Abstract
Tissue homeostasis is controlled by the availability of growth factors, which sustain exogenous nutrient uptake and prevent apoptosis. Although autophagy can provide an alternate intracellular nutrient source to support essential basal metabolism of apoptosis-resistant growth factor–withdrawn cells, antiapoptotic Bcl-2 family proteins can suppress autophagy in some settings. Thus, the role of autophagy and interactions between autophagy and apoptosis in growth factor–withdrawn cells expressing Bcl-2 or Bcl-xL were unclear. Here we show autophagy was rapidly induced in hematopoietic cells upon growth factor withdrawal regardless of Bcl-2 or Bcl-xL expression and led to increased mitochondrial lipid oxidation. Deficiency in autophagy-essential gene expression, however, did not lead to metabolic catastrophe and rapid death of growth factor–deprived cells. Rather, inhibition of autophagy enhanced survival of cells with moderate Bcl-2 expression for greater than 1 wk, indicating that autophagy promoted cell death in this time frame. Cell death was not autophagic, but apoptotic, and relied on Chop-dependent induction of the proapoptotic Bcl-2 family protein Bim. Therefore, although ultimately important, autophagy-derived nutrients appear initially nonessential after growth factor withdrawal. Instead, autophagy promotes tissue homeostasis by sensitizing cells to apoptosis to ensure only the most apoptosis-resistant cells survive long-term using autophagy-derived nutrients when growth factor deprived.
INTRODUCTION
Metazoan cells require growth factor input to maintain metabolism and viability in both development and tissue homeostasis (Raff, 1992; Rathmell et al., 2000). The phosphoinositide 3-kinase (PI3K)-Akt mammalian target of rapamycin (mTOR) pathway is an important signaling mechanism of many growth factors that maintains nutrient uptake and prevents apoptotic cell death (Vivanco and Sawyers, 2002; Wieman et al., 2007). When cells are deprived of appropriate growth factor stimulation, the loss of PI3K-Akt-mTOR signaling, glucose transporter localization on the cell surface, and glucose uptake leads to apoptosis or autophagy (Tsujimoto and Shimizu, 2005; Wieman et al., 2007). Unlike the cell death pathway of apoptosis, autophagy is a dual-function mechanism of bulk cytoplasmic and organelle degradation in lysosomes that can promote cell survival if controlled or can lead to death if excessive. The regulation of autophagy and how it impacts apoptosis and cell fate upon growth factor withdrawal, however, remain uncertain.
Autophagy may influence cell fate upon growth factor deprivation by multiple mechanisms. Regulated autophagy can promote cell survival by providing metabolic fuel for mitochondrial oxidation after lysosomal digestion of intracellular components (Eskelinen, 2005; Lum et al., 2005b). Indeed, cells incapable of apoptosis can survive for an extended time after growth factor withdrawal or nutrient deprivation by utilizing autophagy as a source of nutrients to support basal cell metabolism (Lum et al., 2005a; Degenhardt et al., 2006; Fung et al., 2008). Autophagy may also promote survival by clearance of damaged organelles or protein aggregates in stressed cells (Abedin et al., 2007; Colell et al., 2007). Alternately, autophagy can induce cell death and autophagic death has been observed in cells in which caspases have been inhibited (Yu et al., 2004) and has also been implicated in the death of Drosophila larval salivary glands (Lee and Baehrecke, 2001), the elimination of cells in the inner cell mass of blastocysts (Qu et al., 2007), and the death of insulin-deprived neural stem cells (Yu et al., 2008).
The Bcl-2 family of proteins regulate cell death upon growth factor withdrawal and is critical in the regulation and function of both apoptosis and autophagy. Anti-apoptotic Bcl-2 family proteins such as Bcl-2 and Bcl-xL are up-regulated by growth factors and are frequently overexpressed in cancer (Walensky, 2006). After growth factor withdrawal, proapoptotic BH3-only Bcl-2 family proteins such as Bim and Puma (Bouillet et al., 1999; Erlacher et al., 2006) are induced and bind to antiapoptotic Bcl-2 family proteins, inhibiting their interaction with Bax and Bak and allowing apoptosis to occur (Walensky, 2006). The ratio of antiapoptotic to proapoptotic Bcl-2 family proteins, therefore, is critical in determining the sensitivity to apoptosis. Importantly, antiapoptotic Bcl-2 family proteins can bind the autophagy-essential protein Beclin-1 and can inhibit autophagy in settings of acute nutrient deprivation (Pattingre et al., 2005). This interaction with Beclin-1 is affected by the balance of proapoptotic BH3-only Bcl-2 family proteins (Maiuri et al., 2007; Oberstein et al., 2007). Autophagy induction, however, has been observed in the presence of overexpressed Bcl-2 or Bcl-xL after ischemia (Degenhardt et al., 2006) or DNA damage (Shimizu et al., 2004).
Although it is apparent that autophagy can promote survival of growth factor–deprived cells that are incapable of apoptosis (Lum et al., 2005a; Degenhardt et al., 2006), the metabolic and cell survival role of autophagy in growth factor deprivation of apoptosis-competent cells with normal or moderate expression of Bcl-2 or Bcl-xL was unclear. In this study, we tested the hypothesis that autophagy could be activated after growth factor withdrawal despite Bcl-2 expression and that autophagy influenced both cell metabolism and survival. We found that moderate expression of Bcl-2 or Bcl-xL delayed apoptosis but did not prevent the rapid induction of autophagy upon growth factor withdrawal of hematopoietic cells. Autophagy did provide a source of nutrients, yet these autophagy-derived nutrients were not required for survival within the first week of growth factor withdrawal, and autophagy instead eventually enhanced the rate of apoptotic cell death. Notably, this apoptosis was due to autophagy-dependent transcriptional induction of the BH3-only protein Bim via the stress response transcription factor Chop. Thus, induction of autophagy after growth factor withdrawal can simultaneously contribute to basal cell metabolism and sensitize cells to apoptosis through induction of Bim. Autophagy itself, therefore, is not metabolically essential for short-term survival of growth factor–deprived cells but can regulate the balance of proto antiapoptotic Bcl-2 family proteins. This may contribute to tissue homeostasis by increasing apoptotic stress to ensure that only cells with a high resistance to apoptosis may survive long enough to utilize autophagy as a nutrient source and long-term survival strategy.
MATERIALS AND METHODS
Cell Culture
FL5.12, Ba/F3, and 32D cells were cultured and transfected as described (Zhao et al., 2007). IL3-dependent Bax−/− Bak−/− cells were kindly provided by Drs. Julian Lum and Craig Thompson (University of Pennsylvania, Philadelphia, PA). FL5.12 cells stably expressing Bcl-xL or doxycycline-inducible MyrAkt have been previously described (Plas et al., 2001). M57 (Atg5 Tet-Off) mouse embryonic fibroblasts (MEFs; generously provided by Dr. Noboru Mizushima, Tokyo Medical and Dental University, Bunkyo-Ku, Japan) were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma, St. Louis, MO) with 10% fetal bovine serum and 2 mM l-glutamine. MEFs were treated with 5 μg/ml doxycycline (Sigma) for 4 d to suppress Atg5 expression. For serum starvation, MEFs were washed three times in serum-free DMEM, and then plated in serum-free DMEM with or without doxycycline.
FL5.12 and 32D cells were retrovirally transduced with Bcl-2, and FL5.12 cells expressing Bcl-2 were cloned for moderate and high Bcl-2 expression. Ba/F3 cells were transfected with Bcl-xL or Bcl-2, selected with blasticidin, and cloned. 32D Bcl-2–expressing cells were withdrawn from cytokine for 24 h after retroviral infection and then cytokine was added back to select for Bcl-2–expressing cells. Some cells were treated with chloroquine (CQ; 40 μM, Sigma) or tunicamycin (2 μg/ml, Sigma).
Plasmid Constructs
Human Bcl-2 and Bcl-xL were subcloned into the MSCV-IRES-tNGFR retroviral vector. Green fluorescent protein (GFP)-LC3 from rat (Kabeya et al., 2000) was subcloned into the pMXIRES.2-hCD2 retroviral vector (generously provided by Dr. Youwen He, Duke University, Durham, NC). Short-hairpin RNA interference (shRNAi) hairpins were constructed as previously described (Fox et al., 2003) using the following targeting sequences for Beclin-1, 5′ GAATGTCAGAACTACAAACGCTGTT; Atg12, 5′ GAGCCCCCGTCCTCGGCTGCAGTT; Chop, 5′ GACTTCTCTTGCTCGCCGAGTT; Atg5 (Lum et al., 2005a); Atg7 (Yu et al., 2004); and Bim and GFP (Zhao et al., 2007).
Immunoblots
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor cocktail (BD PharMingen, San Jose, CA) and phosphatase inhibitors (Sigma). Proteins were separated by SDS-PAGE (Bio-Rad, Hercules, CA). Primary antibodies used included the following: mouse anti phospho-Akt Ser473 (Cell Signaling, Beverly, MA); rabbit anti-Akt (Cell Signaling); rabbit anti-phospho mTOR Ser2448 (Cell Signaling); rabbit anti-Bim (BD PharMingen); rabbit anti-Puma (Cell Signaling); rabbit anti-Bax (Cell Signaling); rabbit anti-Bcl-xL (Cell Signaling); rabbit anti-mBcl-2 (BD PharMingen); mouse anti-hBcl-2 (BD PharMingen); mouse anti-GFP (Chemicon International, Temecula, CA); mouse anti-Beclin-1 (BD Transduction Laboratories, Lexington, KY); rabbit anti-LC3 (generously provided by Dr. Tamotsu Yoshimori, National Institute of Genetics, Mishima, Japan); rabbit anti-Actin (Santa Cruz Biotechnology, Santa Cruz, CA); and mouse anti-Actin (Sigma). Secondary antibodies used include: anti-rabbit and anti-mouse horseradish peroxidase–labeled antibodies (BD PharMingen); Alexa-Flour 680 goat anti-rabbit IgG (Invitrogen, Carlsbad, CA); and IRDye 800CW goat anti-mouse IgG (LiCor, Lincoln, NE). Immunoblots were imaged either with ECL-Plus (Amersham Biosciences, Piscataway, NJ), Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL), or Odyssey infrared imaging system (LiCor) and uniformly contrasted. Some images have been digitally rearranged for ease of viewing and rearranged lanes are indicated by spaces. For quantitated immunoblots, levels of proteins under analysis were quantified by normalization to actin and to the control sample.
Electron and Fluorescent Microscopy
For transmission electron microscopy, cell pellets were fixed in 4% gluteraldehyde in 0.1 M sodium cacodylate buffer overnight. Pellets were prepared by the Duke University Pathology Electron Microscopy Facility. Images were obtained with a Philips LS 410 electron microscope (Mahwah, NJ). Electron micrographs are shown at 9700× magnification. For fluorescence microscopy, cells were fixed in 1% paraformaldehyde and PBS and imaged on a LSM410 confocal microscope (Zeiss, Thornwood, NY) and Metamorph software (Universal Imaging, West Chester, PA).
Cell Death and Caspase Activity Analysis
Cell death was assessed in triplicate samples by flow cytometry analysis for propidium iodide exclusion (1 μg/ml; Invitrogen) versus forward scatter. MEFs were trypsinized and resuspended in propidium iodide solution before analysis. For active Bax and propidium iodide DNA analysis for sub-G1 DNA content, cells were fixed in 0.25% paraformaldehyde (Sigma) and permeabilized in 100 μg/ml digitonin (Sigma) and then costained with mouse anti-Bax (Clone 6A7 BD PharMingen), rat anti-mouse FITC (BD PharMingen), and 5 μg/ml propidium iodide. Caspase 3 activity was assessed as previously described (Nutt et al., 2005). Samples were analyzed in triplicate for absorbance at 405 nm. Values given are VMAX of absorbance 1 h after start of reaction. Asterisks denote p < 0.05 by Student's t test.
Glucose Uptake
Glucose uptake was measured as previously described (Wieman et al., 2007), with 5-min incubation of cells with 2-deoxy-d-[3H]glucose (2 μCi/reaction) before separation of internalized glucose from free glucose. Asterisks denote p < 0.05 by Student's t test.
Acyl-Carnitine and Metabolic Analysis
Cells were cultured with 1 μM l-carnitine (Sigma) 36 h before harvest and acyl-carnitines were analyzed as previously described (An et al., 2004). For measurement of β-oxidation, Bcl-xL cells were preincubated with 3H-palmitate (palmitic acid-[9,10-3H] Sigma) for 48 h and then washed free of palmitate. Cells were cultured in the presence or absence of IL3 for 24 h and harvested, and cell lysate was run over a Dowex column (Sigma) to separate 3H-labeled water, which was then measured as an indication of intracellular β-oxidation. Etomoxir (Sigma) was used at a final concentration of 100 μM. Asterisks denote p < 0.05 by Student's t test.
Quantitative Real-Time PCR Analysis
Cells were lysed in Trizol (Invitrogen), and mRNA was extracted by addition of chloroform and subsequent ethanol precipitation. RNA was treated with DNAse (Invitrogen) and then reverse-transcribed to cDNA using the SuperScript II Reverse Transcriptase system (Invitrogen). Quantitative real-time PCR was performed with IQ Sybr-Green Supermix (Bio-Rad) on a Bio-Rad ICycler Real-Time PCR machine. The following primers were used: Bim, GGTCCTCCAGTGGGTATTTCTCTT, ACTGAGATAGTGGTTGAAGGCCTGG; Chop, GGGCCAACAGAGGTCACAC, CTTCATGCGTTGCTTCCCA; Atg5, ATGCGGTTGAGGCTCACTTTA, GCCCAAAACTGGTCAAATCTGTC; Atg7, TTCAGTGCTTTTGACATGAGTGC, CACCTGACTTTATGGCTTCCC; and Atg12, AACAAAGAAATGGGCTGTGGAGCG, TTCCGAGGCCACCAGTTTAAGGAA. All values were normalized to quantitative real-time amplification of the housekeeping gene β2-microglobulin (β2M): ACCGGCCTGTATGCTATCCAGAAA, GGTGAATTCAGTGTGAGCCAGGAT. Each quantitative real-time PCR figure is the average of three independent experiments. Asterisks denote p < 0.05 by Student's t test.
RESULTS
Bcl-2 and Bcl-xL Prevent Cell Death in Growth Factor–deprived Cells Despite Loss of Glucose Uptake
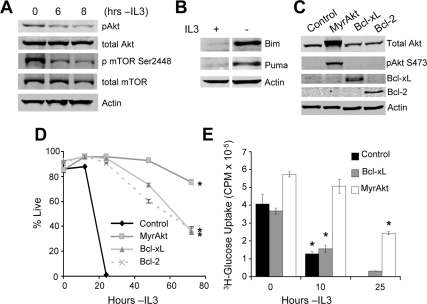
The IL3-dependent lymphoid/myeloid precursor cell line FL5.12 was analyzed in the presence and absence of IL3 to closely examine the molecular changes that may underlie atrophy and cell death after cytokine deprivation (Rathmell et al., 2000). When cells were deprived of growth factor, phosphorylation of Akt and mTOR decreased (Wieman et al., 2007; Figure 1A), and expression of the proapoptotic proteins Bim (Zhao et al., 2007) and Puma increased (Figure 1B). Expression of activated Akt (myristoylated Akt [MyrAkt]), Bcl-xL, or Bcl-2 (Figure 1C) each prevented cell death (Figure 1D). In contrast, glucose uptake was decreased in control and Bcl-xL cells but not in MyrAkt cells after IL3 withdrawal (Figure 1E). This is consistent with previous findings showing the inability of Bcl-xL to affect trafficking of the glucose transporter Glut1 and the ability of Akt to promote trafficking of Glut1 to the cell surface (Wieman et al., 2007).
Figure 1.
Bcl-2 and Bcl-xL prevent cell death in growth factor–deprived cells despite decreased glucose uptake. (A and B) Cells were cultured in IL3 or withdrawn from IL3 for (A) 6 or (B) 10 h and analyzed by immunoblot. (C) The expression of MyrAkt, Bcl-xL, and Bcl-2 was established by immunoblot. Cell survival (D) and glucose uptake (E) were determined at indicated times after IL3 withdrawal. Means of triplicate samples and SDs are shown for survival curves and SEs for glucose uptake. Data shown are representative of three or more experiments. *p < 0.05 by Student's t test of IL3 withdrawn samples compared with control (D) or + IL3 samples (E).
Autophagy Is Induced in Growth Factor–withdrawn Cells Expressing Bcl-xL or Bcl-2
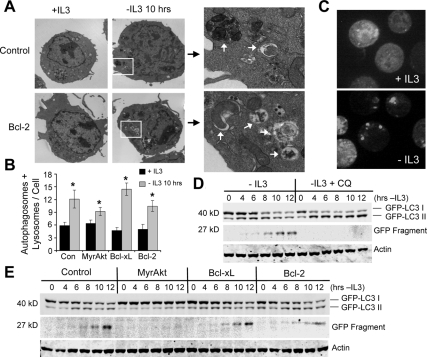
The decrease in mTOR phosphorylation (Figure 1A) and glucose uptake after IL3 withdrawal in control and Bcl-xL–expressing cells (Figure 1E) may have resulted in the induction of autophagy. The appearance of autophagosomes by transmission electron microscopy was, therefore, examined in control cells or cells expressing MyrAkt, Bcl-2, or Bcl-xL in the presence or absence of cytokine for 10 h. Despite the ability of Bcl-2 and Bcl-xL to block autophagy in acute settings (Pattingre et al., 2005), autophagosomes became clearly visible after 10 h without cytokine in both control cells and Bcl-2–expressing cells (Figure 2A). These findings are consistent with reports of autophagy induction even in the presence of overexpressed Bcl-xL or Bcl-2 after serum withdrawal (Maiuri et al., 2007; Oberstein et al., 2007), ischemia (Degenhardt et al., 2006), or DNA damage (Shimizu et al., 2004). Quantitation of autophagosomes and lysosomes in control, MyrAkt-, Bcl-xL–, and Bcl-2–expressing cells revealed a significant induction of autophagy after cytokine deprivation (Figure 2B), though fewer autophagosomes and lysosomes were observed in MyrAkt-expressing cells, consistent with mTOR inhibition of autophagy.
Figure 2.
Bcl-xL and Bcl-2 expression do not prevent induction of autophagy after growth factor withdrawal. (A) Control and Bcl-2–expressing cells were cultured in IL3 or withdrawn from IL3 for 10 h and examined by transmission electron microscopy. Boxed areas are shown in detail on right, and white arrows indicate autophagosomes and autolysosomes. (B) Quantitation of autophagomes and lysosomes in control (Con) and MyrAkt-, Bcl-xL–, and Bcl-2–expressing cells cultured in IL3 or withdrawn from IL3 for 10 h (n = 27 cells each). Means and SEs are shown. *p < 0.05 by Student's t test of IL3 withdrawn samples compared with +IL3 samples. (C) Bcl-xL–expressing cells stably expressing GFP-LC3 were deprived of cytokine for 6 h and examined by fluorescence microscopy for localization of GFP-LC3. (D) Cells expressing GFP-LC3 were deprived of cytokine for indicated times in the absence or presence of chloroquine (CQ), and GFP-LC3 processing was assessed by anti-GFP immunoblot. (E) Control and Myr-Akt–, Bcl-xL–, and Bcl-2–expressing cells with stable GFP-LC3 expression were withdrawn from IL3 for indicated times, and GFP-LC3 processing was observed by immunoblot. Data shown are representative of three or more experiments.
As an independent measure of autophagy, the localization and processing of microtubule-associated protein light chain 3 (MAP-LC3), a small protein involved in autophagosome formation that is processed into an active form when autophagy is induced, was also examined (Tanida et al., 2004). Cells stably expressing GFP-tagged LC3 were withdrawn from IL3 and then analyzed over a 12-h period. Bcl-xL–expressing cells with stably expressed GFP-LC3 showed a characteristic LC3 punctate pattern in the absence of IL3, indicative of autophagy (Figure 2C). In addition, immunoblot analysis revealed that GFP-LC3 I began to decrease in control cells with a concomitant increase in GFP-LC3 II shortly after IL3 withdrawal, and this conversion continued out to 12 h (Figure 2D). A smaller 27-kDa protein, recognized by the anti-GFP antibody, also began to appear by 8 h (Figure 2D). This smaller protein has previously been used as an indicator of autophagy (Hosokawa et al., 2006) and appeared to have arisen because of the lysosomal degradation of GFP-LC3, as the lysosome inhibitor CQ inhibited the appearance of the smaller form of GFP-LC3 and caused a buildup of GFP-LC3 II (Figure 2D). GFP-LC3 processing and cleavage was inhibited in MyrAkt-expressing cells (Figure 2E), yet appeared to occur normally upon growth factor withdrawal in both Bcl-2– and Bcl-xL–expressing cells (Figure 2E). Together, these data show that autophagy is rapidly induced upon growth factor withdrawal, and expression of Bcl-2 or Bcl-xL does not affect this process.
Autophagy Promotes Production and Metabolism of Long-Chain Fatty Acids after Growth Factor Withdrawal
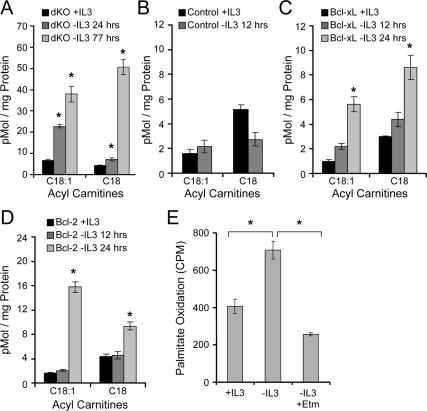
Hematopoietic Bax−/− Bak−/− cells rely on autophagy to provide intracellular nutrients for mitochondrial oxidation after prolonged periods of growth factor withdrawal (Lum et al., 2005a). To determine if Bcl-2– or Bcl-xL–expressing cells also derived metabolic benefit from autophagy in the initial days after IL3 withdrawal, acyl-carnitines were measured in control and MyrAkt-, Bcl-xL–, and Bcl-2–expressing cells. Acyl-carnitines are derived from the covalent linkage of carnitine to amino acids or lipids for transport across the mitochondrial inner membrane and represent carbon chains available for mitochondrial catabolism (An et al., 2004). After 10 h of IL3 withdrawal, small-chain acyl-carnitine levels were not significantly changed (Supplemental Figure S1, A–D). After 1 d, however, Bcl-xL– and Bcl-2– expressing cells (Supplemental Figure S1, C and D, respectively) showed a greater than twofold increase in short carbon carnitine-linked chains. C5-carnitine levels also increased, suggesting elevated amino acid metabolism (Supplemental Figure S1, A, C, and D). Carbon sources available for mitochondrial oxidation, therefore, appeared to increase even in the face of decreased glucose uptake.
The acyl-carnitine profile of IL3 withdrawn Bax−/− Bak−/− cells was characterized to establish how reliance on autophagy may affect metabolism. Similar to Bcl-2– and Bcl-xL–expressing cells, an increase in short chain acyl-carnitines was observed in IL3 deprived Bax−/− Bak−/− cells (data not shown). Importantly, we also observed an increase in both C18:1 (oleate) and C18 (stearate) long chain acyl-carnitines (Figure 3A), suggesting mobilization of fatty acids from intracellular lipids for metabolism, consistent with proposed models of autophagy as a nutrient-generating process (Lum et al., 2005b). C18:1 and C18-carntines did not change at early time points but were also greatly increased in Bcl-xL–and Bcl-2–expressing FL5.12 cells 1 d after IL3 withdrawal (Figures 3, B–D).
Figure 3.
Growth factor withdrawal promotes production and metabolism of long-chain fatty acids. (A–D) IL3-dependent Bax−/− Bak−/− (dKO; A), Control (B), Bcl-xL (C), and Bcl-2 (D) cells were cultured in IL3 or withdrawn from IL3 for indicated times, and C18:1 (oleate) and C18 (stearate) levels were measured by tandem mass spectrometry for acyl-carnitines. (E) Bcl-xL–expressing cells were incubated with 3H-palmitate for 48 h, washed, and cultured in the presence of IL3 or withdrawn from IL3 for 1 d. Generation of tritiated water was measured as an indication of palmitate metabolism. Etomoxir (Etm), a β-oxidation inhibitor, was added to some samples. Means of triplicate samples and SEs are shown. *p < 0.05 by Student's t test of IL3 withdrawn samples compared with +IL3 samples, except where otherwise indicated. Data shown are representative of three or more experiments.
Changes in steady-state levels of acyl-carnitines did not necessarily indicate altered flux of lipids through mitochondrial β-oxidation or if these lipids were derived from autophagy. To directly observe flux, Bcl-xL–expressing cells were labeled with 3H-palmitate, and the rate of β-oxidation was measured. Oxidation of incorporated 3H-palmitate increased upon growth factor withdrawal, and this was blocked by addition of the cartinine palmitoyltransferase and β-oxidation inhibitor etomoxir (Etm, Figure 3E). Thus, although MyrAkt cells continued to uptake and utilize glucose, Bcl-2– or Bcl-xL–expressing cells decreased glucose uptake (Figure 1E) and switched to mitochondrial oxidation of lipids and amino acids after growth factor withdrawal.
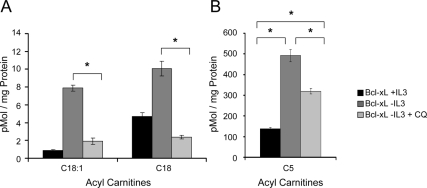
To determine if the source of these long-chain fatty acids was autophagy, chloroquine was added to growth factor deprived Bcl-xL–expressing cells to inhibit lysosomal degradation of intracellular components. Chloroquine eliminated the increase of C18:1 and C18-carnitines (Figure 4A). Interestingly, chloroquine treatment did not fully prevent increased C5-carnitine levels in Bcl-xL–expressing cells after growth factor withdrawal (Figure 4B), suggesting that growth factor–withdrawn cells may maintain some autophagy-independent amino acid metabolism. Therefore, in growth factor–withdrawn cells expressing Bcl-2 or Bcl-xL, autophagy appears to be a critical source of fatty acids but not of amino acids.
Figure 4.
Autophagy serves as a sole source of lipid- but not amino acid–derived metabolites for mitochondrial oxidation after growth factor withdrawal. (A and B) Bcl-xL–expressing cells were cultured in IL3 or withdrawn from IL3 for 1 d. Chloroquine (CQ) was added to some samples 8 h before harvest to inhibit lysosomal degradation pathways. (A) Lipid-derived long-chain– and (B) amino acid–derived short-chain acyl-carnitines were measured by tandem mass spectrometry. Means of triplicate samples and SEs are shown. *p < 0.05 by Student's t test of IL3 withdrawn samples compared with +IL3 samples, except where otherwise indicated.
Despite Nutrient Production, Autophagy Induces Cell Death in Bcl-2– and Bcl-xL–expressing Cells
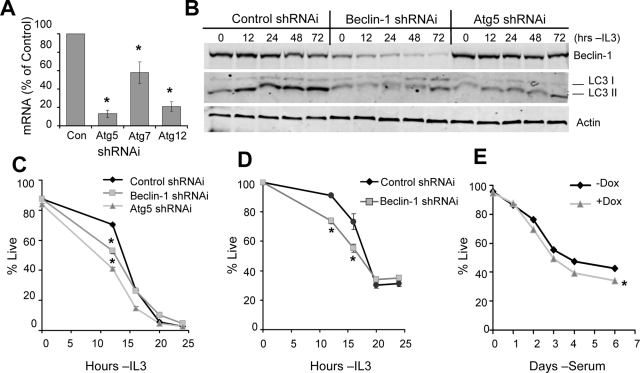
The generation of nutrients for mitochondrial oxidation suggested that autophagy may play a positive role in cell survival after cytokine withdrawal and decreased glucose uptake. To directly examine the role of autophagy in Bcl-2– and Bcl-xL–mediated cell survival, we utilized shRNAi constructs against the autophagy-essential genes Atg5, Atg7, Atg12 (Figure 5A), and Beclin-1 (Figure 5B). Both Beclin-1 and Atg5 shRNAis decreased autophagy when cells were withdrawn from IL3, as indicated by reduced processing of LC3 I to LC3 II (Figure 5B). Control, Beclin-1, and Atg5 shRNAis were first transfected into control cells lacking exogenous Bcl-2 or Bcl-xL expression, and survival after IL3 withdrawal was assessed. Inhibition of autophagy led to more rapid death by 12 h after IL3 withdrawal (Figure 5C). This death was not due to modulation of Bcl-2 family proteins by autophagy, because endogenous Bcl-2 and Bcl-xL were unaffected by autophagy shRNAi after IL3 withdrawal (Supplemental Figure S2). Cells expressing Glut1 and Hexokinase, which can maintain high glucose uptake and increased survival even after growth factor withdrawal (Zhao et al., 2007), also showed increased death shortly after growth factor withdrawal when autophagy was inhibited (Figure 5D). To determine if autophagy affected death of nonhematopoietic cells upon growth factor withdrawal, immortalized MEFs with regulated expression of Atg5 (Hosokawa et al., 2006) were examined upon serum deprivation (Figure 5E). Addition of doxycycline suppressed Atg5 expression (Supplemental Figure S3) and Atg5-deficient MEFs died similarly or slightly faster than PBS control-treated cells after serum withdrawal (Figure 5E). Doxycycline had no effect on wild-type MEFs (data not shown). Together these data suggest that cell death was similar or modestly enhanced shortly after growth factor withdrawal in autophagy-deficient cells, and this death was not due to nutrient deficiency.
Figure 5.
Autophagy supports cell survival in a nutrient-independent manner early after cytokine withdrawal. (A) Efficiency of mRNA suppression for shRNAi against Atg5, Atg7, or Atg12 was determined by quantitative real-time PCR. (B) Bcl-2 cells were transiently transfected with control, Beclin-1, or Atg5 shRNAi, withdrawn from cytokine for up to 48 h, and harvested, and Beclin-1 knockdown efficiency and endogenous LC3 processing were detected by immunoblot. (C and D) Control (C) and Glut1/Hexokinase-expressing (D) cells were transfected with control, Beclin-1, or Atg5 shRNAi, and viability was observed over time after IL3 withdrawal. (E) Atg5 Tet-Off MEFs were treated with PBS or 5 μg/ml doxycycline for 4 d, withdrawn from serum, and analyzed for viability. Means and SDs from triplicate samples are shown. *p < 0.05 by Student's t test of test samples relative to control samples. Data shown are representative of three or more experiments.
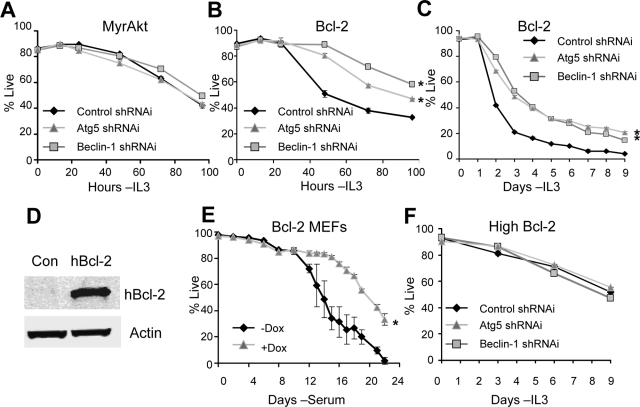
Long-term cell viability upon inhibition of autophagy was next examined in MyrAkt- and Bcl-2–expressing cells. Neither Beclin-1 nor Atg5 shRNAi affected survival in cells expressing MyrAkt (Figure 6A). In contrast, inhibition of autophagy increased survival of cells with moderate Bcl-2 expression at later time points (Figure 6B). Similar results were also observed in cells expressing Bcl-xL (Supplemental Figure S4). Importantly, inhibition of autophagy in cells with moderate Bcl-2–expressing led to a persistent twofold or greater survival advantage lasting more than 1 wk (Figure 6C). Similarly, inhibition of autophagy in Bcl-2–expressing MEFs (Figure 6D) with conditional Atg5 expression led to increased cell survival after serum withdrawal (Figure 6E). Consistent results supporting a prodeath role for autophagy in growth factor withdrawal were observed in the hematopoietic-derived Ba/F3 (Supplemental Figures S5, A–D) and 32D (Supplemental Figure S6, A and B) cell lines. Importantly, high levels of Bcl-2 expression protected cells from this death and inhibition of autophagy provided no further advantage or disadvantage (Figure 6F). Thus, despite decreased glucose uptake and the generation of alternate nutrients by autophagy, autophagy-derived metabolites are not essential to support cell viability early after growth factor withdrawal. Instead autophagy acts to promote death in growth-factor cells expressing moderate levels of Bcl-2 or Bcl-xL.
Figure 6.
Autophagy induces cell death in growth factor–deprived Bcl-2– and Bcl-xL–expressing cells. (A and B) MyrAkt (A) and Bcl-2–expressing (B) cells were transfected with control, Beclin-1, or Atg5 shRNAi, and cell viability was observed over time after IL3 withdrawal. (C) Moderate Bcl-2–expressing cells transfected with control, Beclin-1, or Atg5 shRNAi were withdrawn from cytokine, and viability was tracked over 9 d. (D) Human Bcl-2 was overexpressed in Atg5 Tet-Off MEFs. Protein levels were analyzed by immunoblot. (E) Atg5 Tet-Off MEFs expressing Bcl-2 were treated with PBS or 5 μg/ml doxycycline for 4 d, withdrawn from serum, and analyzed for viability over the indicated time course. (F) Cells with high Bcl-2 expression were transfected with control, Beclin-1, or Atg5 shRNAi, and withdrawn from cytokine, and viability was tracked over 9 d. Means and SDs of triplicate samples are shown. *p < 0.05 by Student's t test of test samples relative to control samples. For death curves, *p < 0.05 by Student's t test for multiple time points for test samples relative to control samples. Data shown are representative of three or more experiments.
Autophagic-induced Cell Death is Apoptotic
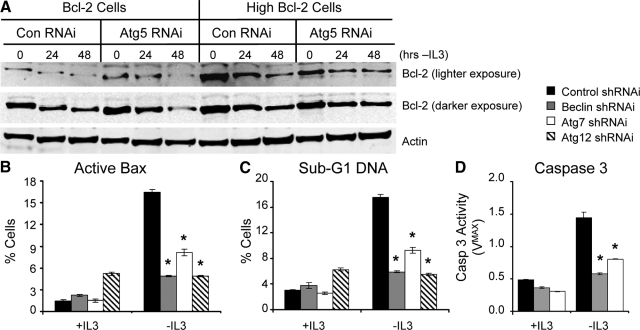
The ability of Bcl-2 levels to modulate autophagy-induced cell death after growth factor withdrawal suggested that cell death may have been apoptotic. Apoptosis induction is regulated by the balance of proapoptotic and antiapoptotic proteins, and autophagy-dependent loss of Bcl-2 may have lowered the threshold for apoptosis. Although Bcl-2 protein levels did decrease after IL3 withdrawal in both Bcl-2– and high Bcl-2–expressing cell lines, this was not influenced by shRNAi knockdown of Atg5 (Figure 7A). To test whether autophagic-induced death after IL3 withdrawal in Bcl-2 cells was indeed apoptotic, Bax conformation change and activation (Figure 7B), degradation of DNA to sub-G1 levels (Figure 7C), and Caspase 3 activity (Figure 7D) were measured after growth factor withdrawal. All three markers of apoptosis were increased after IL3 withdrawal of Bcl-2 cells (Figure 7, B–D), and inhibition of autophagy suppressed these markers (Figure 7, B–D). Together, these data suggest that apoptosis occurs after IL3 withdrawal in cells with moderate Bcl-2 expression and is enhanced in an autophagy-dependent manner.
Figure 7.
Autophagy does not lead to Bcl-2 degradation, but does promote apoptosis in growth factor deprived Bcl-2–expressing cells. (A) Moderate (left) and high (right) Bcl-2–expressing cells were transfected with control or Atg5 shRNAi, withdrawn from cytokine, and analyzed for exogenous human Bcl-2 protein expression. (B and C) Moderate Bcl-2–expressing cells were transfected with Control, Beclin, Atg7, or Atg12 and active Bax (B), and sub-G1 DNA content (C) was analyzed in the presence of IL3 or after 36-h IL3 withdrawal. (D) Caspase 3 activity was measured in Bcl-2–expressing cells transfected with control, Beclin-1, or Atg7 shRNAi in the presence of IL3 or after 24-h IL3 withdrawal. Means and SDs of triplicate samples are shown. *p < 0.05 by Student's t test of test samples relative to control samples. Data shown are representative of three or more experiments.
Autophagy Augments Induction of the Proapoptotic Transcription Factor Chop
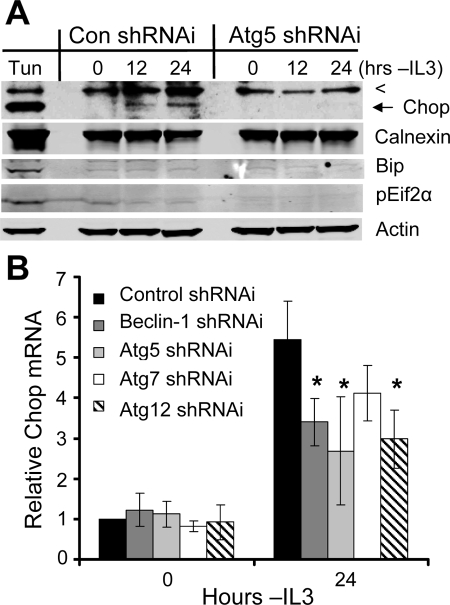
It was not clear how autophagy may lead to apoptosis, but autophagic degradation of intracellular components may have initiated a proapoptotic signaling pathway. In particular, autophagy has been shown to play a role in destruction of unfolded proteins (Hoyer-Hansen and Jaattela, 2007), which may accumulate during the cellular atrophy that occurs after growth factor withdrawal and may affect cell survival. To determine if the unfolded protein response occurred upon growth factor withdrawal and if autophagy affected this process, Bcl-2–expressing cells transfected with control or Atg5 shRNAi were withdrawn from cytokine, and the unfolded protein response and ER stress proteins Calnexin, Bip, and phospho-EIF2α, along with the multistress transcription factor Chop, were observed over time. In contrast to tunicamycin (Tun)-treated cells, which induced all ER stress and unfolded protein response markers examined, Calnexin, Bip, and phospho-EIF2α were not altered upon cytokine withdrawal in cells transfected with control shRNAi (Figure 8A). Chop, however, was induced after growth factor withdrawal (Figure 8A), although at a lower level than after Tun treatment. Importantly, induction of Chop protein was not detectable in autophagy-deficient cells (Figure 8A), suggesting that Chop induction after growth factor withdrawal may occur downstream of autophagy in the absence of a canonical ER stress. Indeed, inhibition of autophagy by shRNAi directed against the autophagy-essential genes Beclin-1, Atg5, Atg7, or Atg12 prevented full induction of Chop mRNA after IL3 withdrawal (Figure 8B). Autophagy contributed to but was not essential for Chop up-regulation, as partial induction of Chop mRNA still did occur regardless of autophagy inhibition.
Figure 8.
Autophagy promotes induction of the proapoptotic transcription factor Chop after growth factor withdrawal. (A) Bcl-2–expressing cells were treated with tunicamycin (Tun) for 12 h or transfected with control (Con) or Atg5 shRNAi and withdrawn from cytokine for the indicated times. Expression of Chop, Calnexin, Bip, and pEif2α was determined by immunoblot. The top line is a nonspecific band (<), and Chop is indicated by an arrow. (B) Bcl-2–expressing cells were transfected with control or shRNAi against autophagy-essential genes, and IL3 was withdrawn for 1 d, and then Chop mRNA levels were analyzed by quantitative real-time PCR. Means and SDs of triplicate independent experiments are shown. *p < 0.05 by Student's t test of test samples relative to control samples. Data shown are representative of three or more experiments.
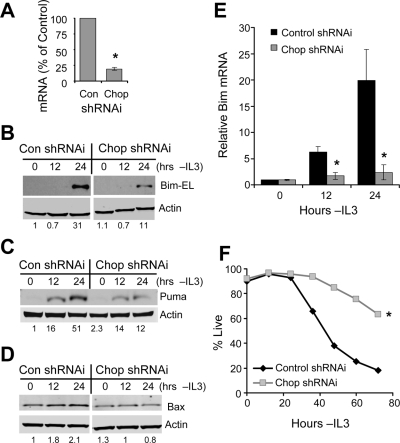
Chop was previously shown to regulate the transcriptional induction of the proapoptotic protein Bim (Puthalakath et al., 2007) and Puma (Ishihara et al., 2007) and may regulate other prodeath factors to cause apoptosis in growth factor–deprived cells. To ascertain the importance of Chop in the induction of proapoptotic proteins upon IL3 withdrawal, Chop expression was suppressed by shRNAi (Figure 9A). This led to a decrease in the induced protein levels of Bim-EL, (Figure 9B), Puma (Figure 9C), and Bax (Figure 9D), all of which may contribute to apoptosis in growth factor–deprived Bcl-2–expressing cells. The alternate splice isoforms of Bim, Bim-L and Bim-S, were not readily detectable by protein blot in exposures shown here. Bim regulation by Chop occurred at least in part by modulation of Bim mRNA levels, as shRNAi inhibition of Chop completely prevented the increase in Bim mRNA after IL3 withdrawal (Figure 9E). Importantly, compared with cells transfected with control shRNAi, suppression of Chop induction by shRNAi greatly increased survival of cells with moderate Bcl-2 expression after growth factor deprivation (Figure 9F). Thus, Chop is induced after IL3 withdrawal and regulates the proapoptotic proteins Bim, Puma, and Bax, and autophagy can partially regulate the induction and activity of Chop.
Figure 9.
Chop is required for induction of proapoptotic Bcl-2 family proteins and cell death upon growth factor withdrawal. (A–D) Bcl-2–expressing cells were transfected with control (Con) or Chop shRNAi and withdrawn from IL3 for the indicated times. (A) Inhibition of Chop expression by shRNAi was tested by quantitative real-time PCR. Induction of Bim-EL (B), Puma (C), and Bax (D) in control (Con) or Chop shRNAi transfected cells was observed by immunoblot. Bim mRNA (E) was harvested, and mRNA levels were determined by quantitative real-time PCR. (F) Bcl-2–expressing cells were transfected with control or Chop shRNAi, withdrawn from IL3, and cell survival was observed over time. Means and SDs of triplicate samples are shown. *p < 0.05 by Student's t test of test samples relative to control samples. For death curves, *p < 0.05 by Student's t test for multiple time points for test samples relative to control samples. Data shown are representative of three or more experiments. For immunoblots, levels of proteins under analysis were quantified by normalization to actin and to the control sample in each panel, and quantitations are shown under each lane.
Autophagy Specifically Regulates the Proapoptotic Protein Bim in a Chop-dependent Manner
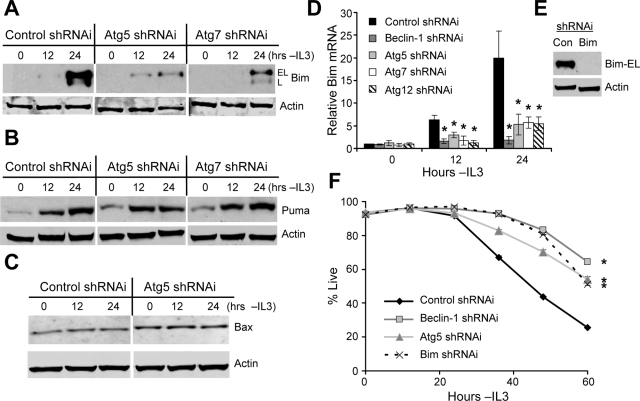
Because Chop was required for induction of Bim, Puma, and Bax after IL3 withdrawal and was itself partially regulated by autophagy, the role of autophagy in regulation of Bcl-2 family proteins was next examined. Bim-EL and Bim-L induction were greatly suppressed by inhibition of autophagy in growth factor–deprived cells (Figure 10A), whereas Puma (Figure 10B) and Bax (Figure 10C) were unaffected. To determine if regulation of Bim was transcriptional, Bcl-2–expressing cells transfected with control shRNAi or shRNAi against autophagy-essential genes were analyzed for Bim mRNA induction by quantitative real-time PCR (Figure 10D). Compared with IL3 withdrawn Bcl-2–expressing cells treated with control shRNAi, cells treated with each autophagy-gene shRNAi showed greatly reduced levels of Bim mRNA both at 12 and 24 h after IL3 deprivation (Figure 10D). Therefore, although Chop may regulate several proapoptotic proteins after growth factor withdrawal, autophagy specifically suppressed Chop-dependent Bim induction or mRNA stabilization (Matsui et al., 2007).
Figure 10.
Autophagy specifically up-regulates the proapoptotic protein Bim in a Chop-dependent manner. (A–C) Bcl-2–expressing cells were transiently transfected with control, Atg5 or Atg7 shRNAi, withdrawn from IL3 for the indicated times, and Bim-EL and Bim-L (A), Puma (B), and Bax (C) levels were determined by immunoblot. (D) Bcl-2–expressing cells were transfected with control, Beclin-1, Atg5, Atg7, or Atg12 shRNAi, and IL3 was withdrawn for indicated times. RNA was harvested, and Bim mRNA levels were analyzed by quantitative real-time PCR. (E and F) Bcl-2 cells were transfected with control (Con), Beclin-1, Atg5, or Bim shRNAi. (E) Inhibition of Bim expression by shRNAi was tested by immunoblot of IL3-deprived cells. (F) Cell viability was observed over time after IL3 withdrawal. Means and SDs from triplicate samples are shown. *p < 0.05 by Student's t test of test samples relative to control samples. For death curves, *p < 0.05 by Student's t test for multiple time points for test samples relative to control samples. Data shown are representative of three or more experiments.
Bim has been shown to be of particular importance in promoting the death of hematopoietic cells (Bouillet et al., 1999; Zhao et al., 2007). To assess the role of Bim in the death of cells with moderate Bcl-2 expression after cytokine withdrawal, cells were transfected with control, Bim (Figure 10E), Beclin-1, or Atg5 shRNAi and were analyzed for death upon growth factor withdrawal. Consistent with autophagic regulation of Bim to promote cell death, inhibition of Bim induction by shRNAi increased survival to a similar degree as inhibition of autophagy (Figure 10F). Therefore, specific autophagic regulation of Chop-dependent Bim induction appears to account for the apoptotic death caused by activation of autophagy in Bcl-2– and Bcl-xL–expressing cells after growth factor withdrawal.
DISCUSSION
Autophagy has been described in various systems to either promote or prevent cell death (Lum et al., 2005b; Levine and Kroemer, 2008). The role that autophagy may play in growth factor withdrawal and its interaction with apoptosis under this condition has been uncertain. In this study, we demonstrate a dual-role for autophagy in survival and death of growth factor–withdrawn hematopoietic cells. Autophagy was induced upon growth factor withdrawal despite expression of Bcl-2 and Bcl-xL and provided apparent metabolic benefits. After nutrient deprivation or prolonged growth factor withdrawal, autophagy-derived nutrients have been shown to be essential to support cell viability of apoptosis-incompetent cells (Lum et al., 2005a; Mathew et al., 2007). Here, however, we found that autophagy-derived metabolites were not required to support survival of growth factor deprived hematopoietic cells within at least a 1-wk period after cytokine withdrawal. Instead, autophagy ultimately promoted apoptosis of growth factor–deprived cells through a Chop-dependent specific regulation of the proapoptotic BH3-only protein Bim. Thus, autophagy appears to contribute to tissue homeostasis by sensitizing cells to apoptosis after growth factor withdrawal, ensuring that only the most apoptosis-resistant cells may survive to utilize autophagy as a long-term survival mechanism.
Bcl-2 family proteins have been shown to interact with the BH3-domain–containing autophagy protein Beclin-1 under amino acid starvation or rapamycin treatment, and this interaction can inhibit induction of autophagy (Pattingre et al., 2005; Liang et al., 2006). However, after serum withdrawal, the BH3-only protein Bad can compete with Beclin-1 for binding to Bcl-2 or Bcl-xL, thus relieving Beclin-1 to initiate autophagy even in the presence of overexpressed Bcl-2 or Bcl-xL (Maiuri et al., 2007; Oberstein et al., 2007). Autophagy in cells overexpressing Bcl-2 or Bcl-xL has also been observed under ischemic conditions (Degenhardt et al., 2006) and after DNA damage (Shimizu et al., 2004). Here, autophagy occurred normally in Bcl-2– and Bcl-xL–expressing cells after IL3 withdrawal. We observed no change in levels of endogenous phospho-Bad in cells after growth factor withdrawal (data not shown), but increased expression of BH3-only proteins such as Bim and Puma may have sufficiently disrupted the Bcl-2/Bcl-xL—Beclin-1 interaction to allow for autophagy induction.
Autophagy can support cell metabolism when exogenous nutrient uptake decreases after growth factor withdrawal (Lum et al., 2005a; Mathew et al., 2007). Indeed, Bax−/− Bak−/− cells eventually come to rely on autophagy to support essential metabolic processes (Lum et al., 2005a). Although autophagy can produce nutrients for survival, direct measurement of nutrients made available by autophagy has not been previously shown. We found that induction of autophagy in growth factor–withdrawn Bcl-2– or Bcl-xL–expressing cells led to increased amino acids (reflected by increased in C5-carnitines; Figure 4B and Supplemental Figure S1) and long-chain fatty acids (reflected by increased C18:1- and C18-carnitines, Figures 3 and 4A; Millington et al., 1990; An et al., 2004). Importantly, although availability of long-chain fatty acids appeared to be wholly dependent on autophagy, amino acids were only partly autophagy dependent. Consistent with a growth factor and autophagy-independent source of nutrients such as amino acids supporting cellular metabolism, cells expressing Bcl-2 were capable of growth factor–independent survival with diminished autophagy for periods of at least a week (Figures 6, C and F). Residual glucose uptake or reliance on amino acid oxidation may have supported cell viability. Indeed, Bax−/− Bak−/− cells maintained a measurable glycolytic flux for at least 5 wk after growth factor withdrawal (Lum et al., 2005a). Cellular atrophy due to proteasomal degradation may have also provided amino acids sufficient to sustain minimal mitochondrial oxidation. Alternatively, increased chaperone-mediated autophagy has been reported after inhibition of macroautophagy, and this pathway may contribute nutrients (Dice, 2007; Kaushik et al., 2008). The role of both autophagy and alternate fuels as nutrient sources upon growth factor withdrawal will be important for future study.
In addition to support of cellular metabolism, autophagy may promote cell viability by degrading damaged intracellular organelles (Colell et al., 2007). Under circumstances where autophagy appeared to promote survival, as was the case in control FL5.12 cells after growth factor withdrawal, the mechanism that supported cell survival was not provision of nutrients to replace lost glucose uptake, as cells expressing Glut1 and hexokinase were equivalently affected by inhibition of autophagy (Figure 5, C and D). In cells expressing Bcl-2 and thus protected from mitochondrial and other organelle damage after cytokine withdrawal, autophagy was not required at these early time points. It is likely, therefore, that autophagy inhibited cell death early after growth factor withdrawal via degradation of damaged organelles.
At later times after growth factor withdrawal, autophagy sensitized cells with moderate Bcl-2 or Bcl-xL expression to apoptosis. Normally, apoptosis is initiated soon after growth factor withdrawal by induction of proapoptotic BH3-only proteins and neutralization of antiapoptotic Bcl-2 family proteins. In cells that strongly resist apoptosis, autophagy may ultimately support long-term survival, (Lum et al., 2005a; Degenhardt et al., 2006; Levine and Kroemer, 2008; Swerdlow et al., 2008), as induction of proapoptotic BH3 only proteins would not be sufficient to induce apoptosis. Indeed, Bax−/− Bak−/− cells are completely resistant to Bim-induced cytotoxicity. Thus, an initial barrier of apoptosis must be circumvented to allow persistent cell survival with autophagy in the absence of growth factors. Our data indicate that through enhanced induction of Bim, autophagy itself increases the apoptotic stress that cells must endure to survive long-term. Supporting this notion, autophagy has previously been shown to lead to apoptosis in some developmental and tissue homeostasis circumstances (Akdemir et al., 2006; Yousefi et al., 2006; Scott et al., 2007; Wang et al., 2008a,b), although the mechanisms have been uncertain. Regulation of the proapoptotic protein Bim by autophagy represents a novel connection between autophagy and apoptosis that may be applicable in a wide variety of instances where cells are growth factor deprived, such as in development and in tissue homeostasis.
Autophagic induction of apoptosis occurred through augmentation of Chop-dependent up-regulation of the proapoptotic BH3-only protein Bim (Figure 9, B and E). Chop is an ER stress-induced transcription factor (Oyadomari and Mori, 2004; Puthalakath et al., 2007), but canonical ER stress and the unfolded protein response did not occur after growth factor withdrawal (Figure 8A). Chop has been shown to respond to multiple cell stresses (Oyadomari and Mori, 2004), and autophagy may initiate or enhance such a pathway. Importantly, inhibition of autophagy prevented only a portion of Chop induction after growth factor withdrawal, and Chop was necessary to induce Bax and Puma in addition to Bim. Autophagy, however, was only necessary for Chop-mediated up-regulation of Bim mRNA. Thus, suppression of Chop by shRNAi resulted in greater survival in Bcl-2– and Bcl-xL–expressing cells withdrawn from growth factor than suppression of Bim expression alone (Figure 9F compared with Figure 10F). Modification of Chop induction and activity by autophagy played an important role in cell survival, but the precise mechanisms remain unclear and may be context specific. In particular, when Bcl-2–expressing MEFs were serum-starved, we were unable to detect autophagy-dependent regulation of Bim, despite enhanced cell death, suggesting that autophagy may promote different proapoptotic signaling pathways in different systems. This is an important area for future study.
Autophagy can be both positive and negative in cell survival, but it remains unclear how these two outcomes are regulated and under what conditions cells may utilize nutrients from autophagy as a survival mechanism. Here, we used nontransformed hematopoietic cells with little or moderate resistance to apoptosis to examine the role of autophagy in growth factor withdrawal. Surprisingly, autophagy enhanced apoptosis by promoting Chop-dependent up-regulation of Bim. Cells or tissues sensitive to expression of Bim may be particularly sensitive to autophagic regulation of Chop and apoptosis. These findings are consistent with the view that apoptosis occurs to prevent cells from surviving long-term in the absence of growth factors (Lum et al., 2005a; Degenhardt et al., 2006; Fung et al., 2008) and further show that autophagy augments this apoptotic stress. In this way, only cells with a highly favorable balance of antiapoptotic to proapoptotic Bcl-2 family members may persist long-term after growth factor withdrawal to utilize alternative fuel sources such as those provided by autophagy. It remains important to further address mechanisms by which apoptosis and autophagy interact to better understand how these two pathways affect both normal and disease processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Tamotsu Yoshimori (National Institute of Genetics, Mishima, Japan), Youwen He (Duke University), Noboru Mizushima (Tokyo Medical and Dental University, Bunkyo-Ku, Japan), and Julian Lum and Craig Thompson (University of Pennsylvania) for generously providing reagents described above, and the Duke University Pathology Electron Microscopy Facility. We also thank Drs. Christopher Newgard, Sally Kornbluth, and Leta Nutt (Duke University) and Eric Baehrecke (University of Massachusetts Medical School, Worcester, MA), as well as Sarah Jacobs and Christina Powers, for helpful comments and insights. This work was supported by the National Cancer Institute Grant RO1 CA123350.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0829) on December 24, 2008.
REFERENCES
- Abedin M. J., Wang D., McDonnell M. A., Lehmann U., Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Akdemir F., et al. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- An J., Muoio D. M., Shiota M., Fujimoto Y., Cline G. W., Shulman G. I., Koves T. R., Stevens R., Millington D., Newgard C. B. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Kontgen F., Adams J. M., Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Colell A., et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice J. F. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- Erlacher M., Labi V., Manzl C., Bock G., Tzankov A., Hacker G., Michalak E., Strasser A., Villunger A. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J. Exp. Med. 2006;203:2939–2951. doi: 10.1084/jem.20061552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E. L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Fox C. J., Hammerman P. S., Cinalli R. M., Master S. R., Chodosh L. A., Thompson C. B. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C., Lock R., Gao S., Salas E., Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol. Biol. Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M., Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Ishihara T., Hoshino T., Namba T., Tanaka K., Mizushima T. Involvement of up-regulation of PUMA in non-steroidal anti-inflammatory drug-induced apoptosis. Biochem. Biophys. Res. Commun. 2007;356:711–717. doi: 10.1016/j.bbrc.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Massey A. C., Mizushima N., Cuervo A. M. Constitutive Activation of Chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Baehrecke E. H. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Lum J. J., Bauer D. E., Kong M., Harris M.H.Li.C., Lindsten T., Thompson C. B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005a;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Lum J. J., DeBerardinis R. J., Thompson C. B. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005b;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Maiuri M. C., et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Asou H., Inaba T. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol. Cell. 2007;25:99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Millington D. S., Kodo N., Norwood D. L., Roe C. R. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J. Inherit. Metab. Dis. 1990;13:321–324. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
- Nutt L. K., Margolis S. S., Jensen M., Herman C. E., Dunphy W. G., Rathmell J. C., Kornbluth S. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A., Jeffrey P. D., Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Plas D. R., Talapatra S., Edinger A. L., Rathmell J. C., Thompson C. B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- Puthalakath H., et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Raff M. C. Social controls on cell survival and cell death. Nature. 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- Rathmell J. C., Vander Heiden M. G., Harris M. H., Frauwirth K. A., Thompson C. B. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell. 2000;6:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Scott R. C., Juhasz G., Neufeld T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Swerdlow S., McColl K., Rong Y., Lam M., Gupta A., Distelhorst C. W. Apoptosis inhibition by Bcl-2 gives way to autophagy in glucocorticoid-treated lymphocytes. Autophagy. 2008;4:1–9. doi: 10.4161/auto.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C. L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Walensky L. D. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ. 2006;13:1339–1350. doi: 10.1038/sj.cdd.4401992. [DOI] [PubMed] [Google Scholar]
- Wang Y., Han R., Liang Z. Q., Wu J. C., Zhang X. D., Gu Z. L., Qin Z. H. An autophagic mechanism is involved in apoptotic death of rat striatal neurons induced by the non-N-methyl-D-aspartate receptor agonist kainic acid. Autophagy. 2008a;4:214–226. doi: 10.4161/auto.5369. [DOI] [PubMed] [Google Scholar]
- Wang Y., Singh R., Massey A. C., Kane S. S., Kaushik S., Grant T., Xiang Y., Cuervo A. M., Czaja M. J. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J. Biol. Chem. 2008b;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieman H. L., Wofford J. A., Rathmell J. C. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell. 2007;18:1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E. H., Lenardo M. J. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu S. W., et al. Autophagic death of adult hippocampal neural stem cells following insulin withdrawal. Stem Cells. 2008;26:2602–2610. doi: 10.1634/stemcells.2008-0153. [DOI] [PubMed] [Google Scholar]
- Zhao Y., et al. Glycogen synthase kinase 3alpha and 3beta mediate a glucose-sensitive antiapoptotic signaling pathway to stabilize Mcl-1. Mol. Cell. Biol. 2007;27:4328–4339. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.