Abstract
The anaphase-promoting complex (APC) is a ubiquitin ligase that controls progression through mitosis by targeting specific proteins for degradation. It is unclear whether the APC also contributes to the control of cytokinesis, the process that divides the cell after mitosis. We addressed this question in the yeast Saccharomyces cerevisiae by studying the effects of APC mutations on the actomyosin ring, a structure containing actin, myosin, and several other proteins that forms at the division site and is important for cytokinesis. In wild-type cells, actomyosin-ring constituents are removed progressively from the ring during contraction and disassembled completely thereafter. In cells lacking the APC activator Cdh1, the actomyosin ring contracts at a normal rate, but ring constituents are not disassembled normally during or after contraction. After cytokinesis in mutant cells, aggregates of ring proteins remain at the division site and at additional foci in other parts of the cell. A key target of APCCdh1 is the ring component Iqg1, the destruction of which contributes to actomyosin-ring disassembly. Deletion of CDH1 also exacerbates actomyosin-ring disassembly defects in cells with mutations in the myosin light-chain Mlc2, suggesting that Mlc2 and the APC employ independent mechanisms to promote ring disassembly during cytokinesis.
INTRODUCTION
Cytokinesis is the complex process by which the cell divides after completing nuclear division. In animal and fungal cells, this process involves the contraction of an actomyosin ring that contains actin, nonmuscle myosin II, and several other structural and regulatory proteins and forms at the division site shortly before division. Although the actomyosin ring has been intensively studied, there remain many questions about the mechanisms that control its assembly, contraction, and disassembly (Robinson and Spudich, 2000; Glotzer, 2005; Eggert et al., 2006; Vavylonis et al., 2008).
The regulation of cytokinesis and the actomyosin ring have been studied extensively in the budding yeast Saccharomyces cerevisiae (Wolfe and Gould, 2005). Preparations for cytokinesis begin in late G1 with the formation of a septin ring at the presumptive bud site (Versele and Thorner, 2005; Iwase et al., 2006). The yeast septins have two functions in cytokinesis: they recruit other cytokinesis proteins to the mother-bud neck beginning in late G1 (Gladfelter et al., 2001), and they act as a diffusion barrier that compartmentalizes cytokinesis factors (Dobbelaere and Barral, 2004). Among the first proteins recruited is Myo1, the sole type-II myosin in S. cerevisiae, which forms a ring at the presumptive bud site shortly before bud emergence (Bi et al., 1998; Lippincott and Li, 1998; Luo et al., 2004). The myosin light chains Mlc1 and Mlc2 also associate with the Myo1 ring by the beginning of mitosis (Luo et al., 2004). The final preparations for cytokinesis occur after the chromosomes have segregated in anaphase, when Iqg1, a member of the IQGAP protein family, is localized to the bud neck. Iqg1 is required for actin-filament formation at the neck, a late (and possibly final) step in assembly of the actomyosin ring (Epp and Chant, 1997; Lippincott and Li, 1998; Shannon and Li, 1999; Osman et al., 2002), as well as for other aspects of cytokinesis unrelated to actomyosin-ring function (Korinek et al., 2000; Nishihama and Pringle, unpublished data).
For successful cell reproduction, cytokinesis must occur after the completion of chromosome segregation in anaphase. A key regulator of anaphase events is the anaphase-promoting complex or cyclosome (APC), an E3 ubiquitin ligase that, together with an E1 (ubiquitin-activating enzyme), an E2 (ubiquitin-conjugating enzyme), and an activator (Cdc20 or Cdh1 in S. cerevisiae), assembles ubiquitin chains on its substrates, thereby triggering their degradation by the 26S proteasome (Peters, 2006; Thornton and Toczyski, 2006; Rodrigo-Brenni and Morgan, 2007). The APC has two major substrates: securin, the destruction of which triggers the metaphase-to-anaphase transition, and the mitotic cyclins, activators of the cyclin-dependent kinase Cdk1 (Cdc28 in S. cerevisiae). APC-mediated cyclin degradation lowers Cdk1 activity and thereby promotes the completion of mitosis and entry into G1.
APC activity is low from S phase to early mitosis and higher in late mitosis and G1 (Peters, 2006; Thornton and Toczyski, 2006; Sullivan and Morgan, 2007). The activator subunit Cdc20 associates with the APC to initiate the metaphase-to-anaphase transition, after which the related activator Cdh1 maintains APC activity in late mitosis and G1. Although securin and the mitotic cyclins are the only substrates whose destruction by the APC is required for cell division (Thornton and Toczyski, 2003), the timely destruction of other substrates is thought to increase the efficiency and robustness of cell-cycle progression. Because Cdh1 activates the APC just before cytokinesis, it is conceivable that degradation of specific APCCdh1 substrates contributes in some way to the control of cytokinesis.
Three APCCdh1 substrates are known to be involved in the control of cytokinesis: polo-related kinases (Cdc5 in S. cerevisiae), vertebrate anillin, and yeast Iqg1 (Charles et al., 1998; Song and Lee, 2001; Echard et al., 2004; Straight et al., 2005; Zhao and Fang, 2005; Yoshida et al., 2006; Ko et al., 2007). These proteins are all required for efficient progression through cytokinesis; thus, their APCCdh1-mediated destruction is not expected to promote cytokinesis but might instead be predicted to help inactivate the cytokinesis machinery as cell division is completed.
To identify potential APC functions in cytokinesis, we used light and electron microscopy to visualize the cytokinesis machinery and cytokinesis structures in apc mutant cells. We found that the actomyosin ring contracts in these mutants but does not disassemble properly, at least in part due to a failure to degrade Iqg1. The APC-mediated degradation of Iqg1 appears to function in parallel with at least one other regulatory pathway to promote the efficient completion of cytokinesis.
MATERIALS AND METHODS
Strains, Plasmids, Growth Conditions, and Genetic Methods
Yeast strains are listed in Table 1 and are in the W303 or S288C genetic background, as indicated. Yeast were grown at 30°C unless otherwise indicated. Glucose, 2%, was used as carbon source except for experiments involving induction of gene expression under GAL promoter control, for which 2% raffinose plus 2% galactose was used. Standard procedures were used for growth of Escherichia coli, genetic manipulations, PCR, and other molecular biological procedures (Guthrie and Fink, 1991).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| AFS34a | a ade2-1 can1-100 ura3-1 leu2-3,112 his3-11,15, trp1-1 | Straight et al. (1996) |
| AFS35a | α ade2-1 can1-100 ura3-1 leu2-3,112 his3-11,15, trp1-1 | A. Straight |
| AFS92 | As AFS34 except bar1Δ | Jaspersen et al. (1998) |
| BY4741b | a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al. (1998) |
| BY4742b | α his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al. (1998) |
| YEF473Ab | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Bi and Pringle (1996) |
| YEF473Bb | α his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Bi and Pringle (1996) |
| YEF1681 | as YEF473A except MYO1-GFP:kanMX6 | Bi et al. (1998) |
| yBRT135-1a | As AFS92 except pds1Δ::LEU2 clb5Δ::HIS3 trp1Δ::SIC1-10X:TRP1 | Thornton and Toczyski (2003) |
| yTMN19 | As AFS35 except apc2Δ::CAN1 pds1Δ::LEU2 clb5Δ::HIS3 trp1Δ::SIC1-10X:TRP1 | Thornton and Toczyski (2003) |
| GT049 | As AFS92 except MYO1-EGFP:His3MX6 | This study |
| GT050 | As AFS92 except IQG1-EGFP:His3MX6 | This study |
| GT052 | As AFS92 except MYO1-EGFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT053 | As AFS92 except IQG1-EGFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT073 | As yTMN19 except MYO1-GFP:URA3 | This study |
| GT074 | As yBRT135-1a except MYO1-GFP:URA3 | This study |
| GT079 | As AFS92 except MYO1-EGFP:His3MX6 cdh1Δ::LEU2 ase1Δ::URA3 | This study |
| GT081 | As AFS92 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 [GAL-SIC1, URA3] | This study |
| GT104 | As AFS92 except MLC2-GFP:His3MX6 | This study |
| GT108 | As AFS92 except MYO1-GFP:His3MX6 [pCDC5-CDC5Δ5-70-myc9, URA3] | This study |
| GT110 | As AFS92 except MLC2-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT124 | As BY4741 except IQG1Δ42-TAP:His3MX6:LEU2 | Ko et al. (2007) |
| GT132 | As AFS92 except MYO1-Cherry:kanMX6 MLC2-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT133 | As BY4741 except MYO1-Cherry:kanMX6 IQG1-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT134 | As BY4741 except MYO1-Cherry:kanMX6 SEC2-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT158 | As YEF473A except MYO1-GFP:kanMX6 mlc2Δ::His3MX6 | This study |
| GT160 | As YEF473A except MYO1-GFP:kanMX6 iqg1Δ::His3MX6 | This study |
| GT165 | As BY4741 except cdh1Δ::LEU2 | This study |
| GT166 | As BY4742 except cdh1Δ::LEU2 | This study |
| GT178c | MYO1-GFP:kanMX6 iqg1Δ::His3MX6 cdh1Δ::LEU2 | GT160 × GT166 |
| GT200 | As BY4742 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| GT203c | MYO1-GFP:kanMX6 mlc2Δ::His3MX6 cdh1Δ::LEU2 | GT158 ×GT165 |
| GT204 | As BY4741 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 spo12Δ::kanMX6 | This study |
| GT208 | As AFS92 except [MET25p-yEGFP3-MLC1, HIS3, CEN/ARSH4] | This study |
| GT209 | As AFS92 except [MET25p-yEGFP3-MLC1, HIS3, CEN/ARSH4] cdh1Δ::LEU2 | This study |
| GT225 | As AFS92 except MYO1-CHERRY:kanMX6 cdh1Δ::LEU2 MET25p-yEGFP3-MLC1, HIS3, CEN6/ARSH4 | This study |
| GT229 | As BY4741 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 clb2Δ::kanMX6 | This study |
| GT235 | As AFS92 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 hsl1Δ::TRP1 swe1Δ::LEU2 | This study |
| GT262 | As BY4741 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 fin1Δ::kanMX6 | This study |
| KO255 | As YEF473A except cdh1Δ::TRP1 | Ko et al. (2007) |
| Myo1-GFP | As BY4741 except MYO1-GFP:His3MX6 | Huh et al. (2003) |
| RNY2349 | As YEF473A except MYO1-GFP:kanMX6 cdh1Δ::TRP1 | This study |
| RNY2356 | As BY4741 except MYO1-GFP:His3MX6 mlc2Δ::kanMX6 | This study |
| RNY2369 | As BY4741 except MYO1-GFP:His3MX6 cdh1Δ::LEU2 | This study |
| RNY2371 | As BY4741 except MYO1-GFP:His3MX6 mlc2Δ::kanMX6 cdh1Δ::LEU2 | This study |
| RNY2373 | As BY4741 except MYO1-GFP:His3MX6 IQG1Δ42-TAP:His3MX6:LEU2 | This study |
| RNY2375 | As BY4741 except MYO1-GFP:His3MX6 mlc2Δ::kanMX6 IQG1Δ42-TAP:His3MX6:LEU2 | This study |
a W303 strain background.
b S288C strain background. Two different S288C strain families were used, derived from strains BY4741/BY4742 or YEF473.
c These strains have a hybrid S288C background resulting from crossing strains of the BY4741/BY4742 and YEF473 backgrounds, as indicated in the Source column.
The following strains and plasmids were kindly provided by other laboratories: strains yBRT135-1a and yTMN19 by David Toczyski (University of California, San Francisco, San Francisco, CA); an hsl1Δ swe1Δ cdh1Δ strain by Mark Solomon (Yale University, New Haven, CT; Burton and Solomon, 2001); a TUS1-GFP strain by David Pellman (Dana Farber Cancer Institute, Boston, MA); a pGFP-MLC1 plasmid by Antonella Ragnini-Wilson (University of Rome, Rome, Italy; Wagner et al., 2002); and mCherry plasmids by Peter Walter (University of California, San Francisco, San Francisco, CA; Shaner et al., 2004). The construction of plasmid YCp111-CDC3-green fluorescent protein (GFP) is described elsewhere (Nishihama and Pringle, unpublished data).
Cell-clustering Assay
To determine cell-cluster indices, cells from an exponential-phase culture were washed with water, sonicated briefly, and observed by differential interference contrast (DIC) microscopy. For each strain, cells were categorized as 1) single or double cell bodies or 2) clusters of three or more cell bodies. The cluster index is the percentage of 400 scored entities that were in category 2.
Light Microscopy of Fixed Cells
Visualization of F-actin, Myo1-GFP, and DNA in the same cells was performed as described (Bi et al., 1998). Exponential-phase cells were pelleted by centrifugation for 30 s and then fixed by resuspending the pellet in ice-cold 70% ethanol and incubating on ice for 10 min. Cells were then incubated at room temperature for 2 min with 20 U/ml (0.66 μM) TRITC-phalloidin (Invitrogen, Carlsbad, CA) in PBS containing 1 mg/ml BSA (Sigma, St. Louis, MO), washed three times with PBS, and resuspended in mounting medium containing 1 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Invitrogen); then 2–3 μl of this suspension was placed on a slide, covered with a coverslip, and then pressed with a weight for 10 min before microscopic examination.
Fluorescence and DIC microscopy were performed using a Zeiss Axiovert 200M microscope equipped with a 63x NA 1.4 oil immersion DIC objective (Plan-Apochromat, Zeiss, Thornwood, NY), an X-cite 120 mercury arc lamp (EXFO, Electro-Optical Engineering, Plano, TX), and an Orca ER camera (Hamamatsu Photonics, Bridgewater, NJ). MetaMorph (Molecular Devices, Sunnyvale, CA) was used for data collection. Seven images in the GFP (750 ms) and Texas Red (50 ms) filters, 1 × 1 binning, were acquired at 0.5-μm intervals along the Z axis and then projected into one image by maximum intensity in Metamorph. The corresponding images were paired with the DAPI and DIC images. Contrast was enhanced using ImageJ (http://rsb.info.nih.gov/ij/) and Photoshop (Adobe Systems, San Jose, CA).
Live-Cell Microscopy
Most live-cell microscopy was performed with the microscope and imaging software described above. For time-lapse studies, exponential-phase cells were grown at room temperature in synthetic complete (SC) medium (supplemented with 0.01% Ade and 0.01% Trp) to minimize background fluorescence. For the Iqg1-GFP experiments in Figure 3, C and D, the cells were first arrested in nocodazole (Sigma; 10 μg/ml) for 3 h at room temperature, washed three times with fresh medium, and then grown for 1 h before beginning time-lapse observations. For microscopy, cells were adhered with concanavalin A (Sigma) to 35-mm glass-bottom Petri dishes (MatTek, Ashland, MA), as follows. 300 μl of 50 μg/ml concanavalin A in PBS, pH 7.4, was incubated on dishes for 10 min. Dishes were washed three times with PBS, pH 7.4, and dried, and 300 μl of culture was added for ≥30 min at room temperature, washed three times with SC medium, and observed at room temperature. Movies lasted 30 min, and images were taken at 1-min intervals (unless otherwise indicated) at multiple stage positions (n = 3–5). For GFP and mCherry, image acquisition ranged from 300 to 750 ms depending on fluorescence intensity with 1 × 1 binning. Maximum projections of the fluorescence images were generated by acquiring seven images at 0.5-μm intervals for each stage position. Power levels of the mercury arc lamp were lowered to minimize phototoxicity.
Figure 3.
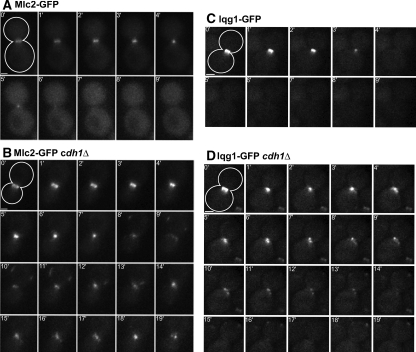
Mlc2 and Iqg1 ring-disassembly defects in APC mutant cells. Strains GT104 (MLC2-GFP; A), GT110 (MLC2-GFP cdh1Δ; B), GT050 (IQG1-GFP; C), and GT053 (IQG1-GFP cdh1Δ; D) were examined by time-lapse microscopy; time point 0 marks the initiation of ring contraction. The cell outlines are drawn in the 0-min panels. Bars, 1 μm.
For the experiments in Figures 5 and 7C, microscopy was performed using a Nikon Eclipse E600-FN microscope with an Apo 100×/1.40 NA oil-immersion objective (Melville, NY), an ORCA-2 cooled-CCD camera and MetaMorph software. Cells were grown to exponential phase at 24°C in appropriate media (Figures 5, A, SC, and B, SC-Leu, and 7C, SC supplemented with 0.01% Ade and 0.01% Trp) and concentrated by centrifugation just before beginning observations. For the time-lapse experiments in Figure 5, cells in liquid medium were pressed gently between slide and coverslip to remove excess medium.
Figure 5.
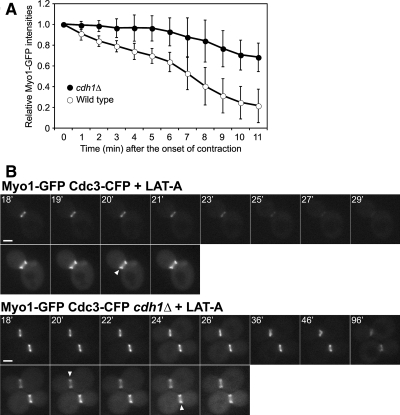
APCCdh1-dependent disassembly of the actomyosin ring during contraction (A) or in its absence (B). Strains YEF1681 (MYO1-GFP) and RNY2349 (MYO1-GFP cdh1Δ) were used. (A) Cells were examined by 4D time-lapse microscopy (see Materials and Methods). The intensities of Myo1-GFP, relative to the intensity measured at the beginning of ring contraction, are plotted for the indicated times (mean ± SD; n = 7 for each strain). All Myo1 rings had completed contraction by the final time point. No corrections were made for possible photobleaching during the course of the experiment; thus, the removal of Myo1 from the ring may have been even more defective in cdh1Δ cells than these data suggest. (B) Transformants containing plasmid YCp111-CDC3-CFP were treated with 200 μM LAT-A at time zero and examined by time-lapse microscopy at 1-min intervals. Top panels, Myo1-GFP; bottom panels, Cdc3-CFP. Arrowheads show the splitting of the septin hourglass structure into two rings, indicating the onset of cytokinesis. Representative images are shown. Bars, 2 μm.
Figure 7.
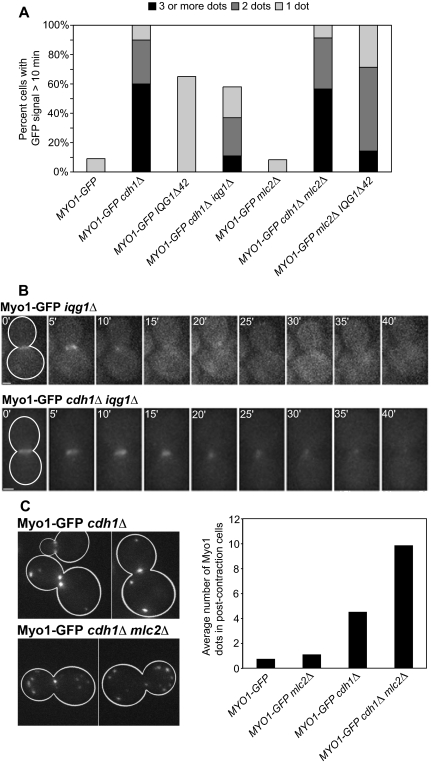
Contributions of Iqg1 destruction and Mlc2 to disassembly of the actomyosin ring. (A) The following strains expressing Myo1-GFP were examined by time-lapse microscopy: Myo1-GFP (wild type), RNY2369 (cdh1Δ), RNY2373 (IQGΔ42), GT178 (cdh1Δ iqg1Δ), RNY2356 (mlc2Δ), RNY2371 (cdh1Δ mlc2Δ), and RNY2375 (mlc2Δ IQG1Δ42). For each strain, 10–61 cells were analyzed; for each cell, the maximum number of Myo1-GFP dots observed at any time >10 min after the completion of ring contraction was recorded. (B) Strains GT160 (MYO1-GFP iqg1Δ) and GT178 (MYO1-GFP iqg1Δ cdh1Δ) were examined by time-lapse microscopy, acquiring images at 5-min intervals to decrease photobleaching of the weak Myo1-GFP signals. Presumably because actin was not recruited to the rings in these iqg1Δ strains, normal ring contraction did not occur. Instead, the Myo1 signal gradually disappeared. Bars, 1 μm. (C) Exponential-phase cultures of strains Myo1-GFP, RNY2356, RNY2369, and RNY2371 (see A) were examined, and the number of Myo1-GFP dots per cell was counted in each cell (17–28 cells per strain) that had completed ring contraction but not cell separation. Representative images of strains RNY2369 and RNY2371 are shown at left.
To quantitate the fluorescence intensities derived from Myo1-GFP rings in Figure 5A, a Z-series of 11 images at 0.3-μm steps was captured at each time point (1-min intervals), from which maximum-projection images were created using MetaMorph. A region box just large enough to enclose the ring was drawn, and the box was also duplicated and placed in a nearby background position. The integrated intensity of the ring box at each time point was measured and recorded using the regional-measurement function of MetaMorph and that of the background box was subtracted. The intensity value for each time point was divided by that at the beginning of contraction to determine relative intensity.
For the experiments in Figure 5B, cells from 100- to 500-μl cultures were resuspended in 10 μl of SC-Leu medium containing 200 μM latrunculin A (LAT-A) or an equal volume of DMSO as a control and examined by time-lapse microscopy at 1-min intervals. Image recording was started between 5 and 10 min after treatment. The Myo1-GFP and Cdc3-CFP images were captured in a single Z plane (cell center) with yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) filter sets, respectively.
For the experiment in Figure 7C, a Z-series of seven images at 0.5-μm steps was captured for GFP fluorescence. Maximum-projection images were created as above and used for counting Myo1-GFP dots at the same intensity scale (dynamic range) for all images.
Image Analysis and Quantification
Ring Contraction.
All quantification was performed manually using ImageJ. Briefly, for each projected stack of images, every cell that showed ring contraction was counted. Only the cells where the start and end of ring contraction were clearly visible were included to calculate the average time of ring contraction.
Ring Disassembly.
For disassembly, only cells in which ring contraction ended with at least 10 min remaining in the movie were included. If a GFP dot was visible ≥10 min after contraction had ended, then it was scored as “GFP visible >10 min.”
Myo1 Patches per Cell.
We counted the maximum number of GFP patches visible at any time ≥10 min after ring contraction had ended. Cells in which no GFP dots were visible ≥10 min after the end of ring contraction were scored as zero.
GFP-mCherry Colocalization.
The colocalization of each GFP and mCherry patch was monitored manually at each 5-min interval. Only cells in which GFP-mCherry colocalization was constant (from the end of ring contraction until the end of the time-lapse) were counted as positives.
Electron Microscopy
Cells growing exponentially in SC medium were examined by transmission electron microscopy after fixation with glutaraldehyde and potassium permanganate, embedding in LR White resin, and staining with uranyl acetate and lead citrate, as described in detail elsewhere (Nishihama and Pringle, unpublished data). Micrographs were obtained using a JEOL (Tokyo, Japan) JEM1230 electron microscope and a Gatan (Pleasanton, CA) model 967 cooled CCD camera and were processed using DigitalMicrograph software (Gatan) and Photoshop (Adobe Systems, San Jose, CA).
RESULTS
APC-mutant Cells Are Defective in Disassembly of the Actomyosin Ring
To determine if APC activity helps control cytokinesis, we examined cells defective in APC function. Cells with cytokinesis defects form chains of cell bodies that remain connected even after brief sonication (Ko et al., 2007). About 3% of exponentially growing wild-type cells were present in clusters of ≥3 cell bodies, whereas in a cdh1Δ strain, the number of clustered cells rose threefold to 9% (Supplemental Figure S1). Thus, APCCdh1 activity appears to be required for efficient cytokinesis.
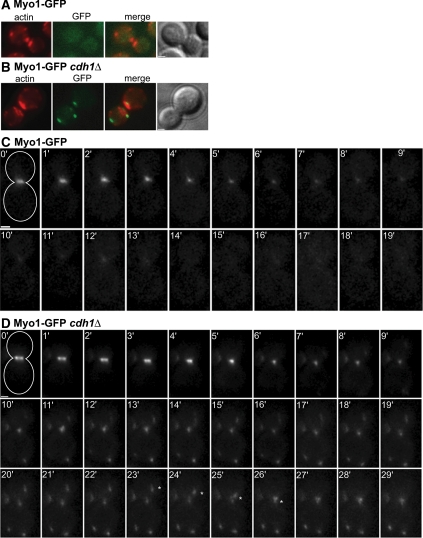
To further explore APCCdh1 function in cytokinesis, we analyzed the localization of GFP-tagged cytokinesis proteins in fixed cdh1Δ cells. We found that the localizations of many proteins (Cdc12, Hof1, Bni1, Bnr1, Cyk3, Mob1, Myo2, and Tus1) were not detectably affected by deletion of CDH1 (data not shown). However, the behavior of the type II myosin Myo1 was strikingly altered. In a wild-type strain, Myo1-GFP was not detectable in cells that had recently completed cytokinesis, as marked by the absence of a Myo1 ring and the presence of large actin patches at the bud neck (Bi et al., 1998; Figure 1A). However, in cdh1Δ cells at the same stage, multiple Myo1-GFP patches were typically present at the neck and elsewhere in the cell and did not colocalize with actin patches (Figure 1B).
Figure 1.
Cdh1 is required for normal disassembly of the actomyosin ring. Myo1-GFP was visualized in wild-type (A and C; strain GT049) and cdh1Δ (B and D; strain GT052) cells. (A and B) Cells were fixed with ethanol and stained with TRITC-phalloidin (red) to visualize actin filaments. The presence of large actin patches at the bud neck indicates recent completion of cytokinesis. (C and D) Time-lapse images were captured at 1-min intervals; time point 0 marks the initiation of ring contraction. The cell outlines are drawn in the 0-min panels. A Myo1-GFP dot moving toward the bud neck in the cdh1Δ cell is marked with an asterisk in panels 23 min–26 min. Bars, 1 μm.
We next analyzed the localization dynamics of Myo1-GFP by time-lapse microscopy. In wild-type cells, the Myo1 ring appeared at the time of bud emergence and remained at the bud neck until cytokinesis, when it contracted to a single dot in an average time of 5.9 ± 0.9 min (Figure 1C; Table 2). Within 10 min after the end of contraction, the Myo1 dot completely disappeared, suggesting that the actomyosin ring had fully disassembled. In cdh1Δ cells, the Myo1 ring also appeared at bud emergence and later contracted to a single dot with similar kinetics (5.8 ± 1.3 min; Figure 1D; Table 2). However, the Myo1 dot did not disappear after the completion of ring contraction. Instead, Myo1-GFP patches remained visible at the site of cytokinesis and elsewhere in the cell (Figure 1D). More extended time-lapse observations showed that these patches could persist throughout G1 and only disappeared as the new Myo1 ring formed in the next cell cycle (Supplemental Figure S2). Many of the persistent Myo1 patches were highly mobile. We tracked their movements and saw that in ∼63% of the cells, a Myo1 patch traveled toward the bud neck and appeared to merge with another patch at that site (Figure 1D, asterisks). In most cases, Myo1 patch movement toward the neck occurred exclusively in the daughter cell. Patch movement was not seen in cells treated with the actin-depolymerizing agent LAT-A, suggesting that these movements depend on F-actin (data not shown).
Table 2.
Ring contraction and disassembly in selected strains
| Strain | Genotype | Ring contraction (min)a | No. of cells | GFP signal >10 min (%) | No. of cells |
|---|---|---|---|---|---|
| GT049 | MYO1-GFP | 5.9 ± 0.9 | 27 | 8 | 25 |
| GT052 | MYO1-GFP cdh1Δ | 5.8 ± 1.3 | 17 | 100 | 21 |
| GT104 | MLC2-GFP | 5.1 ± 1.2 | 14 | 12 | 17 |
| GT110 | MLC2-GFP cdh1Δ | 6.9 ± 1.7 | 15 | 100 | 11 |
| GT208 | GFP-MLC1 | 5.7 ± 1.1 | 24 | 100 | 22 |
| GT209 | GFP-MLC1 cdh1Δ | 6.2 ± 1.6 | 14 | 100 | 19 |
| GT050 | IQG1-GFP | 4.6 ± 1.0 | 7 | 9 | 11 |
| GT053 | IQG1-GFP cdh1Δ | 5.3 ± 1.5 | 8 | 100 | 9 |
| GT229 | MYO1-GFP clb2Δ cdh1Δ | 9.1 ± 1.5 | 16 | 100 | 12 |
| GT235 | MYO1-GFP hsl1Δ swe1Δ cdh1Δ | 7.6 ± 1.7 | 10 | 100 | 11 |
| GT079 | MYO1-GFP ase1Δ cdh1Δ | 6.3 ± 1.0 | 13 | 100 | 18 |
| GT262 | MYO1-GFP fin1Δ cdh1Δ | 8.7 ± 1.5 | 7 | 100 | 9 |
| GT204 | MYO1-GFP spo12Δ cdh1Δ | 5.9 ± 1.0 | 12 | 100 | 15 |
| GT108 | MYO1-GFP CDC5Δ5-70 | 6.1 ± 1.5 | 15 | 6 | 17 |
| GT081 | MYO1-GFP GAL-SIC1 cdh1Δ | 5.1 ± 1.2 | 18 | 95 | 19 |
a Values are mean ± SD.
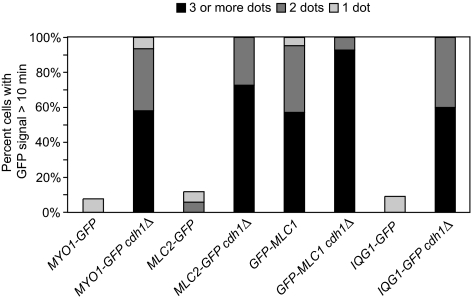
To better quantitate the cdh1Δ phenotype, we counted the numbers of cells with one or more Myo1-GFP patches visible somewhere in the cell at times >10 min after the completion of ring contraction. Only 8% of wild-type cells, but 100% of cdh1Δ cells, contained these Myo1 patches (Figure 2; Table 2). In almost all of the cdh1Δ cells observed, the Myo1 patches remained visible for the duration of the time-lapse observations. We then counted the maximum number of Myo1-GFP patches visible in each cell at any time from 10 min after the end of ring contraction until the end of the time-lapse series. More than 90% of cdh1Δ cells contained two or more patches, whereas none of the wild-type cells had more than one (Figure 2).
Figure 2.
Myo1, Mlc2, Mlc1, and Iqg1 exhibit ring-disassembly defects in APC mutant cells. Strains GT049 (MYO1-GFP), GT052 (MYO1-GFP cdh1Δ), GT104 (MLC2-GFP), GT110 (MLC2-GFP cdh1Δ), GT208 (GFP-MLC1), GT209 (GFP-MLC1 cdh1Δ), GT050 (IQG1-GFP), and GT053 (IQG1-GFP cdh1Δ) were examined by time-lapse microscopy (see Figure 3; Supplemental Figure S4; Table 2). For each cell, the maximum number of GFP patches visible at any time >10 min after the completion of ring contraction was recorded.
The results described above were obtained in the W303 strain background. Similar results were obtained with strains in the S288C background (see Figure 7, below).
To confirm that holoenzyme APC activity, and not just Cdh1, is required for Myo1-ring disassembly, we analyzed cells lacking all APC activity. Because the APC is normally essential for viability, we used a background (pds1Δ clb5Δ SIC110X) in which the APC is not essential (Thornton and Toczyski, 2003) and compared cells that were otherwise wild type to those lacking Apc2, an essential subunit of the APC holoenzyme. In the former strain, as in wild-type cells, the Myo1-GFP signal disappeared within 10 min after the end of ring contraction (Supplemental Figure S3A). In the apc2Δ strain, Myo1 patches persisted for ≥10 min after the end of ring contraction (Supplemental Figure S3B), supporting the hypothesis that the APC, with its activator Cdh1, is required for proper disassembly of Myo1-containing structures after actomyosin-ring contraction.
APCCdh1 Is Required for Myosin Light-Chain and Iqg1 Disassembly
We next investigated whether the APC also regulates the disassembly of other actomyosin-ring components. The “regulatory” myosin light-chain Mlc2 binds to the IQ2 motif of myosin (Luo et al., 2004), and in wild-type cells, its behavior was similar to that of Myo1: the Mlc2-GFP ring contracted in an average time of 5.1 ± 1.2 min (Figure 3A; Table 2) and disappeared within 10 min after the completion of ring contraction in 88% of cells (Figure 2). In cdh1Δ cells, the rate of ring contraction was similar (6.9 ± 1.7 min), but Mlc2-GFP patches persisted in all cells for ≥10 min after the end of ring contraction, and most cells contained ≥3 such patches (Figures 2 and 3B; Table 2).
Mlc1, the “essential” myosin light chain, is a multifunctional protein that associates with Myo1 during cytokinesis and with the type V myosin, Myo2, on secretory vesicles throughout most of the cell cycle (Stevens and Davis, 1998; Wagner et al., 2002; Luo et al., 2004). Because cells carrying Mlc1 tagged at its chromosomal locus were inviable (perhaps because GFP-tagged Mlc1 has reduced function), we analyzed the behavior of GFP-Mlc1 after expression under control of the MET promoter. In wild-type cells, the GFP-Mlc1 ring contracted in an average time of 5.7 ± 1.1 min (Supplemental Figure S4A; Table 2). As described previously (Wagner et al., 2002), GFP-Mlc1 did not disappear after ring contraction but remained visible in all cells >10 min after the end of ring contraction (Figure 2; Supplemental Figure S4A), presumably reflecting its association with Myo2. In cdh1Δ cells, GFP-Mlc1 contracted with wild-type kinetics (6.2 ± 1.6 min; Supplemental Figure S4B; Table 2). As in wild type, GFP-Mlc1 foci remained visible >10 min after the end of ring contraction in all cells, but the number of cells with ≥3 such foci increased from 57% in wild type to 93% in cdh1Δ cells (Figure 2; Supplemental Figure S4B). These data suggest that APCCdh1 activity is involved in disassembling the Myo1-associated subpopulation of Mlc1 complexes after actomyosin-ring contraction.
Finally, we analyzed the IQGAP protein Iqg1, which interacts with the actomyosin ring through an association with Mlc1 (Boyne et al., 2000; Shannon and Li, 2000). In wild-type cells, the Iqg1-GFP ring contracted in an average time of 4.6 ± 1.0 min (Figure 3C; Table 2), and an Iqg1-GFP patch remained visible >10 min after the end of ring contraction in just one of 11 cells examined (Figure 2). In cdh1Δ cells, the rate of ring contraction was similar (5.3 ± 1.5 min; Figure 3D; Table 2), but two or more Iqg1-GFP patches remained visible in all cells >10 min after the end of ring contraction (Figures 2 and 3D). We conclude that Iqg1-ring disassembly, like that of Myo1, Mlc2, and probably Mlc1, depends on APCCdh1.
Myo1 Colocalizes with Mlc2, Iqg1, and Mlc1 in cdh1Δ Cells
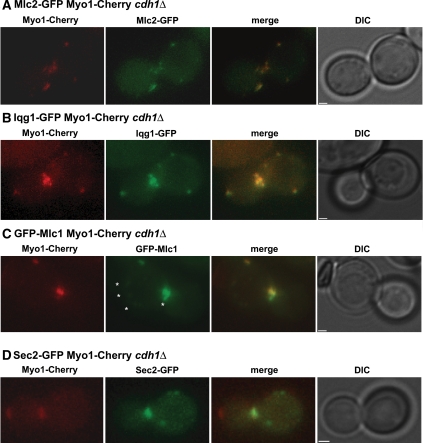
Because Myo1, Mlc2, Mlc1, and Iqg1 are all defective in dispersal in apc mutant cells, we used time-lapse microscopy of double-labeled cells to investigate whether these proteins colocalize in the same patches after ring contraction. In a cdh1Δ strain expressing both Mlc2-GFP and Myo1 tagged with mCherry (Myo1-Cherry), we observed colocalization of the two proteins at all time points in each of eight cells examined (Figure 4A; Supplemental Figure S5; Supplemental Table S1). Similarly, Iqg1-GFP and Myo1-Cherry colocalized at all time points in each of 14 cells examined (Figure 4B; Supplemental Figure S6; Supplemental Table S1).
Figure 4.
Myo1 colocalizes with Mlc2, Mlc1, and Iqg1, but not with Sec2, in cdh1Δ cells. cdh1Δ strains expressing Myo1-Cherry (red) and a GFP-tagged protein were examined by time-lapse microscopy (see Supplemental Figures S5–S8); representative images are shown. Strains used were as follows: GT132 (MLC2-GFP; A), GT133 (IQG1-GFP; B), GT225 (GFP-MLC1; C), and GT134 (SEC2-GFP; D). The asterisks in C indicate faint GFP-Mlc1 foci that do not colocalize with Myo1-Cherry. Bars, 1 μm.
As expected, cells coexpressing GFP-Mlc1 and Myo1-Cherry presented a more complicated picture. GFP-Mlc1 patches were detectable in just eight of 11 cells examined (probably because of variable expression from the MET promoter). In these cells, all Myo1-Cherry patches colocalized with GFP-Mlc1 at every time point (Figure 4C; Supplemental Figure S7; Supplemental Table S1), but some faint GFP-Mlc1 patches did not colocalize with Myo1-Cherry (Figure 4C, asterisks) and presumably represented Myo2-associated protein (see above). To explore this further, we investigated whether Myo1-Cherry colocalized with the secretory-vesicle protein Sec2-GFP. We observed partial colocalization in only one of eight cells undergoing cytokinesis (Figure 4D; Supplemental Figure S8; Supplemental Table S1). These results are consistent with the other evidence that there are both secretory-vesicle– and actomyosin-ring–associated pools of Mlc1 and that the latter, but not the former, dissociates after cytokinesis in an APCCdh1-dependent manner.
APCCdh1 Involvement in Ring Disassembly during Contraction
The actomyosin ring has long been thought to disassemble progressively as it contracts (Schroeder, 1972). Consistent with this model, the total fluorescence intensity of GFP-tagged ring components appeared to decline during contraction in wild-type cells (Figures 1C and 3, A and C). This decline seemed less pronounced in cells lacking Cdh1 (Figures 1D and 3, B and D). Quantitative measurements of Myo1-GFP fluorescence confirmed that cdh1Δ cells have a significant defect in the normal decline in ring fluorescence (Figure 5A). Thus, APCCdh1 is not just required for ring disassembly after contraction but also for the disassembly that occurs during contraction.
To explore further the relationships between ring contraction and APCCdh1-mediated disassembly, we analyzed ring behavior in cells treated with LAT-A. The absence of filamentous actin in these cells is known to prevent Myo1-ring contraction, and the Myo1 ring gradually disassembles in late mitosis, at about the time that cytokinesis would normally occur (Bi et al., 1998). In agreement with these results, we found that Myo1-GFP disappeared within 10 min of septin-ring splitting in 10 of 10 wild-type cells observed (Figure 5B, top). In contrast, in cdh1Δ cells, the Myo1-GFP ring remained visible for the duration of the experiment, for as long as 76 min after septin-ring splitting, in all 19 cells observed (Figure 5B, bottom). APCCdh1 is thus required for ring disassembly even when contraction is prevented.
Defective Completion of Septation in cdh1Δ Cells
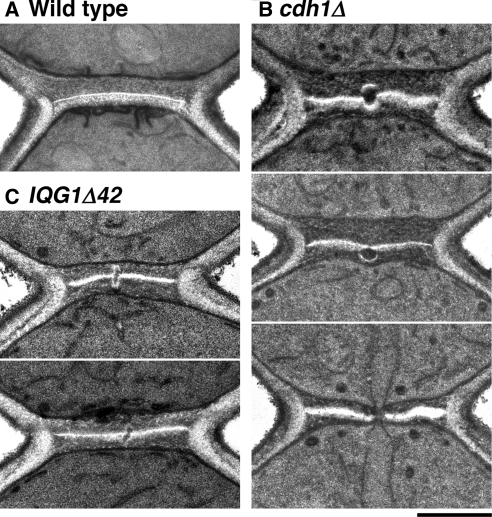
We used electron microscopy to explore further the functions of the APC in cytokinesis. In wild-type cells, ingression of the cleavage furrow takes place concomitantly with actomyosin-ring contraction and formation of the chitinous primary septum of the cell wall (visible as an electron-lucent line in electron micrographs; Vallen et al., 2000; Roh et al., 2002; Cabib, 2004). The process is completed by fusion of the invaginating membranes in the center of the neck and formation of a smooth, continuous disk of primary septum; secondary septa are then deposited on both sides of the primary septum (Figure 6A). In a cdh1Δ mutant, the early stages of cytokinesis and septum formation appeared normal, but the completion of septation was often strikingly abnormal. Among 95 cells examined in which some secondary-septum formation had occurred but cell separation had not begun, 26 (27%) exhibited one of the three types of defective septal structures shown in Figure 6B. Careful examination of an isogenic wild-type strain (YEF473A) revealed that a few cells (five of 100 examined) had similar but less pronounced abnormalities.
Figure 6.
Defects in the final stage of primary-septum formation in cdh1Δ and IQG1Δ42 cells. Strains YEF473A (A, wild type), KO255 (B, cdh1Δ), and GT124 (C, IQG1Δ42) were examined by electron microscopy after growth at 24°C; representative images are shown. Bar, 0.5 μm.
Removal of Several Known APC Targets Does Not Promote Actomyosin-Ring Disassembly
Because the APC is an E3 ubiquitin-protein ligase that mediates the destruction of its substrates by the 26S proteasome, the evidence presented above suggests that the destruction of one or more APC substrates is required for the disassembly of the actomyosin ring during and after its contraction. Clb2, Hsl1, Ase1, Fin1, and Spo12 are all substrates of APCCdh1 (Juang et al., 1997; Burton and Solomon, 2001; Shah et al., 2001; Woodbury and Morgan, 2007). If the degradation of any one of these substrates is required for actomyosin-ring disassembly, then deletion of the corresponding gene might rescue the disassembly defect in cdh1Δ cells. We constructed strains lacking both Cdh1 and one of the candidate substrates and monitored Myo1-GFP disassembly by time-lapse microscopy. However, we observed no rescue of the ring-disassembly defect: in each strain, Myo1-GFP foci persisted for >10 min in 100% of cells in which Myo1 had completed contraction (Table 2).
The Polo-like protein kinase Cdc5 is another APCCdh1 substrate that is involved in regulating cytokinesis. Because Cdc5 is essential for viability, we could not easily test the effects of deleting CDC5. Instead, we analyzed Myo1 behavior in cells expressing an APC-resistant form of Cdc5 that lacks amino acids 5-70 (Charles et al., 1998; Shirayama et al., 1998). In these cells, the Myo1-GFP ring contracted with normal kinetics (6.1 ± 1.5 min; Table 2), and Myo1-GFP patches were not present at times >10 min after the completion of ring contraction (Supplemental Figure S9). Thus, the stabilization of Cdc5 alone is not sufficient to block actomyosin-ring disassembly.
A major function of the APC is to trigger the destruction of mitotic cyclins and thereby reduce Cdk1 activity in late mitosis. Thus, it seemed possible that elevated Cdk1 activity in a cdh1Δ cell might inhibit actomyosin-ring disassembly. Although this possibility seemed unlikely, because the Clb-Cdk1 inhibitor Sic1 accumulates in late mitosis and suppresses Cdk1 activity in cdh1Δ cells (Schwab et al., 1997), we tested it further by overexpressing SIC1 in cdh1Δ cells and analyzing Myo1-GFP ring behavior. Sic1 overproduction did not rescue the defect in actomyosin-ring disassembly, as 95% of the cells contained one or (commonly) more Myo1-GFP patches at times ≥10 min after the completion of ring contraction (Supplemental Figure S10; Table 2). Taken together with the results of CLB2 deletion (see above), this result suggests strongly that APCCdh1 does not promote actomyosin-ring disassembly by reducing Cdk1 activity.
APCCdh1-dependent Degradation of Iqg1 Contributes to Actomyosin-Ring Disassembly
We recently identified Iqg1 as an APCCdh1 substrate (Ko et al., 2007). As Iqg1 is required to recruit actin to the bud neck and to initiate actomyosin-ring contraction, we speculated that its APC-dependent degradation might contribute to actomyosin-ring disassembly. We therefore monitored Myo1-GFP in cells carrying an APC-resistant mutant form of Iqg1, Iqg1Δ42, which lacks the first 42 amino acids that contain its APC-recognition sequence. Single Myo1-GFP patches were observed at times ≥10 min after the completion of ring contraction in 65% of cells (n = 20), compared with 9% of wild-type cells (n = 22; Figure 7A). Thus, the degradation of Iqg1 appears to contribute to actomyosin-ring disassembly. However, the fact that the IQG1Δ42 mutant does not fully phenocopy the cdh1Δ mutant suggests that Iqg1 is not the only APCCdh1 substrate whose degradation is important for ring disassembly.
Electron-microscopic observations supported these conclusions. In 18% (n = 55) of IQG1Δ42 cells examined at the stage when septum formation was largely complete, we observed defects in septum completion that were similar to those seen in cdh1Δ cells (Figure 6C; cf. Figure 6B). In accord with the fluorescence-microscopy observations, the abnormalities in IQG1Δ42 cells typically appeared less severe than those in cdh1Δ cells.
To further assess the importance of Iqg1 destruction in actomyosin-ring disassembly, we analyzed Myo1 behavior in iqg1Δ cells. The Myo1-GFP signal was far less intense in these cells than in wild type, so that the results were less clear than in our other experiments. Consistent with earlier observations (Shannon and Li, 1999), Myo1 ring contraction was not apparent in cells lacking Iqg1; instead, the Myo1 ring faded away over a period of ∼15 min (Figure 7B, top). Few if any Myo1-GFP patches were seen ≥10 min later. In cells lacking both Iqg1 and Cdh1, the Myo1-GFP signal was more readily detected, and it faded away over a period of ∼15–25 min, sometimes after a slow constriction (presumably reflecting the slow closure of the neck by secondary septal material in iqg1Δ cells: Nishihama and Pringle, unpublished results; Figure 7B, bottom). Ten minutes later, persistent Myo1-GFP patches were observed in 59% of cdh1Δ iqg1Δ cells (n = 61), compared with 100% of cdh1Δ cells, and the maximum number of patches per cell was also substantially decreased (Figure 7A). This partial rescue of the cdh1Δ phenotype by the iqg1Δ mutation is consistent with the hypothesis that APC-dependent destruction of Iqg1 contributes to actomyosin-ring disassembly.
The APC Collaborates with Mlc2 in Ring Disassembly
A defect in actomyosin-ring disassembly has also been observed in cells lacking Mlc2 (Luo et al., 2004). To determine if Mlc2 and Cdh1 promote ring disassembly by the same or distinct mechanisms, we compared the disassembly defect in an mlc2Δ cdh1Δ double mutant to the defects in the single mutants. In mlc2Δ single mutant cells, we did not observe a significant defect: as in wild-type, myosin patches were visible in only ∼8% of cells (n = 12) at times ≥10 min after the end of ring contraction (Figure 7A). However, the delay in myosin-ring disassembly observed by Luo et al. (2004) was only 2–8 min after the end of ring contraction, and our methods may not be sensitive enough to detect this defect.
To examine the double mutant, we first used time-lapse microscopy. By this assay, its phenotype was similar to that of the cdh1Δ single mutant (Figure 7A; n = 23). We were concerned, however that our time-lapse protocol might underestimate the long-term accumulation of Myo1 patches, because this protocol scores only those cells in which the completion of ring contraction occurred at some point during the 30-min recorded time frame. Thus, we performed an additional experiment in which we scored an exponential-phase population for the number of Myo1-GFP patches per cell in all cells that had completed ring contraction but not cell separation. With this assay, we found that Myo1-GFP patches accumulated to a twofold higher level in the double mutant than in the cdh1Δ single mutant (Figure 7C). We also found that the double-mutant population contained more clustered cells than either single mutant (18 vs. 5 and 9%), indicating a higher rate of cytokinesis failures (Supplemental Figure S1). Taken together, the data indicate that the mlc2Δ cdh1Δ double mutant has a more severe phenotype than either of the single mutants, suggesting that Mlc2 and Cdh1 promote actomyosin-ring disassembly by mechanisms that are at least partially distinct.
Because deletion of IQG1 partially rescued the defect in actomyosin-ring disassembly in cdh1Δ cells, we next tested whether stabilization of Iqg1 in the absence of Mlc2 would affect ring disassembly. In mlc2Δ IQG1Δ42 cells, Myo1-GFP patches did not disappear after ring contraction in any of the 14 cells examined (Figure 7A), a phenotype much more severe than that of the mlc2Δ (or IQG1Δ42) single mutant. In terms of the numbers of patches per cell, the double-mutant phenotype was less severe than that of a cdh1Δ single mutant (Figure 7A). Nonetheless, the observation that an APC-resistant Iqg1 exacerbates the phenotype of an mlc2Δ mutant further supports the conclusion that APCCdh1 and Mlc2 promote actomyosin-ring disassembly by mechanisms that are at least partially distinct.
DISCUSSION
APCCdh1 Promotes Disassembly of the Actomyosin Ring and Completion of Cytokinesis
In wild-type S. cerevisiae cells completing cell division, the actomyosin ring contracts, disassembles, and does not reform until the following cell cycle. We found that in cells with defective APC function, the actomyosin ring contracts at a normal rate but fails to disassemble efficiently during and after contraction, resulting in the formation of large, mobile patches containing (at least) Myo1, Mlc2, Mlc1, and Iqg1. These patches remain in the cell throughout G1 and disappear around the start of the following cell cycle. Because the proteins found in the patches also associate with each other in wild-type cells (Boyne et al., 2000; Luo et al., 2004), we speculate that the patches contain large macromolecular complexes of Myo1, Iqg1, Mlc2, and Mlc1 and that the activation of the APC in late mitosis normally promotes the dissociation of these complexes.
Electron microscopic analysis of the final stages of cytokinesis revealed defects or discontinuities at the center of the primary septum in cells lacking APCCdh1 activity. It seems likely that these defects are caused by problems in membrane invagination and fusion and/or in primary-septum formation that result when a complex of actomyosin-ring components remains unresolved at the center of the division plane. Thus, the APC may promote successful abscission by ensuring the complete removal of actomyosin-ring components from the division site. Alternatively, the failure to disassemble ring components may be a consequence, rather than the cause, of a defect in membrane behavior or septum formation. However, this interpretation appears less likely because of our identification of the ring component Iqg1 as one of the APC targets relevant to the phenotype observed.
Apparent Independence of Ring Contraction and Disassembly
Schroeder (1972) observed many years ago that the volume of the actomyosin ring decreases during contraction in sea-urchin embryos, suggesting that the ring disassembles as it contracts. Recent evidence suggests that ring disassembly during division depends in part on actin-depolymerizing factor (ADF)/cofilin in fission yeast and animal cells (Gunsalus et al., 1995; Kaji et al., 2003; Nakano and Mabuchi, 2006). Our studies (Figure 5A) indicate that ring components also disassemble progressively during contraction in S. cerevisiae and that APCCdh1-mediated protein destruction is important for this process.
One might have predicted that removal of ring components would be important for normal ring contraction. Our evidence suggests, however, that contraction occurs with normal kinetics when disassembly is largely prevented by deletion of CDH1. Interestingly, shrinkage of the ring during cytokinesis in S. cerevisiae does not depend on conventional actin-myosin–based contractile activity (Lord et al., 2005); instead, it appears to depend largely on the membrane invagination that results from deposition of the primary septum by the chitin synthase Chs2, which is delivered to the bud neck in an actin-dependent manner (VerPlank and Li, 2005; our unpublished data). Our evidence suggests that this process does not depend on removal of ring components.
Not only does ring contraction appear to be independent of disassembly, but the opposite is also true: our studies of LAT-A–treated cells (Figure 5B) revealed that APCCdh1-mediated disassembly can occur in the absence of contraction. Ring contraction and disassembly thus appear to be independent processes. Furthermore, when ADF/cofilin is mutated in fission yeast cells, myosin light chain dissociates from the stabilized actin filaments that are derived from the actomyosin ring (Nakano and Mabuchi, 2006), and our studies indicate that persistent Myo1 patches in cdh1Δ cells do not contain F-actin (Figure 1B). Thus, the depolymerization of actin filaments and the disassembly of nonactin components of the actomyosin ring appear to occur independently.
Actomyosin-Ring Disassembly Involves APCCdh1-mediated Degradation of Iqg1 and Other Substrates
Our evidence suggests that Iqg1 destruction is required for efficient disassembly of the actomyosin ring. First, deletion of IQG1 partially rescued the disassembly defect in cdh1Δ cells. Second, an APC-resistant version of Iqg1 (Iqg1Δ42) caused defects both in actomyosin-ring disassembly and in septation (as observed by electron microscopy) that were similar to, although less severe than, those observed in cdh1Δ cells. In addition, the IQG1Δ42 ring-disassembly defect was strikingly enhanced when an mlc2Δ mutation was also present.
The fact that the ring-disassembly defect in IQG1Δ42 cells is not as severe as that in cdh1Δ cells suggests that there are additional APCCdh1 substrates whose degradation, in combination with that of Iqg1, promotes actomyosin-ring disassembly. One of our challenges for the future is to identify these substrates. One possible candidate is Bud4, which is the closest S. cerevisiae homolog of the metazoan protein anillin, which has been identified as an APC substrate (Zhao and Fang, 2005). We found that Bud4 levels are low in G1, as expected for an APC substrate; however, we were unable to detect Bud4 ubiquitination by APCCdh1 in vitro (data not shown). Further analysis will therefore be necessary to determine if Bud4 is a bona fide APC substrate and if its destruction helps govern actomyosin-ring behavior.
We also considered Myo1, Mlc2, and Mlc1 as potential APC substrates. However, we found that Myo1 and Mlc2 protein levels are constant throughout the cell cycle (data not shown) and that Mlc1-GFP was present in G1 cells (Figure 3C; Wagner et al., 2002). Given that the abundance of previously identified APC substrates declines dramatically in G1, it thus seems unlikely that Myo1, Mlc2, and Mlc1 are APC targets.
APC and the Myosin “Regulatory” Light-Chain Function in Multiple Pathways to Promote Actomyosin-Ring Disassembly
Previous work has indicated that Mlc2 helps promote actomyosin-ring disassembly (Luo et al., 2004), although little is known about the underlying mechanism. Our results suggest that Mlc2 acts in parallel to APCCdh1: the disassembly defect in cdh1Δ mlc2Δ double-mutant cells is more pronounced than those in the single mutants.
Although cdh1Δ cells display a clear defect in actomyosin-ring disassembly, this defect is not lethal, because cdh1Δ cells are viable and most complete cytokinesis and form a new myosin ring early in the next cell cycle. Additional regulatory mechanisms must ensure that myosin and myosin light chains are available and able to localize to the presumptive bud site in G1. The septins clearly contribute to this regulation, because temperature-sensitive septin mutants fail to localize Myo1 to the presumptive bud site (Bi et al., 1998); interestingly, Myo1 in these mutants colocalizes with multiple septin foci around the cell cortex (Roh et al., 2002). Similarly, Myo1 and Iqg1 are mislocalized near the neck in the absence of Shs1, a nonessential component of the septin complex (Iwase et al., 2007).
We conclude that APCCdh1 promotes the dismantling of ring-protein complexes both during and after contraction, in part through the destruction of one subunit of those complexes, Iqg1. In the absence of Cdh1, these protein complexes may dissociate from the division site after contraction but remain aggregated to form persistent protein clusters at the bud neck and in other locations. These protein clusters disappear upon entry into the next cell cycle, suggesting that they are disassembled in late G1 by other mechanisms, which remain unexplored. It therefore appears that multiple mechanisms govern the disassembly of actomyosin-ring components both during and after cytokinesis, enabling efficient assembly of the new actomyosin ring in the following cell cycle.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Toczyski, Mark Solomon, David Pellman, Antonella Ragnini-Wilson, Peter Walter, Erin O'Shea (Harvard University), and Jonathan Weissman (University of California, San Francisco) for strains and plasmids; Peter Walter, Wallace Marshall, Alexander Johnson, and members of their laboratories for the generous use of their light microscopes; and Jon Mulholland and John Perrino at the Stanford University School of Medicine Cell Sciences Imaging Facility for assistance with electron microscopy. We also thank David Toczyski, Patrick O'Farrell, and members of the Morgan laboratory for helpful suggestions and support, Matt Sullivan and Liam Holt for critical reading of the manuscript, and an anonymous reviewer for suggesting an experiment that significantly improved the manuscript. This work was supported by funding from the National Institute of General Medical Sciences (GM53270 to D.O.M. and GM31006 to J.R.P.).
Abbreviations used:
- APC
anaphase-promoting complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0822) on December 24, 2008.
REFERENCES
- Bi E., Maddox P., Lew D. J., Salmon E. D., McMillan J. N., Yeh E., Pringle J. R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Yosuf H. M., Bieganowski P., Brenner C., Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J. Cell Sci. 2000;113:4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- Brachmann R. K., Yu K., Eby Y., Pavletich N. P., Boeke J. D. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17:1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J. L., Solomon M. J. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. The septation apparatus, a chitin-requiring machine in budding yeast. Arch. Biochem. Biophys. 2004;426:201–207. doi: 10.1016/j.abb.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Charles J. F., Jaspersen S. L., Tinker-Kulberg R. L., Hwang L., Szidon A., Morgan D. O. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G. R., Foley E., O'Farrell P. H. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U. S., Mitchison T. J., Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Epp J. A., Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gunsalus K. C., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M. L. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R., editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iwase M., Luo J., Bi E., Toh-e A. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics. 2007;177:215–229. doi: 10.1534/genetics.107.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H. B., Haarer B. K., Caruso C., Tong Z., Pringle J. R., Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Charles J. F., Tinker-Kulberg R. L., Morgan D. O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y.-L., Huang J., Peters J.-M., McLaughlin M. E., Tai C.-Y., Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kaji N., Ohashi K., Shuin M., Niwa R., Uemura T., Mizuno K. Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J. Biol. Chem. 2003;278:33450–33455. doi: 10.1074/jbc.M305802200. [DOI] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Tully G. H., Ostapenko D., Solomon M. J., Morgan D. O., Pringle J. R. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol. Biol. Cell. 2007;18:5139–5153. doi: 10.1091/mbc.E07-05-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek W. S., Bi E., Epp J. A., Wang L., Ho J., Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 2000;10:947–950. doi: 10.1016/s0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- Lippincott J., Li R. Sequential assembly of myosin II, an IQGAP-like protein and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Laves E., Pollard T. D. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 2005;16:5346–5355. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Vallen E. A., Dravis C., Tcheperegine S. E., Drees B., Bi E. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J. Cell Biol. 2004;165:843–855. doi: 10.1083/jcb.200401040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M. A., Konopka J. B., Cerione R. A. Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J. Cell Biol. 2002;159:601–611. doi: 10.1083/jcb.200205084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Robinson D. N., Spudich J. A. Towards a molecular understanding of cytokinesis. Trends Cell Biol. 2000;10:228–237. doi: 10.1016/s0962-8924(00)01747-5. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni M., Morgan D. O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Roh D. H., Bowers B., Schmidt M., Cabib E. The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell. 2002;13:2747–2759. doi: 10.1091/mbc.E02-03-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. E. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J. Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Lutum A. S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shah R., Jensen S., Frenz L. M., Johnson A. L., Johnston L. H. The Spo12 protein of Saccharomyces cerevisiae: a regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics. 2001;159:965–980. doi: 10.1093/genetics/159.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shannon K. B., Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol. Biol. Cell. 1999;10:283–296. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K. B., Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr. Biol. 2000;10:727–730. doi: 10.1016/s0960-9822(00)00539-x. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Lee K. S. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R. C., Davis T. N. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol. 1998;142:711–722. doi: 10.1083/jcb.142.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Field C. M., Mitchison T. J. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D. O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Thornton B. R., Toczyski D. P. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Thornton B. R., Toczyski D. P. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- Vallen E. A., Caviston J., Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J. Q., Hao S., O'Shaughnessy B., Pollard T. D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- VerPlank L., Li R. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 2005;16:2529–2543. doi: 10.1091/mbc.E04-12-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W., Bielli P., Wacha S., Ragnini-Wilson A. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 2002;21:6397–6408. doi: 10.1093/emboj/cdf650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Woodbury E. L., Morgan D. O. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Kono K., Lowery D. M., Bartolini S., Yaffe M. B., Ohya Y., Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- Zhao W. M., Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J. Biol. Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.