaves 2 μm once, three main bands are formed: the 6.3-kb linear monomer, and two forms that are slightly larger and slightly smaller than this. These are formed from “head-to-head” dimeric forms of 2 μm, where the two different Flp-dependent isoforms of 2 μm are linked in tandem (Figure 5B). The size of these forms depends on the position of the restriction site. The 2 μm from siz1Δ siz2Δ strains contained elevated levels of head-to-head linkages (Figure 5B), suggesting that the HMW species may contain head-to-head linkages. We confirmed this by isolating various 2 μm species and analyzing them separately (Figure 5C). Supercoiled and nicked-circular monomers gave rise to only the 6.3-kb band; whereas the HMW form and the supercoiled and nicked-circular dimers all generated a quantity of the 6.3-kb band that was ∼equal to the sum of the higher and lower bands. Thus, these three species contain equal amounts of head-to-head and head-to-tail tandem dimer interactions. This result is consistent with the possibility that the HMW species contains end-to-end copies of the 2 μm sequence.
2 μm in siz1Δ siz2Δ Contains DSBs near FRT
The observation that hyperamplification of 2 μm requires RAD52 suggested that this amplification may occur via repair of DSBs in 2 μm. The restriction analysis of the isolated HMW species also suggested that it may contain DSBs at discrete sites (Figure 5C). Because these breaks in Figure 5C could have been introduced during isolation of this species, we next assayed for DSBs in 2 μm DNA from whole genomic DNA preparations (Figure 6A). This experiment showed that 2 μm DNA from siz1Δ siz2Δ strains, but not from wt, contains low levels of apparent DSBs at discrete sites. Importantly, these DSBs occur close to the FRT target sites for Flp. As illustrated in Figure 6A, DSBs at one or the other FRT site, combined with restriction enzyme cleavage at a single site, should generate four bands. For Bsu36I, AseI, and EagI, some of the bands were not visible, probably because they were obscured by other bands. However, for PstI, all four bands were visible. Calculations based on band sizes estimated that the breaks are within the inverted repeat sequence ∼100 base pairs from FRT, toward the FLP and REP2-containing side of the plasmid. Experiments to map these breaks more precisely suggested that there are actually DSBs at two sites within the inverted repeat sequence, one at FRT and one ∼130 base pairs away toward the FLP side of the plasmid (Figure 6B). The most obvious sequence feature near this second site is a poly-(dAdT) sequence. It is not clear why breaks would form at this second site.
The observation that DSBs occur near the FRT sites predicted that formation of these DSBs may depend on Flp activity. This was the case, because a siz1Δ siz2Δ strain containing 2 μm with inactive Flp did not contain the bands representing DSBs near FRT (Figure 6C). In addition to these discrete bands, the siz1Δ siz2Δ samples also contained a continuous smear of 2 μm signal that was primarily below the major bands but also continued above them (Figures 5 and 6). This suggests that 2 μm in this mutant contains breaks at random sites and possibly also branched structures or features that inhibit cleavage by restriction enzymes.
The above results suggested a model for 2 μm hyperamplification in which SUMO pathway mutants develop DSBs at or near FRT sites, and that repair of these DSBs by HRR leads to formation of the HMW aberrant species. A simple version of this model would predict that the high levels of DSBs should be present in siz1Δ siz2Δ even when RAD52 is absent. However, this was not the case. The siz1Δ siz2Δ top1Δ rad52Δ strain contained much lower levels of DSBs than siz1Δ siz2Δ top1Δ, indicating that accumulation of these apparent DSBs depends on RAD52 (Figure 6D). The critical experiment in testing this hypothesis is the comparison between top1Δ rad52Δ and siz1Δ siz2Δ top1Δ rad52Δ. We also could not detect a clear difference in the amount of these bands between these strains. Thus, these experiments do not support the idea that SUMO mutants contain long-lived Flp-dependent DSBs in the absence of RAD52.
A Possible Role for BIR in Formation of the HMW Species
These experiments instead suggest that the apparent DSBs near FRT are a characteristic of the HMW 2 μm species. This might occur if breaks take place primarily within this species. However, an alternative interpretation of these results is that these bands may not represent DSBs, but rather the free ends of a linear portion in the HMW species. This would give a similar band pattern. The model of 2 μm amplification proposed by Futcher would not involve free DNA ends at discrete sites, but other possible mechanisms would. Collision of a RF (or two converging RFs) with a Flp-DNA covalent intermediate would generate a DSB with Flp covalently bound to the 3′ end on one side (Figure 7). This break might be repaired by BIR: the end of the break with the free 3′ end could invade an intact circular copy of 2 μm, establishing a rolling circle-like structure, where the RF progresses around the circular template plasmid, generating a linear tail containing tandem copies of 2 μm. This model predicts that genes involved in BIR should be required for formation of the HMW species. We found that siz1Δ siz2Δ top1Δ mutants also lacking singly any of RAD50, RAD51, RAD54, RAD55, RAD57, RAD59, RDH54/TID1, or XRS2 all still contained the HMW species, unlike the corresponding rad52Δ mutant, which entirely lacks this species (Figure 8A; Supplemental Figure S1B). Next we quantified the relative level of the HMW species in a subset of these mutants as well as in rad51Δ rad59Δ and rad54Δ rdh54Δ double mutants, which eliminate the two known BIR pathways, and in pol32Δ, which lacks a subunit of DNA polymerase Δ that is required for RAD51-dependent BIR (Lydeard et al., 2007). This experiment showed ∼50% reductions of the HMW species in pol32Δ and in the rad51Δ rad59Δ and rad54Δ rdh54Δ double mutants (Figure 8A). The reduction in the rad59Δ single mutant was also significant. Thus, the genes involved in known BIR pathways are partially responsible for the amplification of 2 μm in siz1Δ siz2Δ mutants (see Discussion).
Figure 7.
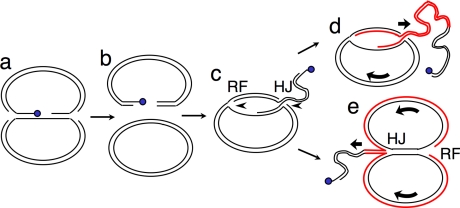
Model for formation of 2 μm rolling circle replication intermediate via BIR. (a) Flp (blue) engaged in a covalent intermediate with DNA at FRT is encountered by one or both replication forks (RFs), resulting in (b) a species containing a DSB where one end has Flp covalently linked to the 3′ end, whereas the other 3′ end is free. (c) The HRR pathway catalyzes invasion of the free 3′ end into an intact circular copy of 2 μm, generating a new RF. Depending on the mechanism by which BIR occurs (there are several models), the RF could be followed by a Holliday junction (HJ) (shown) or only the 3′end/leading strand could invade, forming a D loop (not shown). (d) Progression of the RF, accompanied by branch migration of the HJ (shown) or progression of the D loop followed by separate lagging strand synthesis (not shown) would give rise to rolling circle replication. (e) If the RF progressed more rapidly than the HJ, it would replicate the circular template until RF progression is impeded by the HJ. RF progression might then promote branch migration of the HJ. Red lines indicate DNA synthesized by the BIR-derived RF. Curved arrows indicate direction of rotation of rolling circle.
The model that 2 μm hyperamplification is initiated by a DSB containing covalently bound Flp also suggests that mutants that hyperamplify 2 μm might contain elevated levels of Flp covalently bound to DNA. However, we were unable to detect this covalent intermediate (not shown).
We were also interested in whether the HMW species consists of a continuously replicating species that might contain active RFs outside of normal S phase, or whether it comprises mostly dead-end products that are replicated only during S phase. Analysis of DNA isolated from cells that had been arrested in G2/M by treatment with nocodazole or in G1 with α factor showed that the HMW species persists through the cell cycle (Figure 8B). We next tested for ongoing replication of 2 μm during nocodazole arrest but were unable to detect incorporation of bromodeoxyuridine (BrdU) into 2 μm DNA in nocodazole-arrested cells (Supplemental Figure S1C). These results suggest that at least most of the HMW species is not a continuously replicating rolling circle.
The HMW Species contains ssDNA
One prediction of the BIR model proposed here is that the HMW species might contain ssDNA. This is because the 5′ ends of DSBs or free ends are resected by exonucleases to produce a single-stranded 3′ tail for strand invasion during HRR (Harrison and Haber, 2006). Replication intermediates also contain ssDNA. To quantify ssDNA in 2 μm, we used the PCR-based locus-specific QAOS method (quantitative analysis of ssDNA; Booth et al., 2001; see Materials and Methods; Supplemental Figure S2). This experiment showed that the fraction of the tested site in 2 μm DNA that is single-stranded in siz1Δ siz2Δ is ∼5–10-fold higher than in wt. ssDNA was elevated on both strands (see Materials and Methods). More importantly, the level of ssDNA was about sixfold higher in the HMW species than in siz1Δ siz2Δ-derived supercoiled monomer. Thus, the HMW species contains elevated levels of ssDNA. Because ssDNA activates checkpoint signaling (Harrison and Haber, 2006), this result provides a possible explanation for why SUMO pathway mutants exhibit 2 μm–dependent checkpoint-mediated cell cycle delays.
DISCUSSION
Our data suggest that SUMO attachment to Flp recombinase prevents Flp from causing DNA damage. We propose that when Flp is not sumoylated, Flp-dependent damage is repaired by the cellular HRR pathway, ultimately leading to formation of a HMW species containing multiple tandem copies of the 2 μm sequence. This aberrant process is associated with the elevated levels of 2 μm DNA in SUMO pathway mutants. There is no reason to think that normal Flp-dependent 2 μm amplification in wt cells occurs by the same mechanism. The SUMO pathway could also affect 2 μm behavior via additional mechanisms.
The effect of SUMO on Flp appears to be mediated by Slx5·Slx8. The most likely explanation for this is that Slx5·Slx8 targets sumoylated Flp for ubiquitin-dependent proteolysis. However, we have not been able to detect a difference in the levels of Flp ubiquitylation or degradation between wt and siz1Δ siz2Δ or slx5Δ or slx8Δ mutants, using overexpressed Flp (not shown). One explanation for this would be if Flp can be ubiquitylated and degraded via multiple pathways, where SUMO-dependent degradation accounts for a relatively small fraction, at least when Flp is overexpressed. For example, SUMO- and Slx5·Slx8-dependent ubiquitylation might target only DNA-bound Flp. This model would explain the result that Flp causes DNA damage in SUMO pathway mutants, because increased residence of Flp at the DNA would increase the chance that the Flp-DNA covalent intermediate would be encountered by a RF and converted to a double-strand break. This model would also explain why overexpression of Flp in wt cells did not have the same effect as reducing Flp sumoylation (Figure 4), because overexpressed Flp could still be removed from the DNA by the SUMO pathway.
Several observations suggest that amplification of 2 μm in SUMO pathway mutants may occur via BIR (Figures 5, 6, and 8). However, BIR mutants such as rad51Δ rad59Δ had only an approximately twofold effect on levels of the HMW species, indicating that the two known BIR pathways (RAD51-dependent and RAD51-independent) are not absolutely required for formation of the HMW species. This could suggest 1) that BIR is occurring through a third mechanism, 2) that the requirements for BIR are relaxed when 2 μm is the template, or 3) that the HMW species can form by a mechanism other than BIR. rad51Δ rad59Δ and rad54Δ rdh54Δ eliminate most, but not all, BIR (McEachern and Haber, 2006; Llorente et al., 2008), indicating that there are BIR mechanisms that do not require these pairs of genes. Furthermore, the presence of a circular template might reduce the need for some features of the BIR mechanism. There are several models for how BIR takes place, some of which involve formation of a Holliday junction (HJ) behind the RF (McEachern and Haber, 2006; Llorente et al., 2008). This HJ could either be resolved or could branch-migrate behind the RF. BIR on a circular template would not require either of these mechanisms to deal with the HJ, because the RF could travel around the template until it came up behind the HJ, where it might “push” branch migration of the HJ (Figure 7E). A non-BIR mechanism could also produce the structure in Figure 7E: during plasmid replication, one RF could stall at Flp-bound FRT and regress, forming a “chicken foot” HJ-like structure. If the fork from the other side of the plasmid then passed the FRT site, it might push this regressed fork/HJ backward, extruding a linear tail, where the end represents the site where the fork initially stalled. If the HMW species were formed this way, the end would not be precisely at the Flp cleavage site in FRT, but would be nearby.
Another model that would explain formation of tandem copies of the 2 μm DNA sequence is the double rolling circle model (Futcher, 1986). However, this model does not account for the presence of apparent DSBs near FRT or for the participation of the HRR pathway in 2 μm hyperamplification in SUMO mutants. Both the single and double rolling circle models predict formation of primarily head-to-tail linkages between the tandem copies of 2 μm, if the circular template is a 2 μm monomer. This contrasts with our observation of equal levels of head-to-head and head-to-tail linkages in the HMW species. This discrepancy could be explained either if the template circle is a dimer or larger or if Flp activity randomizes the linkages after formation of the tandem multimer.
We did not detect evidence of ongoing replication of 2 μm in G2/M-arrested cells, suggesting that high levels of active RFs are not present in 2 μm throughout the cell cycle. The rolling circle intermediate proposed here could meet several fates. One is that Flp-dependent recombination between the circle and the tail could redirect the RF, and any HJ, off the end of the tail, generating a linear molecule. There could also be a cellular mechanism that destroys residual RFs late in the cell cycle. The absence of high levels of ongoing replication is consistent with a model where initiation of 2 μm hyperamplification is relatively rare, but that the aberrant species formed is replicated every cell cycle during S phase, so that the species persists in lineages where it develops. This model is consistent with the “nibbled” colony growth of cir+ SUMO pathway mutants, where some cir+ lineages grow well for many generations, indicating that these cells only rarely generate the species that causes growth arrest (Dobson et al., 2005). The species predicted by our model would trigger checkpoint responses even if they are replicated only during S phase, because they would contain free ends without telomeres.
The SUMO pathway has complex effects on genome stability in S. cerevisiae and directly affects diverse aspects of DNA metabolism, including DNA repair (Geiss-Friedlander and Melchior, 2007). However, our work suggests that for 2 μm, the major effect of the SUMO pathway is to prevent Flp-dependent DNA damage, not to regulate the repair mechanism, as has been proposed (Burgess et al., 2007). There is a slight discrepancy between our results, showing that mutants such as rad59Δ have a modest effect on accumulation of the HMW species in siz1Δ siz2Δ top1Δ (Figure 8), and a previous result showing that these mutants strongly suppress the 2 μm–related colony growth defects of the slx8Δ mutant (Burgess et al., 2007). It is likely that this discrepancy is explained by complete loss of 2 μm in the slx8Δ experiment. In our hands, slx5Δ and slx8Δ strains rapidly self-select for loss of 2 μm (not shown). rad59Δ also increased 2 μm loss in siz1Δ siz2Δ top1Δ (not shown). These observations emphasize the importance of using cir° versions of these SUMO pathway mutants for most experiments. Not only do cir+ versions of these mutants show apparent genome instability phenotypes that are actually secondary effects of 2 μm accumulation (Figure 2), but they also display confusing effects resulting from spontaneous loss of the plasmid. Crossing the resulting cir° mutants to cir+ strains accentuates this confusion because it generates mutant segregants that have, at least temporarily, reacquired 2 μm and therefore grow more poorly than the parental cir° strain.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Jayaram for the α-Flp Ab and J. Diffley (Cancer Research UK) for plasmids. We also thank M. Adkins for advice on chloroquine gels and E. Wickstrom, C. Scott, M. King, and E. Alnemri for use of their equipment. H.R.S. was supported by National Research Service Award T32-DK07705. This work was supported in part by MCB-0820228 from the National Science Foundation and in part by a Reapplication Enhancement Award from Thomas Jefferson University.
Abbreviations used:
- 2 μm
yeast 2-μm circle plasmid
- BIR
break-induced replication
- DSB
double-strand break
- EtBr
ethidium bromide
- FRT
Flp recombination target
- GFP
green fluorescent protein
- HJ
Holliday junction
- HMW
high molecular weight
- HRR
homologous recombination repair
- NPC
nuclear pore complex
- QAOS
quantitative analysis of single-stranded DNA
- RF
replication fork
- ssDNA
single-stranded DNA
- wt
wild-type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0659) on December 24, 2008.
REFERENCES
- Adkins M. W., Tyler J. K. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Smith J. A., Seidman J. G., Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 2000. [Google Scholar]
- Booth C., Griffith E., Brady G., Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Volkert F. C. Circular DNA plasmids of yeasts. In: Broach J. R., Jones E. W., Pringle J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 1. Plainview, NY: Cold Spring Harbor Press; 1991. pp. 297–331. [Google Scholar]
- Burgess R. C., Rahman S., Lisby M., Rothstein R., Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. L., Reindle A., Johnson E. S. Misregulation of 2 μm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 2005;25:4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. L., Silver H. R., Xiong L., Belichenko I., Adegite C., Johnson E. S. Topoisomerase I-dependent viability loss in Saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics. 2007;177:17–30. doi: 10.1534/genetics.107.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Levin N. A., Sadoff B. U., Fink G. R. The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1993;90:7637–7641. doi: 10.1073/pnas.90.16.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Pickett A. J., Velmurugan S., Pinder J. B., Barrett L. A., Jayaram M., Chew J. S. The 2 μm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 2005;25:4299–4310. doi: 10.1128/MCB.25.10.4299-4310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher A. B. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Grainge I., Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol. Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- Harrison J. C., Haber J. E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Hay R. T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42:169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Ii T., Fung J., Mullen J. R., Brill S. J. The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle. 2007a;6:2800–2809. doi: 10.4161/cc.6.22.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Mullen J. R., Slagle C. E., Brill S. J. Stimulation of in vitro sumoylation by Slx5-Slx8, evidence for a functional interaction with the SUMO pathway. DNA Repair. 2007b;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M., Mehta S., Uzri D., Voziyanov Y., Velmurugan S. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog. Nucleic Acid. Res. Mol. Biol. 2004;77:127–172. doi: 10.1016/S0079-6603(04)77004-X. [DOI] [PubMed] [Google Scholar]
- Johnson E. S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster D. A., Palle K., Bot E. S., Bjornsti M. A., Dekker N. H. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 2007;178:813–827. doi: 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. K., Liu L. F. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- Llorente B., Smith C. E., Symington L. S. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- Lydeard J. R., Jain S., Yamaguchi M., Haber J. E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Ma W., Resnick M. A., Gordenin D. A. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Haber J. E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;5:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. A., Scarpa M., Rossi N., Cesareni G. Antagonistic controls regulate copy number of the yeast 2 mu plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., Brown G. W., Varela E., Hediger F., Gasser S. M., Krogan N. J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 2007;18:2912–2923. doi: 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Prado F., Gonzalez-Barrera S., Aguilera A. RAD52-dependent and -independent homologous recombination initiated by Flp recombinase at a single FRT site flanked by direct repeats. Mol. Gen. Genet. 2000;263:73–80. doi: 10.1007/pl00008677. [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Murray A. W., Szostak J. W. Roles of the 2 microns gene products in stable maintenance of the 2 microns plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Storici F., Bruschi C. V. Involvement of the inverted repeat of the yeast 2-micron plasmid in Flp site-specific and RAD52-dependent homologous recombination. Mol. Gen. Genet. 2000;263:81–89. doi: 10.1007/pl00008678. [DOI] [PubMed] [Google Scholar]
- Sun H., Leverson J. D., Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K., et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., Hochstrasser M. The yeast Hex3·Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu C. Y., Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.